Background: The post-transcriptional regulation of interleukin-6 (IL-6) production is critical for immune homeostasis.
Results: Monocyte chemotactic protein-induced protein 1 (MCPIP1) and MCPIP4 form a complex but they act independently in regulation of IL-6 mRNA degradation.
Conclusion: MCPIP1 and MCPIP4 may additively contribute to control IL-6 production.
Significance: The study may help to understand the mechanisms by which MCPIP1 protein family control immune homeostasis.
Keywords: inflammation, interleukin, mRNA decay, protein domain, protein-protein interaction, RNase
Abstract
It was recently demonstrated that MCPIP1 is a critical factor that controls inflammation and immune homeostasis; however, the relationship between MCPIP1 and other members of this protein family is largely unknown. Here, we report that MCPIP1 interacts with MCPIP4 to form a protein complex, but acts independently in the regulation of IL-6 mRNA degradation. In an effort to identify MCPIP1-interacting proteins by co-immunoprecipitation (Co-IP) and mass-spec analysis, MCPIP4 was identified as a MCPIP1-interacting protein, which was further confirmed by Co-IP and mammalian two-hybrid assay. Immunofluorescence staining showed that MCPIP4 was co-localized with MCPIP1 in the GW-body, which features GW182 and Argonaute 2. Further studies showed that MCPIP1 and MCPIP4 act independently in regulation of IL-6 mRNA degradation. These results suggest that MCPIP1 and MCPIP4 may additively contribute to control IL-6 production in vivo.
Introduction
Monocyte chemotactic protein-induced protein 1 (MCPIP1,2 also known as ZC3H12A and Regnase-1) is a prototype member of a novel protein family, which includes MCPIP1, MCPIP2, MCPIP3, and MCPIP4 (1, 2). The unique feature of the MCPIP1 protein family is characterized by a CCCH-zinc finger motif located in the middle region of these proteins (1, 2). We have identified the whole CCCH-zinc finger-containing protein family in both human and mouse genomes (3). There are ∼60 CCCH-zinc finger proteins in both human and mouse genomes. Most CCCH-type zinc finger proteins are involved in RNA metabolic pathways such as splicing, polyadenylation, and mRNA decay (3). For example, tristetraprolin, Roquin, and ZAP are well-studied CCCH-zinc finger-containing proteins that target mRNA degradation through different mechanisms (4–9). MCPIP1 is an endonuclease and selectively destabilizes mRNAs that encode certain inflammatory cytokines such as IL-6 and IL-12 (2, 10). Through this central mechanism, MCPIP1 serves as an essential regulator in inflammatory cell activation and immune homeostasis (2). MCPIP1 knock-out mice developed spontaneous inflammatory diseases accompanied by splenomegaly, lymphadenopathy, and multi-organ inflammation especially in the lungs (2, 11, 12). T cell-specific deletion of MCPIP1 produces pathogenic T cells with hyperactivated phenotypes as well as autoimmune diseases (13).
MCPIP1 is a multi-domain-containing protein that includes an ubiquitin association domain (UBA) at the N terminus, a putative NYN-RNase domain, followed by a CCCH-zinc finger domain (ZF), and a proline-rich domain (PRD) at the C terminus (11). A recent study compared the crystal structure of the putative NYN-RNase domain with other reported RNase proteins and suggested that MCPIP1 is a functional RNase (14). The mRNA targets of MCPIP1 nuclease are now expanding to c-Rel, IL-2, ICOS, Ox40, TNFR2, GATA3, and MCPIP1 self mRNA (13, 15, 16). MCPIP1 promotes their degradation by targeting their 3′-untranslated region (3′-UTR) (10, 13). MCPIP1 specifically recognized a stem-loop structure on the 3′-UTR of its substrate mRNAs (10).
MCPIP4 (also known as ZC3H12D, TFL, and p34) was originally reported as a putative tumor suppressor that is deregulated in transformed follicular lymphoma (17–19). Similar to MCPIP1, MCPIP4 is also remarkably induced by Toll-like receptor activation in macrophages and overexpression of MCPIP4 also represses inflammatory activation of macrophages (20). The role of MCPIP4 in vivo seems overlapped but less important than MCPIP1. For example, MCPIP4-null mice showed pretty normal phenotypes under normal condition, but exhibited more activated lymphocytes upon stimulation (21).
In this study, we first found that MCPIP1 interacts with MCPIP4 to form a protein complex but they act independently in regulate IL-6 mRNA degradation, suggesting that MCPIP1 and MCPIP4 may additively contribute to control IL-6 production in vivo.
Materials and Methods
Cells
HEK293, COS-7, HeLa, and RAW264.7 cells were obtained from the American Type Culture Collection. These cells were grown as a monolayer in DMEM (Invitrogen) containing 10% FBS, 2 mm l-glutamine, with 100 units/ml penicillin and 100 μg/ml streptomycin in 5.0% CO2. HEK293-MCPIP1 stable cell line was established by lentiviral transduction of HEK293 with a GFP-MCPIP1-expressing construct and maintained in complete medium with 200 μg/ml of G418 and 0.25 μg/ml of puromycin.
