Abstract
One of the most fundamental questions in the control of gene expression in mammals is how the patterns of epigenetic modifications of DNA are generated, recognized, and erased. This includes covalent cytosine methylation of DNA and its associated oxidation states. An array of AdoMet-dependent methyltransferases, Fe(II)- and α-ketoglutarate-dependent dioxygenases, base excision glycosylases, and sequence-specific transcription factors is responsible for changing, maintaining, and interpreting the modification status of specific regions of chromatin. This review focuses on recent developments in characterizing the functional and structural links between the modification status of two DNA bases 5-methylcytosine and thymine (5-methyluracil).
Keywords: DNA-binding protein, DNA demethylation, DNA enzyme, DNA methylation, DNA methyltransferase, DNA-protein interaction, transcription factor, DNA 5mC oxidation, Tet dioxygenases, base excision DNA glycosylases, modification-dependent and sequence-specific transcription factors
Introduction
In the DNA of higher organisms, cytosine exists in several chemical forms, including unmodified cytosine (C), 5-methylcytosine (5mC),2 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (1–5). These forms are genetically equivalent in terms of base-pairing and protein-coding, but differ in how they interact with macromolecules and influence gene expression. There is much interest in the effects of these modifications in epigenetic regulation, in development and differentiation, in neuron function, and in diseases. In general, the modifications (or “marks”) are added to cytosine in situ, following its incorporation into DNA in the unmodified form. DNA methyltransferases (Dnmt) convert certain cytosines to 5mC, usually within the sequence context CpG (6, 7) or CpA (8). A subset of these 5mC residues is then converted to 5hmC, 5fC, and 5caC in consecutive Fe(II)- and α-ketoglutarate-dependent oxidation reactions by the ten-eleven translocation (Tet) dioxygenases (2–4). The Tet dioxygenases are widely distributed across the eukaryotic tree of life, from mammals to the amoeboflagellate Naegleria gruberi (9), mushroom (Coprinopsis cinerea) (10), and honey bee (Apis mellifera) (11). High-throughput methods for characterizing 5mC oxidation states at single base resolution are becoming available, so our understanding of 5mC oxidation is expected to develop rapidly. In this minireview, we focus on the mechanisms of generating, recognizing, and possibly erasing 5mC and its oxidative forms in DNA.
Tet Proteins Are 5-Methylpyrimidine Dioxygenases, but Not Demethylating Enzymes
Chromatin regulates transcriptional processes through post-synthetic modifications of both of its components: DNA and histones (Fig. 1a). Much remains to be learned about how the combination of these modifications (or lack thereof) facilitates or silences transcription. One broad theme has emerged that a web of interactions tightly coordinates the modification of a segment of DNA and its associated histones, affecting local chromatin structure and determining the functional states. The Tet enzymes belong to a family of Fe(II)- and α-ketoglutarate-dependent dioxygenases that also includes the Jumonji domain-containing histone lysine demethylases, the N-methyl nucleic acid demethylase including Escherichia coli AlkB and its mammalian homologs, and many others. To comprehend the unique features of Tet-driven reaction products, we first briefly review the mechanisms of other dioxygenases considered as demethylases of amino (N)-methylation. The N-demethylation reaction catalyzed by most of these enzymes involves the transient formation of a N-hydroxymethyl intermediate followed by the spontaneous (non-enzymatic) release of formaldehyde.
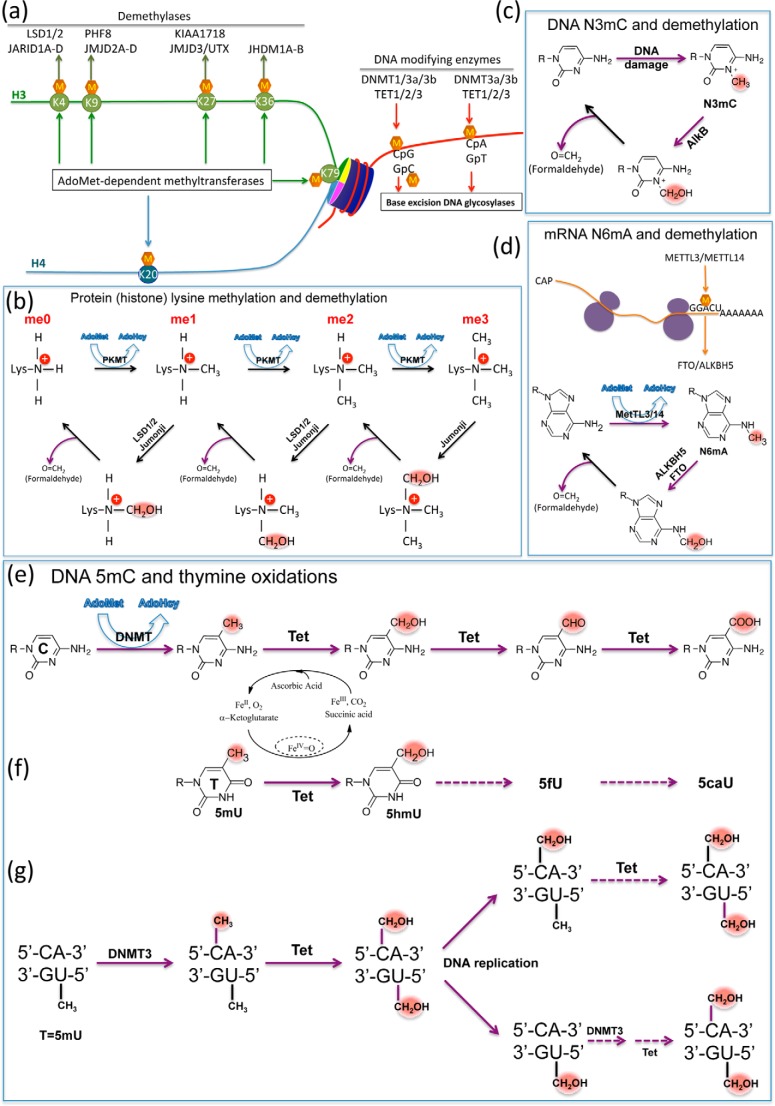
FIGURE 1.
General methylation and demethylation in histone lysines and nucleic acids. a, examples of post-synthetic methylation of both components of a nucleosome: DNA and histones. Major histone lysine methylation occurs at five residues on H3 (green) and one on H4 (blue), and primary enzymes responsible for demethylation are shown. DNA modifications occur at CpG and non-CpG (i.e. CpA) dinucleotides. b, overview of protein lysine methylation by AdoMet-dependent methyltransferases (top) and demethylation reactions catalyzed by LSD1/2 and Jumonji dioxygenases (bottom). The hydroxymethyl intermediate (N-CH2OH) decomposes to release formaldehyde and the (one methyl group reduced) demethylated lysine. c, demethylation of N3-methylcytosine (N3mC) by AlkB dioxygenase (involved in the direct reversal of alkylation damage) results in the production of formaldehyde and unmodified cytosine. d, model of the mRNA N6-adenine methylation by methyltransferase-like METTL3/14 heterodimer, generating N6mA, and demethylation reaction by ALKBH5 and fat mass and obesity-associated protein (FTO) (two of the nine human homologs of AlkB), resulting in a release of formaldehyde and unmodified adenine. e, Tet dioxygenases convert 5mC to 5hmC, 5fC, and 5caC in three consecutive Fe(II)- and α-ketoglutarate-dependent oxidation reactions without release of formaldehyde. f, Tet dioxygenases convert thymine (5mU) to 5hmU, and potentially to 5fU and 5caU, without release of formaldehyde. g, Dnmt3A and Dnmt3B can methylate the cytosine in the context of the CpA/TpG dinucleotide. Tet dioxygenases can oxidize both 5mC and T (5mU). The diagram shows the potential fate of a single CpA/TpG site that is fully hydroxymethylated during DNA replication. After strand synthesis, the hemi-hydroxymethylated (5hmC/T) site could be modified by Tet enzymes to become fully hydroxymethylated (top). On the other hand, the hemi-hydroxymethylated (C/5hmU) site would require two reactions, methylation by Dnmt3 and oxidation by Tet, to become fully hydroxymethylated (bottom).