Plasmids
MCPIP1-GFP, HA-MCPIP1, Flag-MCPIP1, and its mutants were described previously (11). Flag-MCPIP4 was generated via PCR and cloned into the NotI/XbaI sites of pCMV-MAT-tag-Flag1 (Sigma). pEGFP-MCPIP1 and pEGFP-MCPIP4 were constructed via PCR and cloned into the EcoRI/SalI sites of pEGFP-C1 (Clontech). The deletion mutants of pEGFP-MCPIP1 and pEGFP-MCPIP4 were constructed by PCR using pEGFP-MCPIP1 or pEGFP-MCPIP4 as template with corresponding primers. HA-MCPIP4 was generated via PCR and inserted into pCMV4–3HA vector by HindIII and XbaI. pBIND-MCPIP1, pBIND-MCPIP4, pACT-MCPIP1, and pACT-MCPIP4 were constructed via PCR and cloned into the EcoRV/NotI sites of pBIND vector (Clontech) or pACT vector (Clontech). Truncated regions corresponding to 1–457, 1–300, 300–457, and 325–457 of MCPIP1 or 1–356, 357–527, 1–258, 259–527,259–356, 279–356, and del(357–456) of MCPIP4 were generated by PCR using pACT-MCPIP1 or pBIND-MCPIP4 as template with corresponding primers. MCPIP4 (D94N) point mutant was generated by using the Stratagene site-directed mutagenesis method. pcDNA3-Flag-hZC3H12B and pcDNA3-Flag-hZC3H12C were kindly provided by Dr. Hiroshi Suzuki (University of Tokyo, Japan) and described previously (22). The luciferase reporter containing IL6–3′-UTR (1–403) was a gift from Dr. Keith L. Kirkwood (Medical University of South Carolina, Ref. 23). The pRL-TK is a Renilla luciferase reporter from Promega. pGL3-Control is a luciferase reporter without IL6–3′-UTR from Promega. pEGFP-C1 was from Clontech, pBIND, pACT, pBIND-ID, and pACT-MyoD were from Promega.
Reagents
Rabbit anti-MCPIP1 polyclonal antibody was from Genetex, GW182 antibody were purchased from Santa Cruz Biotechnology. Mouse anti-Zc3h12d (MCPIP4) monoclonal antibody was kindly provided by Dr. T. Matsui (20, 21). Rabbit anti-MCPIP4 antibody was purchased from Proteintech. EGFP and actin antibodies were purchased from Cell Signaling Technology Inc. Bovine pancreatic RNase A, Flag, and HA antibodies, anti-Flag M2 affinity gel and anti-HA immunoprecipitation kit and doxycycline were purchased from Sigma. Dual-Luciferase Reporter Assay system was purchased from Promega.
Identification of MCPIP1-interacting Proteins by Co-IP and Mass-Spec Analysis
HEK293 cells were transfected with empty pCMV-Flag vector or pCMV-Flag-MCPIP1 and incubated for 48 h. Transfected cells were lysed in a buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, and protease inhibitor mixture (Roche). Cell lysates were precleared with mouse immunoglobulin G (IgG)-agarose and incubated with 1 μl of mouse anti-Flag M2-agarose beads (Sigma) on ice for 2 h. After an extensive wash with the wash buffer containing 50 mm Tris-HCl, pH 7.4, and 150 mm NaCl, proteins bound to the beads were then eluted into 1× sodium dodecyl sulfate (SDS) running buffer by heating at 95 °C for 5 min. The proteins were separated on a 10% SDS-PAGE and stained by Sypro Ruby. Stained bands were excised out, and proteins were identified by LTQ-orbitrap-velos mass spectrometer.
Confocal Microscopic Analysis
COS-7 cells seeded on glass coverslips were transfected with pEGFP-C1, pEGFP-MCPIP1, or pEGFP-MCPIP4 in a 6-well plate using Lipofectamine 2000. After transfection for 24 h, the cells were washed twice with cold PBS, fixed with 4% paraformaldehyde (pH 7.4) in PBS for 20 min, permeabilized with 0.2% Triton X-100 in PBS for 15 min, and then stained with DAPI (Life Technologies) to visualize the nuclear DNA. The cell images were recorded with a LEICA laser-scanning confocal microscope.
Co-immunoprecipitation
HEK293 Cells were transfected with Flag-MCPIP4 and HA-MCPIP1 by the calcium precipitation. After 24 h, the cells were lysed with cold CelLytic M lysis buffer (Sigma) with protease inhibitors including 1 mm PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin. The lysate supernatant was pre-cleared by incubating the cell lysates with protein A-agarose beads (Invitrogen) for 60 min at 4 °C with gentle agitation, and then incubating with 25 μl of the anti-Flag M2 or anti-HA-agarose beads at 4 °C for 4 h with gentle mixing. Samples were extensively washed two times using wash buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.05% Triton X-100) with protease inhibitors. The precipitates were treated with or without 100 units/ml of RNase A (Sigma) at room temperature for 30 min. The agarose beads were washed three times with the wash buffer. The immunoprecipitates were eluted from the beads with 100 μl of loading buffer, resolved by 12% SDS-PAGE, and analyzed by immunoblotting with Flag or HA antibodies. Membranes were developed using an enhanced chemiluminescence (ECL) detection system (GE Healthcare).
Immunofluorescence
HEK293 and HeLa cells were transfected with the expression plasmids as indicated. After 24 h, the transfected cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.2% Triton X-100 in PBS for 20 min, and then incubated in BlockAid™ Blocking Solution (Molecular Probes) for 1 h at room temperature. Then the cells were incubated with primary antibody or control IgG diluted in PBS at 4 °C overnight. The cells were briefly rinsed with PBS, incubated with Alexa Fluor® 488 or Alexa Fluor® 594 fluorescent-labeled secondary antibody (Life Technologies) for 1 h at room temperature. After intensive wash with PBS, the slides were mounted by cold Vectashield Hard Set mounting medium with DAPI, and visualized by a Nikon C1 plus confocal.
Mammalian Two-hybrid Assay
To analyze the interaction between MCPIP1 and MCPIP4 in mammalian cells, HEK293 cells were co-transfected with various pBIND fusion expression plasmids, pACT expression plasmids, and the luciferase reporter pG5luc (Promega) in a 24-well plate using Lipofectamine 2000. 24 h after transfection, the transfected cells were harvested, washed twice with PBS, and lysed with 1× Passive Lysis Buffer (Promega). Luciferase assay was performed using the Dual-Luciferase Reporter Assay system (Promega) according to the manufacturer's instructions. All the experiments were carried out in triplicate, and the luciferase value was determined using a GloMax® 96 Microplate Luminometer (Promega).
Transfection
Transient transfection into RAW264.7 cells was performed by electroporation following the manufacturer's instruction (Amaxa). Briefly, RAW264.7 cells were grown to confluence in DMEM medium supplemented with 10% FBS. Cells were collected and washed once with DMEM medium and resuspended with the electroporation buffer (Amaxa). After electroporation, the cells were plated on 6-well plates, and the transfection efficiency was monitored by fluorescent microscopy.
Short Interference RNA
The plasmids encoding the short hairpin RNA (shRNA) targeting to mouse MCPIP1 (1# AGCGAGGCCACACAGATATTA: 2# GCTATGATGACCGCTTCATTG: 3# TGGTCTGAGCCGTACCCATTA: 4# CTGTGTACAGAGGCGAGATTT) and MCPIP4 (1# GCTCATGTTCTCCTTTGTAAA: 2# CACCTTACAGAGATAAGATTC: 3# CTTAGGAGACAGGTTCATAAC: 4# GATACTCCTATCAGAGAGCAA) as well as its negative control (CAACAAGATGAAGAGCACCAA) were purchased from Sigma. The plasmids were transfected into Raw264.7 cells by electroporation (Amaxa) following the manufacturer's instruction. 48 h later, the cells were treated with 20 ng/ml of Pam3CSK4 (a Toll-like receptor 2 agonist) for 6 h. The cells were then harvested, and RNA was isolated for QPCR.
Quantitative Real-time PCR (QPCR)
After removing the genomic DNA using DNase I (Ambion), 2 μg of total RNA was reverse-transcribed to cDNA using a high-capacity cDNA reverse transcription kit (Life Technologies). QPCR was performed with StepOne Plus real-time PCR system (ABI) using SYBR Green master mix (ABI). Forty cycles were conducted as follows: 95 °C for 30 s, 60 °C for 30 s, preceded by 1 min at 95 °C for polymerase activation. Primer sequences for all genes we measured in this report are available upon request. Quantification was performed by the delta cycle time method, with β-actin used for normalization.
Protein Isolation and Western Blot
Protein isolation and Western blot were essentially performed as described previously (1).
Statistics
Data are expressed as mean ± S.D. For comparison between two groups, the unpaired Student's test was used. For multiple comparisons, analysis of variance followed by unpaired Student's test was used. A value of p < 0.05 was considered significant.
Results
Identification of MCPIP4 as a MCPIP1-interacting Protein
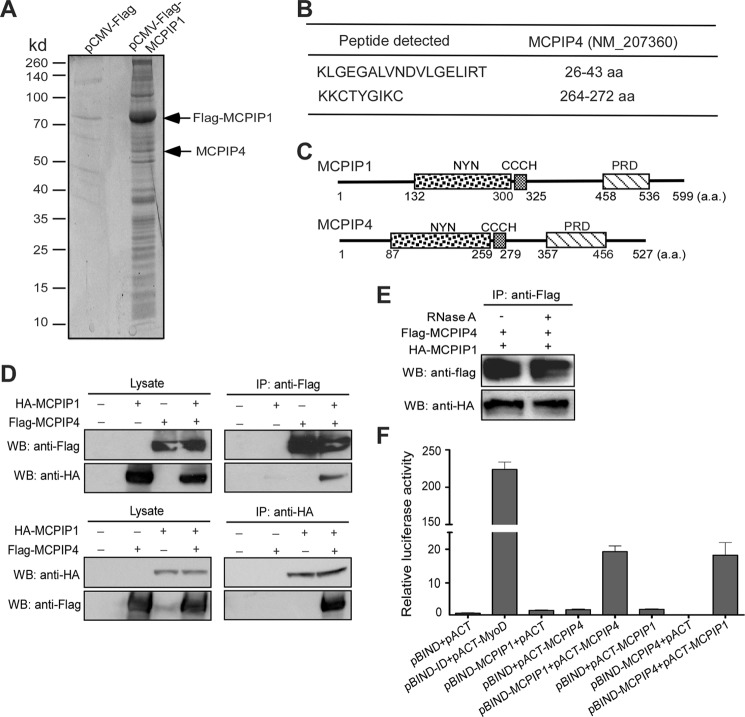
Previously, we and others have demonstrated that MCPIP1 is crucial to control inflammation and immune homeostasis (10, 11, 24–26). However, the molecular mechanisms need to be further elucidated. To identify the interacting proteins that may involve in MCPIP1-mediated repression of inflammation and immunity, we performed immunoprecipitation (IP) followed with mass-spec (MS) analysis. Flag-tagged MCPIP1 was transiently expressed in HEK293 cells, and MCPIP1-bound proteins were immunoprecipitated with anti-Flag M2-agarose beads. After extensive wash, eluted proteins were separated on a 10% SDS-PAGE and stained by Sypro Ruby. Stained bands were excised out, and proteins were identified by LTQ-orbitrap-velos mass spectrometer. One protein band with an apparent molecular mass of 58 kDa was repeatedly identified in the IP assay compared with the lysate from control HEK293 cells transfected with pCMV-Flag vector (Fig. 1A). MS analysis identified this protein as MCPIP4 (ZC3H12D) (Fig. 1B). Other proteins identified in the assay will be described elsewhere.