In the case of protein lysine demethylation, the FAD-dependent amino oxidases LSD1/2 and Fe(II)- and α-ketoglutarate-dependent Jumonji dioxygenases (12) generate a hydroxymethyl intermediate for each reaction that subsequently decomposes to release a formaldehyde spontaneously (without additional enzymatic activities) and the demethylated lysine (Fig. 1b) (with one methyl group reduced). The same basic mechanism applies to demethylation of N-methylated nucleic acids (e.g. N3-methylcytosine, N3-methylthymine, and N1-methyladenine) by the AlkB family of DNA repair enzymes (13, 14) (Fig. 1c) as well as the recently described demethylation of N6-methyladenine (N6mA) in mRNA by ALKBH5 and fat mass and obesity-associated protein (FTO) (15, 16) (Fig. 1d). In fact, during the search for enzymes capable of reversing methylated lysines in histones, it was initially hypothesized that Fe(II)- and α-ketoglutarate-dependent dioxygenases might reverse lysine methylation via a mechanism similar to that of AlkB (17), and the purification of JHDM1, the first identified Jumonji domain-containing histone demethylase, used a biochemical assay based on the detection of formaldehyde, one of the predicted reaction products (18).
In stark contrast, formaldehyde is not released during Tet-mediated hydroxylation of 5-methylcytosine (Fig. 1e). Unlike methylation/demethylation of monoamines, the main mechanistic problem in methylation and demethylation of carbons is that the C5 atom of the cytosine ring is an inert carbon. DNA cytosine methyltransferases solve this problem by flipping the target base into a concave active site and forming a transient covalent adduct at cytosine C6 (19, 20). However, the 5-hydroxymethyl modification at C5 (5hmC) does not change the nature of the C5-CH3 carbon-carbon bond and thus either stays as a stable modification or is further converted to 5fC and 5caC in consecutive Tet-mediated oxidation reactions, generating stable modifications further away from 5mC. That Tet-mediated 5hmC remains a stable mark, rather than serving as an intermediate in direct demethylation, is supported by the observation that 5mC loss in the paternal genome, immediately after fertilization during mouse development, is accompanied by concurrent increase in 5hmC (21).
Very recently, it has been shown that Tet enzymes are also active on thymine, generating 5-hydroxymethyluracil (5hmU) paired with adenine that could be specifically bound by potential protein readers (22, 23) (Fig. 1f). This is distinct from G:5hmU, the deamination product of G:5hmC. Like 5mC, thymine contains a methyl group at C5, and thus the Tet-mediated hydroxylation of 5-methyluracil (thymine) provides an opportunity to establish a pseudo-symmetric, modified CpA/TpG dinucleotide (Fig. 1g). CpA/TpG could be considered an intrinsically “hemi-methylated” DNA element. Methylation at CpA sites by Dnmt3 would then generate a “fully methylated” mCpA/TpG dinucleotide (Fig. 1f). Indeed, the levels of mCpA/TpG undergo dynamic changes during differentiation of the germ line and in brain development from fetus to young adult (24, 25). Some transcription factors recognize 5mCpG and TpG in the same way (26) (see below). Perhaps TpG dinucleotides are selected for when it is advantageous for a particular DNA sequence to be treated as if it were permanently (hemi)-methylated. Further modification by Tet enzymes on both 5mC and T would generate fully modified CpA/TpG sites with 5-hydroxymethyl or even 5-formyl and 5-carboxyl modification on both 5mC and T (23). Such fully modified CpA/TpG might even be maintained during DNA replication through the combined activity of Dnmt3 and Tet enzymes (Fig. 1g). A maintenance methylation function has been proposed for both Dnmt3a and Dnmt3b (27). Assuming that hemi-hydroxymethylated and hemi-methylated sites are substrates for modification by Tet proteins, there is also the potential for regenerating a fully modified hydroxymethylated site, recapitulating the parental DNA state after replication (Fig. 1g).
DNA methylation patterns are reprogrammed at two distinct stages during early vertebrate development: in the zygote and early embryo when DNA methylation patterns contributed by the sperm and egg are largely erased and subsequently re-established, and again during germ cell development, giving rise to germ cell-specific patterns that will be carried into the next generation (21). The Tet enzymes and oxidized 5mC derivatives, presumably through their role as intermediates in 5mC turnover (see below), play a critical role in both processes. Although Tet1 and Tet2 single knock-out mice are viable and reproduce normally (28–30), the majority of Tet1 and Tet2 double knock-out mice die perinatally, and female survivors exhibit reduced fertility (31), implying some functional redundancy between Tet1 and Tet2. In contrast, deletion of Tet3 leads to neonatal lethality (32), and loss of all three Tet enzymes restricts normal differentiation of mouse embryonic stem cells (33). Consistent with a critical role of Tet3 in early embryonic reprogramming, there are high levels of Tet3 and 5hmC in the zygote (34) and conditional deletion of Tet3 in the oocyte results in aberrant hydroxylation and delayed demethylation of the paternal genome upon fertilization, leading to impaired development and reduced fertility (32). It is interesting to note that although Dnmt3a (but not Dnmt3b or Dnmt1) is expressed in both the male and the female pronucleus of the mouse zygote, Tet3 (but not Tet1 or Tet2) is only expressed in the male pronucleus, as observed in an earlier study (32). Thus, Dnmt3a and Tet3 selectively converge in the male pronucleus and coincide with the replication-independent loss of 5mC and gain of 5hmC that occur in the paternal genome in the early mouse embryo (34, 35). However, more recent work indicates that there may also be low levels of Tet3 expressed in the female pronucleus (36, 37), suggesting that 5mC oxidation may also play a role in the erasure of the oocyte-specific DNA methylation in the zygote. Together, the accumulated data speak to a role for both active (via Tet3) and passive (via DNA replication) demethylation in the zygotes, in the early embryo, and in primordial germ cells (38).
Despite the widespread role of 5mC in the regulation of vertebrate gene expression, the situation may be different for invertebrates. For example, there is currently no evidence for the presence of 5mC in Drosophila melanogaster (39). Nevertheless, the Drosophila genome harbors a well conserved homolog of the mammalian Tet gene, the function of which is currently unknown (40). It is possible that Drosophila Tet may oxidize other methylated bases, such as thymine in DNA or 5mC in tRNA (generated by Dnmt2 (41)) or N6mA in mRNA or DNA. A very recent publication (100) has appeared demonstrating that Drosophila Tet catalyzes demethylation of N6mA in DNA both in vivo and in vitro as well as catalyzing 5mC in vitro.
Structures of Tet Dioxygenases in Complex with 5mC DNA
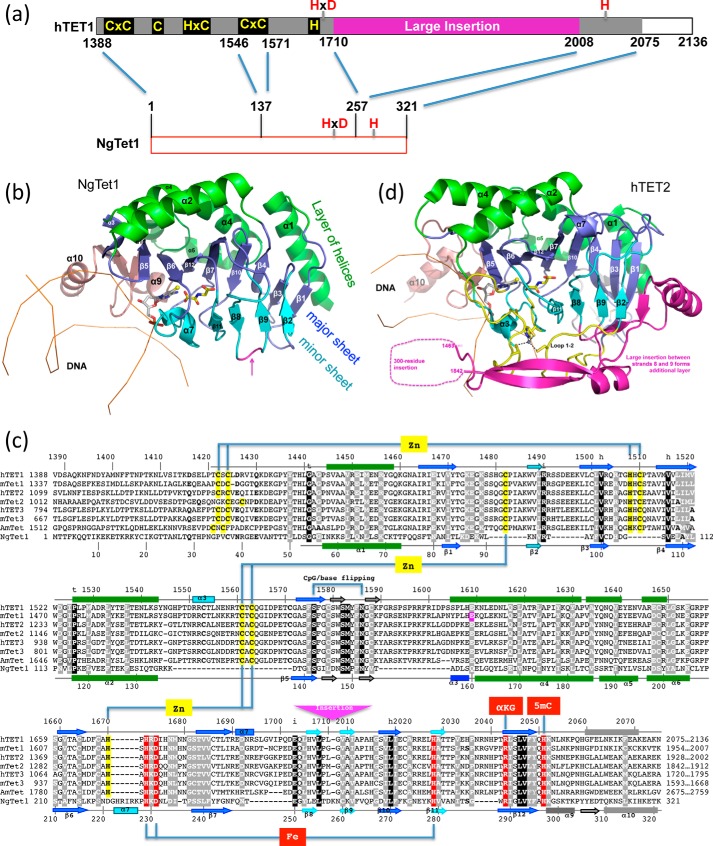
Two x-ray structures are currently available for Tet enzymes: the catalytic domain of human TET2 (42) and a Naegleria Tet-like dioxygenase, NgTet1 (9). Like DNA methyltransferases, Tet enzymes use a base-flipping mechanism to access 5mC. Structurally, NgTet1 represents the core structure of the catalytic domain of the mammalian Tet enzymes (Fig. 2a). Like other structurally characterized α-ketoglutarate-dependent dioxygenases, NgTet1 has a core double-stranded β-helix fold that binds Fe(II) and α-ketoglutarate (Fig. 2b). Two twisted β-sheets (a four-stranded minor sheet and an eight-stranded major sheet) pack together with five helices on the outer surface of the major sheet to form a three-layered structure (Fig. 2b). The unequal number of strands of the two sheets creates the active site located asymmetrically on the side of the molecule where the extra strands of the major sheet are located.
FIGURE 2.
Structures of the Tet enzymes. a, schematic representation of human Tet1 (hTet1) C-terminal catalytic domain and NgTet1. b, structure of NgTet1-DNA complex (Protein Data Bank (PDB) 4LT5). The NgTet1 protein folds in a three-layered jelly-roll structure. c, sequence alignment of human TET1, TET2, and TET3 (NP_085128.2, NP_001120680.1, and NP_001274420.1), mouse Tet1, Tet2, and Tet3 (NP_081660.1, NP_001035490.2, and NP_898961.2), honey bee (A. mellifera) AmTet (GB52555 in BeeBase OSGv3.2), and NgTet1 (XP_002667965.1). d, structure of human TET2-DNA complex (PDB 4NM6). The secondary structure elements are labeled according to NgTet1 structure (panel b). Note the large insertion in human TET2 between strands 8 and 9 (magenta), which is indicated by a magenta arrow in panel c.
There is considerable sequence conservation within the catalytic core among Tet enzymes from different species (Fig. 2c). When compared with NgTet1, mammalian Tet enzymes (human and mouse Tet1–3) and the honey bee Tet protein have eight insertions and one deletion scattered throughout the catalytic core domain (Fig. 2c). Two zinc ion centers, each coordinated by three cysteines and one histidine, bring together the loop inserted between strands 3 and 4 to the N-terminal region, and the three loops between strands 1 and 2, helix 2 and strand 5, and strands 6 and 7, respectively. These intramolecular interactions likely confer stability to the molecule. Invariant residues are involved in structural integrity, hydrophobic core, DNA binding, base-specific interactions, and Fe(II) and α-ketoglutarate binding.
The mammalian and honey bee Tet proteins have their catalytic domains located in the C-terminal part of the proteins with an atypical insertion of ∼300 residues between strands β8 and β9, which is not found in other α-ketoglutarate-dependent dioxygenases, nor in NgTet1. The insertion separates the two halves of the ferrous binding motif, HXD … H. Interestingly, the large insertion in Tet1 shares significant sequence similarity to the C-terminal domain of RNA polymerase II (43), suggesting that it might impart a regulatory function. Additionally, in human TET2, part of this insertion forms a fourth layer on the outer surface of the minor sheet (Fig. 2d).
Recognition of 5mCpG and TpG and Their Oxidized Derivatives
DNA 5mC is a major epigenetic signal that acts to regulate chromatin structure and ultimately gene expression. The oxidized derivatives, 5hmC, 5fC, and 5caC, might also act as distinct epigenetic signals. These modifications protrude into the major groove of DNA, the primary recognition surface for proteins, and change its atomic shape and pattern of electrostatic charge. In principle, such changes can alter the way in which proteins bind to their recognition sequences in DNA by strengthening the interactions, weakening them, or abolishing them altogether. This, in turn, can modulate gene expression and control cellular metabolism and is believed to be one of the principal mechanisms underlying epigenetic processes such as differentiation, development, aging, and disease.