FIGURE 1.
Identification of MCPIP4 as a MCPIP1-interacting protein. A, HEK293 cells were transfected with Flag-MCPIP1 or pCMV-Flag vector. After 24 h, cell lysates were immunoprecipitated by anti-Flag M2-agarose beads. After wash, the immunoprecipitates were separated by 10% SDS-PAGE and stained by Spyro Ruby; B, stained bands were excised out and analyzed by LTQ-orbitrap-velos mass-spectrometer. Two fragments targeted on MCPIP4 amino acid sequence were indicated. C, scheme of MCPIP1 and MCPIP4 protein domains. D, HA-tagged MCPIP1 and Flag-tagged MCPIP4 expression vectors were co-transfected into HEK293 cells. Cell lysates were prepared and incubated with either anti-HA or anti-Flag beads to precipitate MCPIP1 and MCPIP4, respectively. The cell lysates and immunoprecipitates were subjected to Western blot analysis with anti-HA or anti-Flag antibodies. E, HA-tagged MCPIP1 and Flag-tagged MCPIP4 expression vectors were co-transfected into HEK293 cells. Cell lysates were incubated with anti-Flag beads to precipitate MCPIP4. The immunoprecipitates were treated with RNase A (100 unit/ml) for 30 min and then subjected to Western blot analysis with anti-HA or anti-Flag antibodies. F, mammalian two-hybrid assay for the interaction of MCPIP1 and MCPIP4. Different combinations of pBIND- and pACT-derived expression vectors were co-transfected with a reporter containing five Gal4 binding sites upstream of a minimum promoter-drive luciferase gene into HEK293 cells. The luciferase activity was measured using a dual-luciferase assay system. Data are presented as mean ± S.D., n = 4.
As reported previously, MCPIP1 is a protein containing multiple domains (2, 11). As shown in Fig. 1C, the NYN-RNase domain (133–300) and CCCH-zinc finger (305–325) are located at the middle, and both are critical for its RNase activity (2). A proline-rich domain (PRD) is located at its C terminus, without known function. MCPIP4 also has similar RNase, CCCH-zinc finger, and proline-rich domains (Fig. 1C).
To confirm the interaction of MCPIP1 with MCPIP4, HA-tagged MCPIP1 and Flag-tagged MCPIP4 were co-transfected into HEK293 cells, and Co-IP of cell lysates with either anti-Flag or anti-HA antibodies were performed. Both cell lysate input and immunoprecipitates were then analyzed by Western blotting using anti-Flag and anti-HA antibodies. As shown in Fig. 1D, immunoprecipitation with antibodies targeting either MCPIP1 or MCPIP4 results in pull-down of both proteins, confirming their interaction. To exclude the possibility that the interaction of MCPIP1 with MCPIP4 is mediated by RNA, the immunoprecipitates were treated with or without RNase A, and then detected by Western blot with anti-Flag or anti-HA. As shown in Fig. 1E, the interaction of MCPIP1 with MCPIP4 was not affected by RNase A treatment.
To further confirm the interaction of MCPIP1 with MCPIP4, we performed mammalian two-hybrid assay. First, we inserted the gene fragments encoding MCPIP1 and MCPIP4 into the vector pACT containing the herpes simplex virus VP16 activation domain or the pBIND vector containing the yeast Gal4 DNA-binding domain to generate fusion proteins. Then, we transiently co-transfected these vectors into HEK293 cells with a luciferase reporter pG5luc. As positive control, pBIND-ID and pACT-MyoD were co-transfected with the reporter into HEK293 cells, the reporter activity were increased by ∼225-fold over the negative control. As shown in Fig. 1F, either pBIND-MCPIP1 co-transfected with pACT-MCPIP4 or pBIND-MCPIP4 co-transfected with pACT-MCPIP1 resulted in luciferase activity increase by more than 20-fold. Taken together, these data confirm the MS result that MCPIP1 interacts with MCPIP4 in mammalian cells.