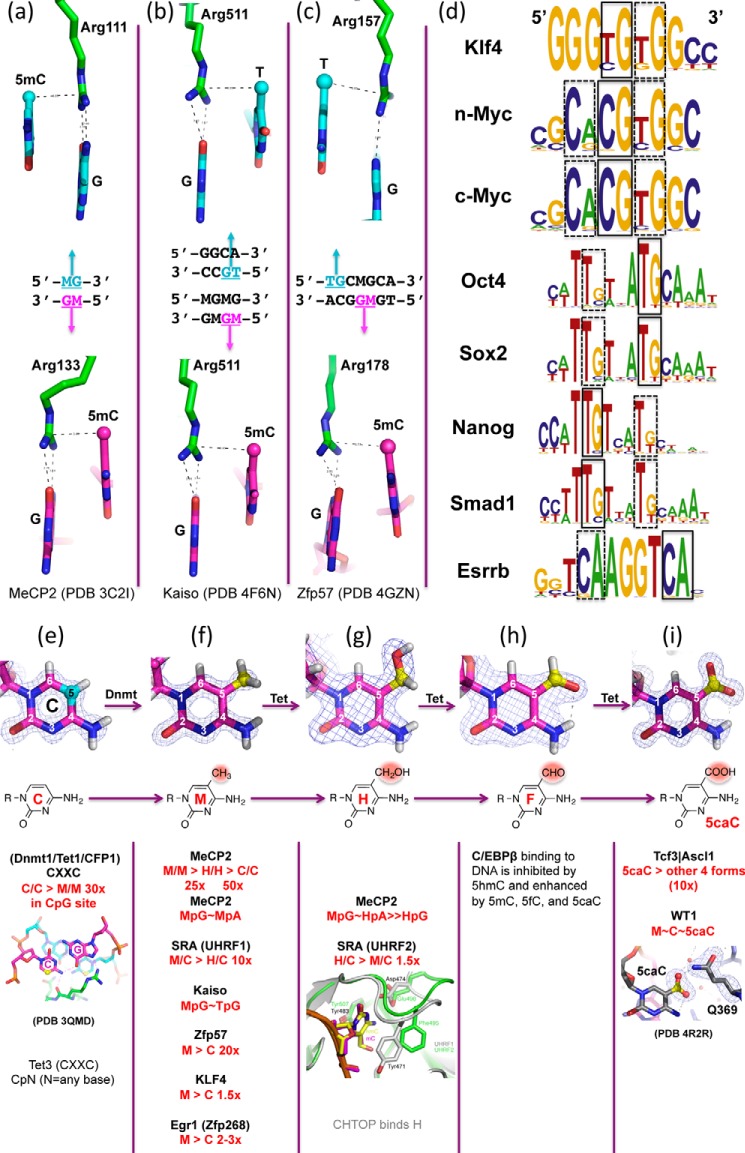
Three well characterized classes of mammalian proteins interact with DNA in a methylation-dependent manner. Methyl-binding domain proteins (MBDs) recognize fully methylated CpG sequences in which both DNA strands contain 5mC. The MBD domain of MeCP2 binds the symmetrical, fully methylated CpG site using two arginines; each hydrogen bonds to one guanine (44). The two arginines are also engaged in van der Waals contacts with the methyl group of the neighboring 5′-5mC of the same DNA strand and form two “5mC-Arg-G triads” (Fig. 3a) to symmetrically bind the fully methylated CpG palindromic duplex. Very recent work suggests that MeCP2 binds methylated CpA sites with similar affinity to that of fully methylated CpG (45, 46). Likewise, the MBD domain of MBD4 binds dinucleotides containing fully methylated 5mCpG/5mCpG and G:T mismatches occurring in the CpG context (5mCpG/TpG) (47). In all three examples, there are two methyl groups present symmetrically (5mCpG/5mCpG, 5mCpA/TpG, and 5mCpG/TpG), suggesting that the MBD proteins simultaneously recognize the two methyl groups on opposite strands.
FIGURE 3.
A methyl-Arg-G triad forms during recognition of methyl-CpG and TpG dinucleotides in double-stranded DNA. a, MeCP2 forms two 5mC-Arg-G triads to bind the palindromic fully methylated CpG duplex symmetrically (PDB 3C2I). b, Kaiso recognizes either a specific unmethylated DNA element containing a TpG dinucleotide (top) or a methylated CpG dinucleotide (bottom) (PDB 4F6M and 4F6N). In both cases, a methyl-Arg-G triad is involved. c, Zfp57 uses a pair of methyl-Arg-G triads to recognize the TpG dinucleotide on the top strand and a methyl-CpG on the bottom strand (PDB 4GZN). A third methyl-Arg-G triad recognizes the TpG on the bottom strand (not shown). d, example of transcription factor recognition sequences containing a CpA/TpG site (taken from Ref. 93). e–i, examples of protein domains with specificity for unmodified cytosine (e), 5mC = M (f), 5hmC = H (g), 5fC = F (h), or 5caC (i). The Tet3 CXXC domain binds to an unmodified cytosine in any sequence context (94), which is distinct from the CXXC domains of MLL (95), CFP1 (96), and Dnmt1 (97), which are restricted to unmodified CpG sites. CHTOP (chromatin target of PRMT1) binds to 5hmC (98). The modification of the CG dinucleotide (underlined) of TGACGCAA to 5mC, 5fC, or 5caC enhances DNA binding by the basic leucine zipper protein C/EBPβ (CCAAT-enhancer-binding protein β), whereas modification to 5hmC inhibits binding of C/EBPβ (99). The carboxylation of cytosine (5caC) in a CpG dinucleotide adjacent to the consensus recognition sequence (underlined) of the basic-helix-loop-helix transcription factor proteins Tcf3/Ascl1 (CGCANNTG) enhanced binding of the heterodimer by ∼10-fold (64).
The SET and RING finger-associated (SRA) domain of UHRF1 recognizes hemi-methylated CpG sequences containing 5mC in only one strand, such as those that arise during DNA replication (48). UHRF1 binds 5hmC DNA with >10-fold weaker affinity than 5mC DNA (49), whereas the SRA domain of UHRF2 has a slightly stronger affinity for 5hmC DNA than 5mC DNA by a factor of ∼1.5 (50). Recently, it was suggested that UHRF2 can directly and specifically bind A:5hmU (relative to A:T) in vitro (22). The SRA domain uses a DNA recognition mode vastly different from that of MBD and zinc finger (ZnF) proteins (see below). Rather, the SRA domain, as a non-enzymatic, sequence-specific DNA-binding domain, utilizes a base-flipping mechanism to interact with DNA, similar to that of DNA-modifying enzymes such as DNA methyltransferases, DNA glycosylases, and Tet dioxygenases.
Almost 20 years ago, Holliday argued that “sequences longer than CpG would be necessary for the regulation of gene expression by methylation” (51). Indeed, the recognition of some sequence-specific transcription factors is blocked by cytosine methylation (for example, CCCTC-binding factor (CTCF) (52, 53)). In contrast, certain Cys2-His2 (C2H2) ZnF proteins bind preferentially to DNA when CpG sites embedded within their recognition sequences are methylated (54). The structures of five ZnF domains bound to 5mC-containing DNA have been solved, including the transcription factors Kaiso, Zfp57, Krüppel-like factor 4 (Klf4), growth response protein 1 (Egr1), and Wilms tumor protein 1 (WT1) (55–58). Kaiso recognizes either methylated CpG dinucleotides (59) or an unmodified sequence with a TpG in the place of 5mCpG (60). Structures of the ZnF domain of Kaiso revealed that Arg-511 of Kaiso interacts with the 5mCpG and TpG dinucleotides in a similar fashion (55), forming a methyl-Arg-G triad (26) (Fig. 3b), just like that described above for the two MBD arginines. The triad is maintained for TpG because the thymine methyl group is in the equivalent position (5-carbon) to that in 5mC. Similarly, Zfp57 uses a pair of methyl-Arg-G triads to recognize 5mCpG and TpG dinucleotides on the two strands, respectively (Fig. 3c).
Klf4 shows the strongest binding to fully methylated DNA, with slightly higher affinity (∼1.5-fold) than that of the unmodified DNA, and each oxidation event, from 5mC to 5hmC to 5fC to 5caC, results in progressively weaker binding (by factors of ∼2, 3, and 6, respectively) (57). Like Kaiso, the consensus-binding sequence element for Klf4 contains either CpG, which can be methylated, or TpG, which is intrinsically methylated on one strand (Fig. 3d). Klf4 is one of the four Yamanaka reprogramming factors (61), and along with the other three (Myc, Oct4, and Sox2), all contain TpG/CpA in their consensus recognition sequences (Fig. 3d). It may not be a coincidence that non-CG (mainly CpA) methylation disappears upon induced differentiation of embryonic stem cells and is restored in induced pluripotent stem cells resulting from expression of the reprogramming factors (24), which recognize CpA-containing sequences.