MCPIP1 and MCPIP4 Are Co-localized in GW-body
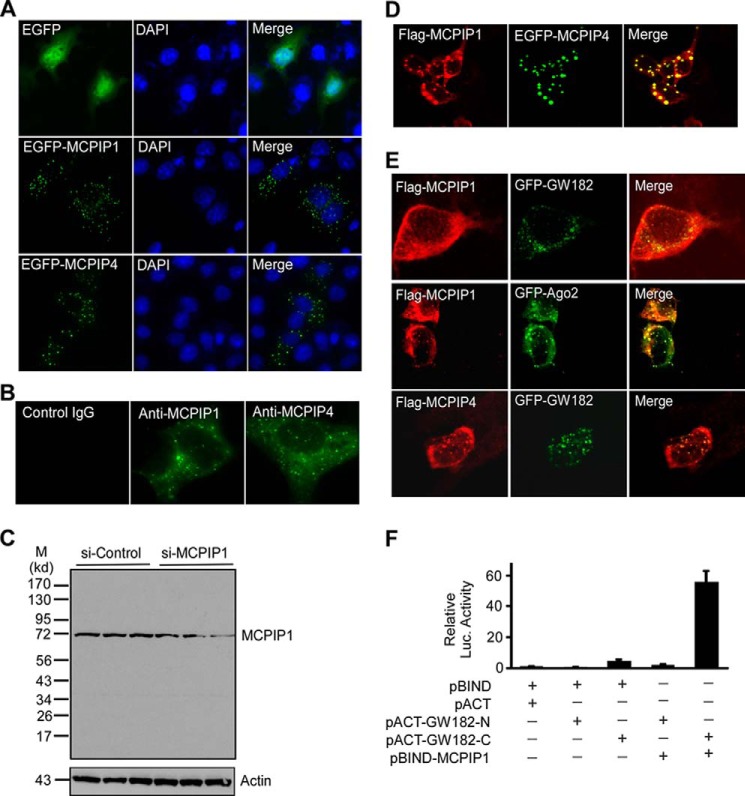
To examine whether MCPIP1 and MCPIP4 are co-localized in cells, GFP-MCPIP1 or GFP-MCPIP4 was transiently transfected into COS-7 cells, and protein localization was visualized by fluorescence microscopy. Results showed both MCPIP1 and MCPIP4 formed granule-like structures in the cytoplasm (Fig. 2A). To further examine whether endogenous MCPIP1 and MCPIP4 also form granule-like structures, Raw264.7 cells were stimulated with PamCSK4 for 6 h to induce the expression of MCPIP1 and MCPIP4. Cells were then stained with the primary anti-MCPIP1 (Genetex) or MCPIP4 (kindly provided by Dr. Matsui) or control IgG, and then stained with fluorescence-labeled anti-IgG. As shown in Fig. 2B, both endogenous MCPIP1 and MCPIP4 also formed granule-like structure in cytoplasm. To determine the specificity of anti-MCPIP1, Raw264.7 cells were transfected with small interference RNA for control (si-Control) or MCPIP1 (si-MCPIP1). 24 h later, transfected cells were stimulated with PamCSK4 for 6 h. MCPIP1 expression in the cells was determined by Western blot with anti-MCPIP1 (Genetex). As shown in Fig. 2C, MCPIP1 protein was detected as a single band around 72 kDa (migrated behind theoretical protein size). The specificity of anti-MCPIP4 monoclonal antibody was determined previously (20, 21). To determine whether MCPIP1 and MCPIP4 are co-localized in cells, HeLa cells were co-transfected with Flag-MCPIP1 and EGFP-MCPIP4 and visualized by staining with anti-Flag and Alexa Fluor596-labeled second antibody. As shown in Fig. 2D, the MCPIP1-granules and MCPIP4-granules were nicely overlapped in the cytoplasm. To characterize the identity of MCPIP1-granules, we previously performed immunostaining with the antibodies against different organelles and granule markers. MCPIP1-granules were not overlapped with mitochondria, Golgi network, lysosome, endosome, and stress granule, etc. (27). However, as shown in Fig. 2E, MCPIP1-granules were partly overlapped with GW182 and Argonaute 2 (Ago2), which are the markers of the GW-body (28). Interestingly, MCPIP4 was also overlapped with GW182 (Fig. 2E). Next, we determined whether MCPIP1 can interact with GW182 or Ago2 using the mammalian two-hybrid assay. As shown in Fig. 2F, when pBIND-MCPIP1 and pACT-GW182-C were co-transfected with the reporter pG5luc, luciferase activity was increased by 55-fold compared with the control group, suggesting that MCPIP1 is associated with the C terminus of GW182. The same strategy was used to determine whether MCPIP1 is also associated with Ago2 and showed that MCPIP1 did not associate with Ago2 (data not shown).
FIGURE 2.
MCPIP1 and MCPIP4 are co-localized in GW-body. A, COS-7 cells were transfected with the plasmids encoding EGFP or EGFP-MCPIP1 or EGFP-MCPIP4. The cells were visualized by confocal microscopy. The nuclei were stained by DAPI. B, RAW264.7 cells were stimulated with Pam3CSK4 for 6 h and then fixed and incubated with the primary ant-MCPIP1 or anti-MCPIP4 or control IgG. Then cells were visualized by Alexa Fluor488-labeled second antibody, and images were taken by confocal microscopy. C, RAW264.7 cells were transfected with small interference RNA for control (si-Control) or MCPIP1 (si-MCPIP1) for 24 h and then stimulated with PamCSK4 for 6 h. Three independent samples from control or MCPIP1 knocking down groups were loaded as indicated. The MCPIP1 protein level in the cell lysates was detected by Western blot with a MCPIP1 antibody (Genetex). Actin was probed as a loading control. D, HeLa cells were co-transfected with the plasmids encoding Flag-MCPIP1 and EGFP-MCPIP4. The cells were labeled with anti-Flag and visualized by Alexa Fluor594-labeled second antibody, and images were taken by confocal microscopy. E, Flag-MCPIP1 or Flag-MCPIP4 were transiently co-transfected with GFP-GW182 or GFP-Ago2 into HeLa cells. After 24 h, cells were fixed and stained with anti-Flag and visualized by confocal fluorescence microscopy. F, different combinations of expression plasmids as indicated were co-transfected with pG5luc reporter into HEK293 cells. 24 h later, cell lysates were prepared for analysis of luciferase activity. Data are presented as mean ± S.D., n = 4.
Mapping the Interacting Domains of MCPIP1 and MCPIP4
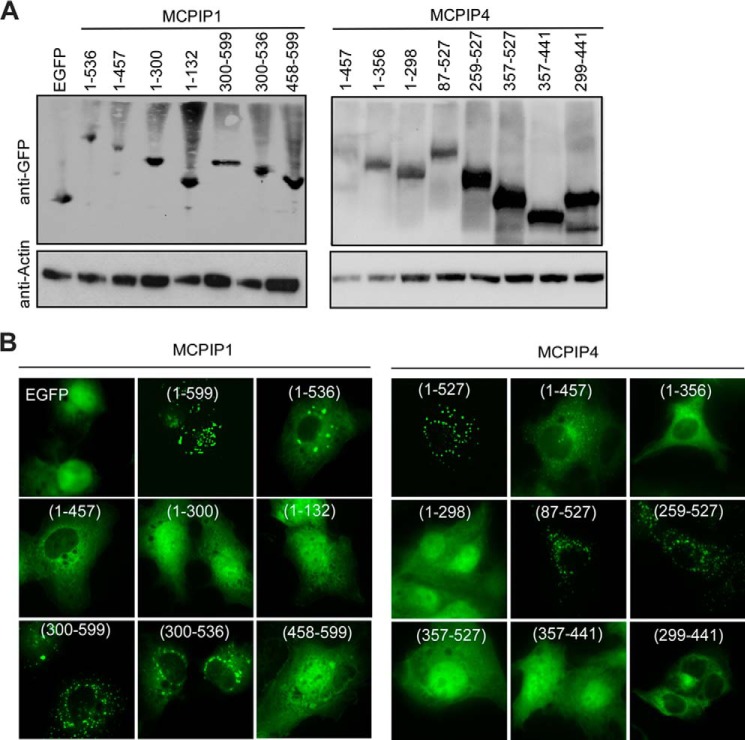
After establishing that full-length MCPIP1 and full-length MCPIP4 associate in vivo, we asked which domains could be responsible for the observed interactions. For that reason, we performed mammalian two-hybrid assay. First, we inserted the gene fragments encoding the serial deletions of MCPIP1 and MCPIP4 into the vector pACT or the pBIND (Fig. 3A). Then, we transiently co-transfected these vectors into HEK293 cells with the reporter pG5luc. As shown in Fig. 3B, 1–356 and 259–527 of MCPIP4 have similar binding ability compared with full-length MCPIP4. However, 1–258 and 357–527 of MCPIP4 lost the binding ability with MCPIP1. These results suggest that the region 259–356 of MCPIP4 is required for interaction with MCPIP1. On the other hand, 1–457 of MCPIP1 has a similar binding ability with MCPIP4 compared with full-length MCPIP1, whereas 1–300 of MCPIP1 lost its binding ability with MCPIP4. These results suggest that the region 301–457 of MCPIP1 is crucial for interaction with MCPIP4. To further examine whether 259–356 of MCPIP4 or 301–457 of MCPIP1 is sufficient for their interaction, we performed similar transfection experiments in HEK293 cells. As shown in Fig. 3C, both 259–356 of MCPIP4 and 301–457 of MCPIP1 were sufficient for their interaction. Moreover, further removal of their CCCH-zinc finger did not affect their interaction (Fig. 3C).
FIGURE 3.
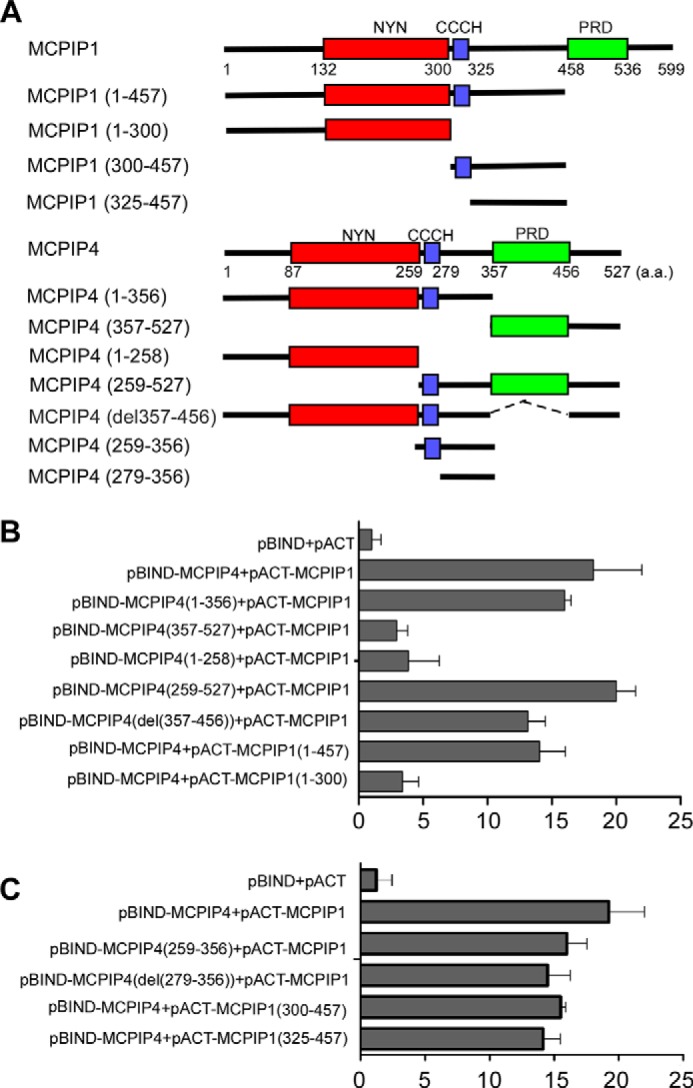
Mapping the interaction domains of MCPIP1 and MCPIP4. A, scheme for generation of the expression constructs encoding the serial deletions of MCPIP1 and MCPIP4 as indicated. B and C, mammalian two-hybrid assay for the interaction of MCPIP1 and MCPIP4. Different combinations of pBIND- and pACT-derived expression vectors were co-transfected with a reporter containing five Gal4 binding sites upstream of a minimum promoter-drive luciferase gene into HEK293 cells. The luciferase activity was measured using a dual-luciferase assay system. Data are presented as mean ± S.D., n = 4.