Both Egr1 and WT1 bind the same consensus DNA sequence containing two CpG sites and display high affinity for the sequence with C or 5mC but much reduced affinity when 5hmC or 5fC was present, indicating that they differentiate primarily between the oxidized and unoxidized 5mC, rather than methylated C from unmethylated C (58). 5caC affected the two proteins differently, abolishing binding by Egr1 but not by WT1. This difference can be ascribed to electrostatic interactions in the binding sites. In Egr1, a negatively charged glutamate conflicts with the negatively charged carboxylate of 5caC, whereas the corresponding glutamine of WT1 interacts with this group favorably (Fig. 3i). It is interesting to note that WT1 physically interacts with Tet2 (62, 63), either by recruiting Tet2 to its target genes and/or by binding the products of Tet2 enzymatic activity. Additional examples of cytosine modification-specific effects on DNA-binding factors (including the stem cell factor Tcf3 (64)) are shown in Fig. 3.
Active DNA Demethylation via Base Excision
Initial interest in the mammalian Tet proteins primarily centered around the hypothesis that oxidized 5mC could serve as an intermediate in one or more DNA demethylation pathways wherein the oxidized 5mC derivatives (5hmC, 5fC, and 5caC) are replaced with normal cytosine. There are several mechanisms by which oxidation of 5mC could mediate DNA demethylation. The first is a DNA replication-dependent passive demethylation (37, 65–67). The hemi-hydroxymethylated CpG site (5hmCpG/CpG) is a poor substrate for the maintenance methyltransferase Dnmt1 (49) and thus unlikely to be methylated by Dnmt1 after replication (68). As noted above, a maintenance methylation function has been proposed for Dnmt3a and Dnmt3b (27) and, in vitro, the Dnmt3a-Dnmt3L and Dnmt3b-Dnmt3L complexes have approximately equal activities on hemi-methylated and hemi-hydroxymethylated CpG substrates (49). Thus, the prospect that 5mC or 5hmC might be restored after replication through a distinct (Dnmt1-independent) mechanism remains a formal possibility (see Fig. 1g).
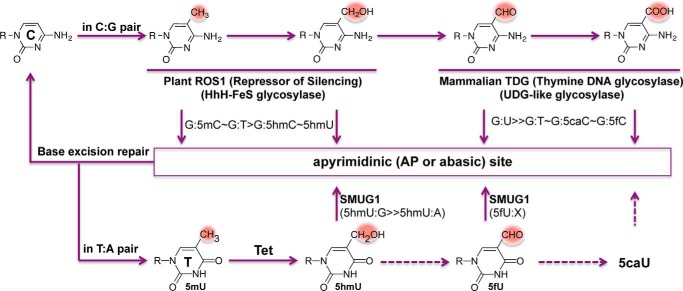
The second mechanism is an active DNA demethylation/demodification of 5mC and its derivatives that involves DNA glycosylases and base excision repair. Arabidopsis ROS1 and mammalian TDG are the two DNA glycosylases currently implicated in this process, which involves the removal of the modified cytosine base, subsequent processing by the base excision repair machinery, and ultimately the incorporation of an unmodified cytosine. The DNA glycosylases impart specificity to this reaction; ROS1 excises 5mC and 5hmC but not 5fC and 5caC (69–71), whereas TDG removes 5fC and 5caC but not 5mC and 5hmC (4, 72–74) (Fig. 4).
FIGURE 4.
DNA glycosylases involved in removing modified bases from native Watson-Crick base pairs. ROS1 excises 5mC or 5hmC from its native base pairing with guanine; TDG excises 5fC or 5caC from base pairing with guanine; and SMUG1 excises 5hmU or 5fU from its native base pairing with an adenine. The base excision repair pathway removes the resulting abasic site and restores the unmodified C or T status.
Arabidopsis ROS1 is a multi-domain bifunctional DNA glycosylase/lyase, which excises 5mC and 5hmC as well as thymine and 5hmU (i.e. the deamination products of 5mC and 5hmC) when paired with a guanine, leaving an apyrimidinic site that is subsequently incised by the lyase activity (75). ROS1 is slow in base excision and fast in apyrimidinic lyase activity in vitro (71, 76), indicating that the recognition of pyrimidine modifications might be a rate-limiting step or that other cofactors might be needed to stimulate ROS1 enzymatic activity (in a manner similar to that described for Dnmt3a activity stimulation by Dnmt3L (77)). Mammalian DNA glycosylases that excise 5mC or 5hmC have not been identified, but such activities have been reported (78, 79).
Mammalian TDG excises the mismatched base from G:X mismatches, where X is uracil, thymine, or 5hmU. These are, respectively, the deamination products of cytosine, 5mC and 5hmC. In addition, TDG excises the Tet protein products 5fC and 5caC when paired with a guanine, but not 5hmC or 5mC. It is worth noting that ROS1 is inactive on G:U (71, 76, 80), whereas TDG, although named as a thymine DNA glycosylase, has much faster activity on G:U mismatches (74, 81). Furthermore, the structurally related E. coli mismatch uracil glycosylase (eMUG) can excise 5caC and 5fC as well (74, 82). Both 5fC and 5caC exhibit an intrabase hydrogen bond between their formyl or carboxyl oxygen atoms, respectively, and the adjacent cytosine N4 exocyclic amine nitrogen atom, both in the free form (83) and in the protein-bound form (58) (Fig. 3, h and i, top panels). It is unknown whether this intrabase hydrogen bond contributes to the previously observed mutagenic potential of 5fC and 5caC in vivo and in vitro (84, 85). Both DNA and RNA polymerases sometimes misincorporate adenine opposite of the 5fC or 5caC, whereas little misincorporation is observed on 5mC- and 5hmC-containing templates. It is possible that the tendency for mismatches to arise opposite 5fC and 5caC (but not 5mC and 5hmC) is somehow correlated with differential excision by TDG. It is still not clear whether the differential excision of 5hmC versus 5fC and 5caC by TDG lies in the early steps of intrahelical interrogation to detect the C5 modification and the initiation of base flipping, or in the post-flipping steps arising from differences in the active-site transition states of the enzyme-bound complexes. It is possible that TDG/eMUG might recognize or even promote a mismatch-like “wobble” pair of G:5fC and G:5caC (with the aid of the intrabase hydrogen bond) and turn them into substrates, whereas ROS1 is sensitive to pyrimidine modifications at C5 position.
In light of the recent observation that 5hmU could also be the product of Tet-mediated thymine hydroxylation (22, 23), it is interesting to note that SMUG1 (named after single-strand-specific monofunctional uracil-DNA glycosylase) can excise 5hmU when mispaired with a guanine as well as paired with an adenine (86) or 5fU in any base pair context (87). In addition, members of the NEIL family of DNA glycosylases have been found to bind oxidized 5mC derivatives 5hmC, 5fC, or 5caC (88) and 5hmU (22), and partially rescue the loss of TDG in a gene reactivation assay (89).
Perspective
Although it is well accepted that DNA methylation patterns are replicated in a semi-conservative fashion during cell division via the selective recognition of hemi-methylated CpG dinucleotides at DNA replication forks, one of the unresolved fundamental questions is how, and indeed whether, the pattern of oxidized 5mC derivatives is similarly “inherited” at CpG and CpA sites. Many transcription factors (e.g. Klf4 and MeCP2) recognize consensus-binding elements, containing either CpG, which can be methylated, or TpG (or CpA), which is intrinsically methylated on one strand and can be methylated on the other strand and further modified on both strands. Transcription factors may have adapted to respond to different states of cytosine modification. These states can affect binding affinity, and so gene activity could plausibly be controlled on a much finer scale by these modifications than a simple “on” or “off.” This hints, perhaps, at new levels of subtlety and versatility in epigenetic regulatory processes.