The Interaction Domain of MCPIP1 and MCPIP4 Is Required, but Not Sufficient for Their Granule-like Structure Formation
To determine whether the interaction domain of MCPIP1 and MCPIP4 is required for their granule-like structure formation, we mapped the domains responsible for the granule-like structure. First, we inserted the gene fragments encoding serial deletions of MCPIP4 and MCPIP1 into pEGFP-C1 vector. These vectors were transfected into HEK293 cells, and the correct expression of the gene fragments were determined by Western blot analysis (Fig. 4A). Then we transfected the vectors into HeLa cells, and the protein localization was visualized by fluorescence microscopy. As shown in Fig. 4B, the region 300–536 of MCPIP1 and the region 259–457 of MCPIP4 are responsible for granule-like structure formation. These regions include their CCCH-zinc finger motif, interaction domain, and proline-rich domain, which may contribute to their granule-like structure formation.
FIGURE 4.
Identification of the domains required for specific cellular localization of MCPIP1 and MCPIP4. A, expression vectors containing EGFP-fused truncations of MCPIP1 or MCPIP4 were transiently transfected into HEK293 cells. The proper expression of these constructs was determined by Western blot analysis with anti-EGFP. The same membranes were probed with anti-actin to show equal loading. B, expression constructs of EGFP or EGFP-fused truncations of MCPIP1 or MCPIP4 were transiently transfected into HeLa cells. The cellular localization of EGFP or EGFP-fused proteins were visualized by confocal microscopy.
Co-expression of MCPIP1 and MCPIP4 Enhances the Repression on the Reporter of IL-6 3′-UTR
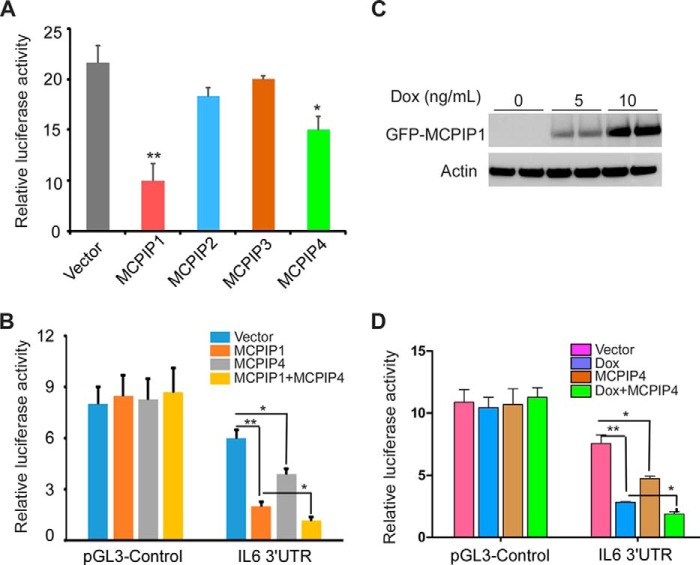
It was reported that MCPIP1 is an RNase that destabilizes a set of mRNAs, including IL-6 and IL-12, through cleavage of their 3′-UTRs (2, 10). The role of MCPIP4 remains unknown. The results described above suggest that MCPIP4 may functionally associate with MCPIP1. To test this idea, we first transfected the expression plasmids encoding MCPIP1/2/3/4 with the reporter of IL-6 3′-UTR into HEK293 cells, respectively. The results showed that MCPIP1 is the most potent member to repress IL-6 3′-UTR. MCPIP4 has a moderate effect on the reporter, whereas MCPIP2 and MCPIP3 have subtle effects on this reporter (Fig. 5A). Next, we co-transfected MCPIP1 or/and MCPIP4 with the reporter of IL-6 3′-UTR into HEK293 cells. As shown in Fig. 5B, overexpression of MCPIP1 or/and MCPIP4 had no effect on the pGL3-control reporter without IL-6 3′-UTR. However, both MCPIP1 and MCPIP4 repressed the reporter activity of IL-6 3′-UTR. Co-expression of MCPIP1 and MCPIP4 enhanced the repression on the reporter of IL-6 3′-UTR (Fig. 5B). In another experiment, MCPIP1-inducible HEK293 stable cells were co-transfected with MCPIP4 and the reporter of IL-6 3′-UTR or control. 24 h post-transfection, cells were induced with 10 ng/ml of doxycycline for 24 h to induce MCPIP1 expression. As shown in Fig. 5D, overexpression of MCPIP1 or MCPIP4 alone significantly repressed the reporter activity of IL-6 3′-UTR, but did not affect the reporter activity of the pGL3-control. Co-expression of MCPIP1 and MCPIP4 had an enhanced effect on the IL-6 3′-UTR. The inducible expression of MCPIP1 in the cell line upon Dox treatment was determined by Western blot with anti-GFP antibody (Fig. 5C).