Finally, members of base excision DNA glycosylases remove the modified cytosine and thymine bases. ROS1/DME, TDG/eMUG, and SMUG1 excise modified bases from “natural” base pairs, but have little sequence specificity for the surrounding DNA context. Another enzyme, R.PabI from Pyrococcus abyssi, initially identified as a restriction enzyme, was recently determined to be a sequence-specific adenine DNA glycosylase (90). It is therefore possible that modification-dependent and sequence-specific DNA glycosylases might exist to recognize the modification within specific sequences, similar to the family of modification-dependent restriction endonucleases (91). Methylpurine glycosylases (MPGs) from E. coli, Saccharomyces cerevisiae, and human have been reported to remove normal bases (G>A>C∼T) from DNA (92). Methylpurine glycosylase protein was recently identified as a 5hmC binder in mouse embryonic stem cells and adult mouse brain using a DNA pulldown approach combined with quantitative mass spectrometry (88). Thus, not all DNA base excision glycosylases are strictly involved in mismatch DNA repair; some actually excise modified bases with proper Watson-Crick base pairing to accomplish DNA demodification. Moreover, intriguing recent work showing that the demethylation of male and female pronuclear DNA is unaffected by the deletion of TDG from the zygote (36) suggests that other demethylation mechanisms (or other DNA glycosylases) must exist downstream of Tet3-mediated oxidation.
This work was supported by National Institutes of Health Grants GM049245-21, GM105132-02 (subcontract via Yu Zheng), and DK094346-01 (to X. C.) and CA077337 and CA132065 (to P. M. V.), and in part by developmental funds from the Winship Cancer Institute of Emory University Cancer Center Support Grant (Grant P30CA138292). This is the third article in the Thematic Minireview series “Metals in Biology: α-Ketoglutarate/Iron-dependent Dioxygenases.” The authors declare that they have no conflicts of interest with the contents of this article.
- 5mC
- 5-methylcytosine
- 5hmC
- 5-hydroxymethylcytosine
- 5fC
- 5-formylcytosine
- 5caC
- 5-carboxylcytosine
- 5hmU
- 5-hydroxymethyluracil
- 5mU
- 5-methyluridine
- 5fU
- 5-fluorouracil
- 5caU
- 5-carboxyl-uracil (isoorotate)
- Dnmt
- DNA methyltransferase(s)
- MBD
- methyl-binding domain protein
- SRA
- SET and RING finger-associated
- ZnF
- zinc finger
- TDG
- thymine-DNA glycosylase
- eMUG
- E. coli mismatch uracil glycosylase
- N6mA
- N6-methyladenine
- mCpA
- methylated CpA
- mCpG
- methylated CpG
- NgTet1
- Naegleria Tet-like dioxygenase.
References
- 1. Kriaucionis S., Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., He C., Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. He Y. F., Li B. Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C. X., Zhang K., He C., Xu G. L. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pfaffeneder T., Hackner B., Truss M., Münzel M., Müller M., Deiml C. A., Hagemeier C., Carell T. (2011) The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Ed. Engl. 50, 7008–7012 [DOI] [PubMed] [Google Scholar]
- 6. Bestor T., Laudano A., Mattaliano R., Ingram V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: the carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203, 971–983 [DOI] [PubMed] [Google Scholar]
- 7. Okano M., Xie S., Li E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219–220 [DOI] [PubMed] [Google Scholar]
- 8. Gowher H., Jeltsch A. (2001) Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J. Mol. Biol. 309, 1201–1208 [DOI] [PubMed] [Google Scholar]
- 9. Hashimoto H., Pais J. E., Zhang X., Saleh L., Fu Z. Q., Dai N., Corrêa I. R., Jr., Zheng Y., Cheng X. (2014) Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506, 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L., Chen W., Iyer L. M., Hu J., Wang G., Fu Y., Yu M., Dai Q., Aravind L., He C. (2014) A TET homologue protein from Coprinopsis cinerea (CcTET) that biochemically converts 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine. J. Am. Chem. Soc. 136, 4801–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wojciechowski M., Rafalski D., Kucharski R., Misztal K., Maleszka J., Bochtler M., Maleszka R. (2014) Insights into DNA hydroxymethylation in the honeybee from in-depth analyses of TET dioxygenase. Open Biol. 4, 140110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi Y. G., Tsukada Y. (2013) The discovery of histone demethylases. Cold Spring Harb. Perspect. Biol. 5, a017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trewick S. C., Henshaw T. F., Hausinger R. P., Lindahl T., Sedgwick B. (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178 [DOI] [PubMed] [Google Scholar]
- 14. Falnes P. Ø., Johansen R. F., Seeberg E. (2002) AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419, 178–182 [DOI] [PubMed] [Google Scholar]
- 15. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y. G., He C. (2011) N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., Vågbø C. B., Shi Y., Wang W. L., Song S. H., Lu Z., Bosmans R. P., Dai Q., Hao Y. J., Yang X., Zhao W. M., Tong W. M., Wang X. J., Bogdan F., Furu K., Fu Y., Jia G., Zhao X., Liu J., Krokan H. E., Klungland A., Yang Y. G., He C. (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trewick S. C., McLaughlin P. J., Allshire R. C. (2005) Methylation: lost in hydroxylation? EMBO Rep. 6, 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 19. Klimasauskas S., Kumar S., Roberts R. J., Cheng X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357–369 [DOI] [PubMed] [Google Scholar]
- 20. Wu J. C., Santi D. V. (1987) Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 262, 4778–4786 [PubMed] [Google Scholar]
- 21. Li E., Zhang Y. (2014) DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 6, a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfaffeneder T., Spada F., Wagner M., Brandmayr C., Laube S. K., Eisen D., Truss M., Steinbacher J., Hackner B., Kotljarova O., Schuermann D., Michalakis S., Kosmatchev O., Schiesser S., Steigenberger B., Raddaoui N., Kashiwazaki G., Müller U., Spruijt C. G., Vermeulen M., Leonhardt H., Schär P., Müller M., Carell T. (2014) Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 10, 574–581 [DOI] [PubMed] [Google Scholar]
- 23. Pais J. E., Dai N., Tamanaha E., Vaisvila R., Fomenkov A. I., Bitinaite J., Sun Z., Guan S., Corrêa I. R., Jr., Noren C. J., Cheng X., Roberts R. J., Zheng Y., Saleh L. (2015) Biochemical characterization of a Naegleria TET-like oxygenase and its application in single molecule sequencing of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 112, 4316–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., Edsall L., Antosiewicz-Bourget J., Stewart R., Ruotti V., Millar A. H., Thomson J. A., Ren B., Ecker J. R. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lister R., Mukamel E. A., Nery J. R., Urich M., Puddifoot C. A., Johnson N. D., Lucero J., Huang Y., Dwork A. J., Schultz M. D., Yu M., Tonti-Filippini J., Heyn H., Hu S., Wu J. C., Rao A., Esteller M., He C., Haghighi F. G., Sejnowski T. J., Behrens M. M., Ecker J. R. (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Y., Zhang X., Blumenthal R. M., Cheng X. (2013) A common mode of recognition for methylated CpG. Trends Biochem. Sci. 38, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones P. A., Liang G. (2009) Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10, 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dawlaty M. M., Ganz K., Powell B. E., Hu Y. C., Markoulaki S., Cheng A. W., Gao Q., Kim J., Choi S. W., Page D. C., Jaenisch R. (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z., Cai X., Cai C. L., Wang J., Zhang W., Petersen B. E., Yang F. C., Xu M. (2011) Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118, 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moran-Crusio K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C., Figueroa M. E., Vasanthakumar A., Patel J., Zhao X., Perna F., Pandey S., Madzo J., Song C., Dai Q., He C., Ibrahim S., Beran M., Zavadil J., Nimer S. D., Melnick A., Godley L. A., Aifantis I., Levine R. L. (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dawlaty M. M., Breiling A., Le T., Raddatz G., Barrasa M. I., Cheng A. W., Gao Q., Powell B. E., Li Z., Xu M., Faull K. F., Lyko F., Jaenisch R. (2013) Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gu T. P., Guo F., Yang H., Wu H. P., Xu G. F., Liu W., Xie Z. G., Shi L., He X., Jin S. G., Iqbal K., Shi Y. G., Deng Z., Szabó P. E., Pfeifer G. P., Li J., Xu G. L. (2011) The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 [DOI] [PubMed] [Google Scholar]
- 33. Dawlaty M. M., Breiling A., Le T., Barrasa M. I., Raddatz G., Gao Q., Powell B. E., Cheng A. W., Faull K. F., Lyko F., Jaenisch R. (2014) Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 29, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wossidlo M., Nakamura T., Lepikhov K., Marques C. J., Zakhartchenko V., Boiani M., Arand J., Nakano T., Reik W., Walter J. (2011) 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241. [DOI] [PubMed] [Google Scholar]
- 35. Iqbal K., Jin S. G., Pfeifer G. P., Szabó P. E. (2011) Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 108, 3642–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo F., Li X., Liang D., Li T., Zhu P., Guo H., Wu X., Wen L., Gu T. P., Hu B., Walsh C. P., Li J., Tang F., Xu G. L. (2014) Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–458 [DOI] [PubMed] [Google Scholar]
- 37. Shen L., Inoue A., He J., Liu Y., Lu F., Zhang Y. (2014) Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15, 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arand J., Wossidlo M., Lepikhov K., Peat J. R., Reik W., Walter J. (2015) Selective impairment of methylation maintenance is the major cause of DNA methylation reprogramming in the early embryo. Epigenetics Chromatin 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raddatz G., Guzzardo P. M., Olova N., Fantappié M. R., Rampp M., Schaefer M., Reik W., Hannon G. J., Lyko F. (2013) Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl. Acad. Sci. U.S.A. 110, 8627–8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dunwell T. L., McGuffin L. J., Dunwell J. M., Pfeifer G. P. (2013) The mysterious presence of a 5-methylcytosine oxidase in the Drosophila genome: possible explanations. Cell Cycle 12, 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goll M. G., Kirpekar F., Maggert K. A., Yoder J. A., Hsieh C. L., Zhang X., Golic K. G., Jacobsen S. E., Bestor T. H. (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398 [DOI] [PubMed] [Google Scholar]
- 42. Hu L., Li Z., Cheng J., Rao Q., Gong W., Liu M., Shi Y. G., Zhu J., Wang P., Xu Y. (2013) Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell 155, 1545–1555 [DOI] [PubMed] [Google Scholar]
- 43. Upadhyay A. K., Horton J. R., Zhang X., Cheng X. (2011) Coordinated methyl-lysine erasure: structural and functional linkage of a Jumonji demethylase domain and a reader domain. Curr. Opin. Struct. Biol. 21, 750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ho K. L., McNae I. W., Schmiedeberg L., Klose R. J., Bird A. P., Walkinshaw M. D. (2008) MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol. Cell 29, 525–531 [DOI] [PubMed] [Google Scholar]
- 45. Guo J. U., Su Y., Shin J. H., Shin J., Li H., Xie B., Zhong C., Hu S., Le T., Fan G., Zhu H., Chang Q., Gao Y., Ming G. L., Song H. (2014) Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gabel H. W., Kinde B., Stroud H., Gilbert C. S., Harmin D. A., Kastan N. R., Hemberg M., Ebert D. H., Greenberg M. E. (2015) Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Otani J., Arita K., Kato T., Kinoshita M., Kimura H., Suetake I., Tajima S., Ariyoshi M., Shirakawa M. (2013) Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4. J. Biol. Chem. 288, 6351–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hashimoto H., Horton J. R., Zhang X., Cheng X. (2009) UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics 4, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hashimoto H., Liu Y., Upadhyay A. K., Chang Y., Howerton S. B., Vertino P. M., Zhang X., Cheng X. (2012) Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 40, 4841–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou T., Xiong J., Wang M., Yang N., Wong J., Zhu B., Xu R. M. (2014) Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol. Cell 54, 879–886 [DOI] [PubMed] [Google Scholar]
- 51. Holliday R. (1996) DNA methylation in eukaryotes: 20 years on. in Epigenetic Mechanisms of Gene Regulation, pp. 5–27, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., Tilghman S. M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 [DOI] [PubMed] [Google Scholar]
- 53. Renda M., Baglivo I., Burgess-Beusse B., Esposito S., Fattorusso R., Felsenfeld G., Pedone P. V. (2007) Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 282, 33336–33345 [DOI] [PubMed] [Google Scholar]
- 54. Sasai N., Nakao M., Defossez P. A. (2010) Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res. 38, 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buck-Koehntop B. A., Stanfield R. L., Ekiert D. C., Martinez-Yamout M. A., Dyson H. J., Wilson I. A., Wright P. E. (2012) Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc. Natl. Acad. Sci. U.S.A. 109, 15229–15234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Y., Toh H., Sasaki H., Zhang X., Cheng X. (2012) An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 26, 2374–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu Y., Olanrewaju Y. O., Zheng Y., Hashimoto H., Blumenthal R. M., Zhang X., Cheng X. (2014) Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 42, 4859–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hashimoto H., Olanrewaju Y. O., Zheng Y., Wilson G. G., Zhang X., Cheng X. (2014) Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 28, 2304–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Prokhortchouk A., Hendrich B., Jørgensen H., Ruzov A., Wilm M., Georgiev G., Bird A., Prokhortchouk E. (2001) The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15, 1613–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Daniel J. M., Spring C. M., Crawford H. C., Reynolds A. B., Baig A. (2002) The p120ctn-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 30, 2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 62. Rampal R., Alkalin A., Madzo J., Vasanthakumar A., Pronier E., Patel J., Li Y., Ahn J., Abdel-Wahab O., Shih A., Lu C., Ward P. S., Tsai J. J., Hricik T., Tosello V., Tallman J. E., Zhao X., Daniels D., Dai Q., Ciminio L., Aifantis I., He C., Fuks F., Tallman M. S., Ferrando A., Nimer S., Paietta E., Thompson C. B., Licht J. D., Mason C. E., Godley L. A., Melnick A., Figueroa M. E., Levine R. L. (2014) DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Reports 9, 1841–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y., Xiao M., Chen X., Chen L., Xu Y., Lv L., Wang P., Yang H., Ma S., Lin H., Jiao B., Ren R., Ye D., Guan K. L., Xiong Y. (2015) WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol. Cell 57, 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Golla J. P., Zhao J., Mann I. K., Sayeed S. K., Mandal A., Rose R. B., Vinson C. (2014) Carboxylation of cytosine (5caC) in the CG dinucleotide in the E-box motif (CGCAG|GTG) increases binding of the Tcf3|Ascl1 helix-loop-helix heterodimer 10-fold. Biochem. Biophys. Res. Commun. 449, 248–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Inoue A., Zhang Y. (2011) Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Inoue A., Shen L., Dai Q., He C., Zhang Y. (2011) Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21, 1670–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kubosaki A., Tomaru Y., Furuhata E., Suzuki T., Shin J. W., Simon C., Ando Y., Hasegawa R., Hayashizaki Y., Suzuki H. (2012) CpG site-specific alteration of hydroxymethylcytosine to methylcytosine beyond DNA replication. Biochem. Biophys. Res. Commun. 426, 141–147 [DOI] [PubMed] [Google Scholar]
- 68. Valinluck V., Sowers L. C. (2007) Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 67, 946–950 [DOI] [PubMed] [Google Scholar]
- 69. Gong Z., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T., David L., Zhu J. K. (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 [DOI] [PubMed] [Google Scholar]
- 70. Ponferrada-Marín M. I., Parrilla-Doblas J. T., Roldán-Arjona T., Ariza R. R. (2011) A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic Acids Res. 39, 1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hong S., Hashimoto H., Kow Y. W., Zhang X., Cheng X. (2014) The carboxy-terminal domain of ROS1 is essential for 5-methylcytosine DNA glycosylase activity. J. Mol. Biol. 426, 3703–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maiti A., Drohat A. C. (2011) Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang L., Lu X., Lu J., Liang H., Dai Q., Xu G. L., Luo C., Jiang H., He C. (2012) Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 8, 328–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hashimoto H., Hong S., Bhagwat A. S., Zhang X., Cheng X. (2012) Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 40, 10203–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li Y., Córdoba-Cañero D., Qian W., Zhu X., Tang K., Zhang H., Ariza R. R., Roldán-Arjona T., Zhu J. K. (2015) An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis. PLoS Genet. 11, e1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ponferrada-Marín M. I., Roldán-Arjona T., Ariza R. R. (2009) ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 37, 4264–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jia D., Jurkowska R. Z., Zhang X., Jeltsch A., Cheng X. (2007) Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vairapandi M., Duker N. J. (1993) Enzymic removal of 5-methylcytosine from DNA by a human DNA-glycosylase. Nucleic Acids Res. 21, 5323–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cannon S. V., Cummings A., Teebor G. W. (1988) 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem. Biophys. Res. Commun. 151, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 80. Morales-Ruiz T., Ortega-Galisteo A. P., Ponferrada-Marín M. I., Martínez-Macías M. I., Ariza R. R., Roldán-Arjona T. (2006) DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. U.S.A. 103, 6853–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bennett M. T., Rodgers M. T., Hebert A. S., Ruslander L. E., Eisele L., Drohat A. C. (2006) Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc. 128, 12510–12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Moréra S., Grin I., Vigouroux A., Couvé S., Henriot V., Saparbaev M., Ishchenko A. A. (2012) Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 40, 9917–9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Szulik M. W., Pallan P. S., Nocek B., Voehler M., Banerjee S., Brooks S., Joachimiak A., Egli M., Eichman B. F., Stone M. P. (2015) Differential stabilities and sequence-dependent base pair opening dynamics of Watson-Crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine. Biochemistry 54, 1294–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kamiya H., Tsuchiya H., Karino N., Ueno Y., Matsuda A., Harashima H. (2002) Mutagenicity of 5-formylcytosine, an oxidation product of 5-methylcytosine, in DNA in mammalian cells. J. Biochem. 132, 551–555 [DOI] [PubMed] [Google Scholar]
- 85. Kellinger M. W., Song C. X., Chong J., Lu X. Y., He C., Wang D. (2012) 5-Formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 19, 831–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wibley J. E., Waters T. R., Haushalter K., Verdine G. L., Pearl L. H. (2003) Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol. Cell 11, 1647–1659 [DOI] [PubMed] [Google Scholar]
- 87. Knaevelsrud I., Slupphaug G., Leiros I., Matsuda A., Ruoff P., Bjelland S. (2009) Opposite-base dependent excision of 5-formyluracil from DNA by hSMUG1. Int. J. Radiat. Biol. 85, 413–420 [DOI] [PubMed] [Google Scholar]
- 88. Spruijt C. G., Gnerlich F., Smits A. H., Pfaffeneder T., Jansen P. W., Bauer C., Münzel M., Wagner M., Müller M., Khan F., Eberl H. C., Mensinga A., Brinkman A. B., Lephikov K., Müller U., Walter J., Boelens R., van Ingen H., Leonhardt H., Carell T., Vermeulen M. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 [DOI] [PubMed] [Google Scholar]
- 89. Müller U., Bauer C., Siegl M., Rottach A., Leonhardt H. (2014) TET-mediated oxidation of methylcytosine causes TDG or NEIL glycosylase dependent gene reactivation. Nucleic Acids Res. 42, 8592–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Miyazono K., Furuta Y., Watanabe-Matsui M., Miyakawa T., Ito T., Kobayashi I., Tanokura M. (2014) A sequence-specific DNA glycosylase mediates restriction-modification in Pyrococcus abyssi. Nat. Commun. 5, 3178. [DOI] [PubMed] [Google Scholar]
- 91. Cohen-Karni D., Xu D., Apone L., Fomenkov A., Sun Z., Davis P. J., Kinney S. R., Yamada-Mabuchi M., Xu S. Y., Davis T., Pradhan S., Roberts R. J., Zheng Y. (2011) The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl. Acad. Sci. U.S.A. 108, 11040–11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Berdal K. G., Johansen R. F., Seeberg E. (1998) Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J. 17, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 94. Xu Y., Xu C., Kato A., Tempel W., Abreu J. G., Bian C., Hu Y., Hu D., Zhao B., Cerovina T., Diao J., Wu F., He H. H., Cui Q., Clark E., Ma C., Barbara A., Veenstra G. J., Xu G., Kaiser U. B., Liu X. S., Sugrue S. P., He X., Min J., Kato Y., Shi Y. G. (2012) Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 151, 1200–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Allen M. D., Grummitt C. G., Hilcenko C., Min S. Y., Tonkin L. M., Johnson C. M., Freund S. M., Bycroft M., Warren A. J. (2006) Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 25, 4503–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xu C., Bian C., Lam R., Dong A., Min J. (2011) The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat. Commun. 2, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Song J., Rechkoblit O., Bestor T. H., Patel D. J. (2011) Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Takai H., Masuda K., Sato T., Sakaguchi Y., Suzuki T., Suzuki T., Koyama-Nasu R., Nasu-Nishimura Y., Katou Y., Ogawa H., Morishita Y., Kozuka-Hata H., Oyama M., Todo T., Ino Y., Mukasa A., Saito N., Toyoshima C., Shirahige K., Akiyama T. (2014) 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 9, 48–60 [DOI] [PubMed] [Google Scholar]
- 99. Khund Sayeed S., Zhao J., Sathyanarayana B. K., Golla J. P., Vinson C. (2015) C/EBPβ (CEBPB) protein binding to the C/EBP|CRE DNA 8-mer TTGC|GTCA is inhibited by 5hmC and enhanced by 5mC, 5fC, and 5caC in the CG dinucleotide. Biochim Biophys Acta 1849, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., Yin R., Zhang D., Zhang P., Liu J., Li C., Liu B., Luo Y., Zhu Y., Zhang N., He S., He C., Wang H., Chen D. (2015) N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906 [DOI] [PubMed] [Google Scholar]