FIGURE 5.
Co-expression of MCPIP1 and MCPIP4 enhanced the repression on the reporter of IL-6 3′-UTR. A, expression plasmids of MCPIP1/2/3/4 or an empty vector were co-transfected with the luciferase reporter of IL-6 3′-UTR into HEK293 cells. A control reporter vector pRL-TK was also transfected to normalize the values. After 24 h, cell lysates were prepared, and the luciferase activity was measured by dual luciferase assay system. Data are presented as mean ± S.D., n = 4, *, p < 0.05; **, p < 0.01 versus control group. B, expression plasmids of MCPIP1 or/and MCPIP4 were co-transfected with the reporter of IL-6 3′-UTR or the pGL3-control reporter into HEK293 cells. After 24 h, the cell lysates were prepared, and the luciferase activity was measured by dual luciferase assay system. Data are presented as mean ± S.D., n = 4, *, p < 0.05; **, p < 0.01 versus vector group. C, inducible MCPIP1-stable expressed HEK293 cell line was treated with 0, 5, or 10 ng/ml of doxycycline for 24 h. The inducible expression of MCPIP1 in the cells was determined by Western blot analysis with anti-GFP antibody. Actin was probed as a loading control. D, inducible MCPIP1-stable cells were transiently co-transfected with the expression plasmid of MCPIP4 or empty vector and the reporter of IL-6 3′-UTR or the pGL3-control reporter and followed by treatment with or without 10 ng/ml of doxycycline for 24 h. The cell lysates were prepared, and the luciferase activity was measured by dual luciferase assay system. Data are presented as mean ± S.D., n = 4, *, p < 0.05; **, p < 0.01 versus vector group.
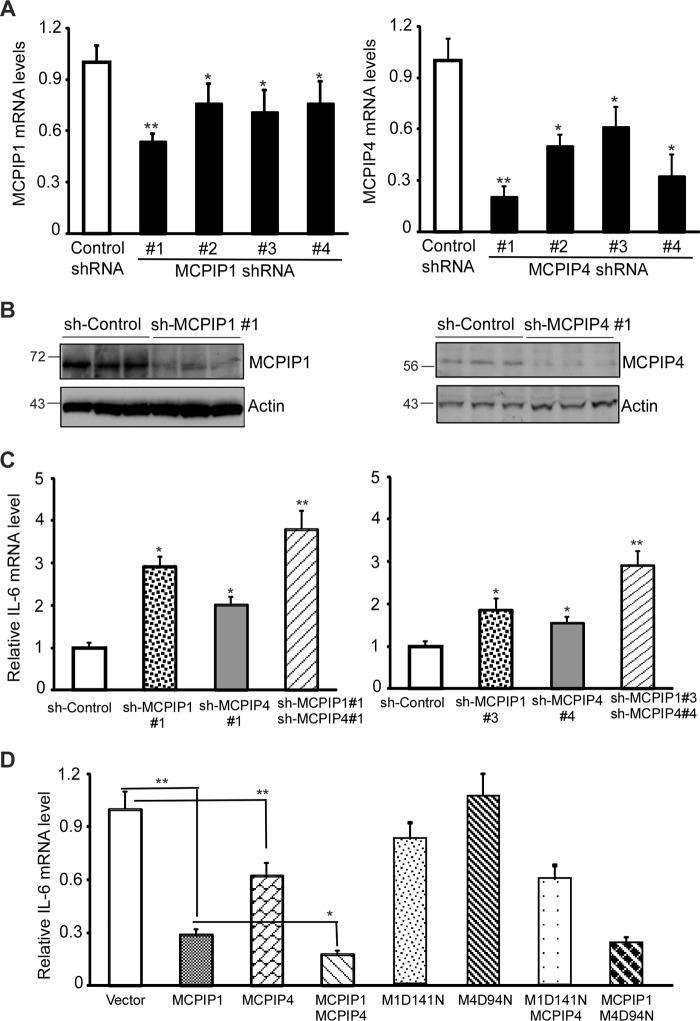
MCPIP1 and MCPIP4 Additively Contribute to Control the IL-6 mRNA Levels in Activated Macrophages
To further compare the functional importance of MCPIP1 and MCPIP4 in inflammatory cytokine production, we purchased the plasmids encoding short hairpin RNA (shRNA) targeting to different positions of mouse MCPIP1 and MCPIP4 mRNA from Sigma. To test their efficacy to knockdown the expression of MCPIP1 and MCPIP4 in activated macrophages, we transfected these plasmids into RAW264.7 cells, a murine macrophage cell line, by electroporation (Amaxa). After 48 h, the transfected cells were treated with 20 ng/ml of Pam3CSK4 (an agonist of Toll-like receptor 2) for 6 h to induce the expression of both MCPIP1 and MCPIP4 (20, 29). The mRNA levels of MCPIP1 and MCPIP4 were examined by QPCR. As shown in Fig. 6A, the shRNA#1 for MCPIP1 more efficiently knocked down MCPIP1 expression and the shRNA#1 for MCPIP4 more efficiently knocked down MCPIP4 expression in activated macrophages. The efficiency of sh-MCPIP1#1 and sh-MCPIP4#1 was further examined by Western blot. As shown in Fig. 6B, the protein levels of MCPIP1 and MCPIP4 were efficiently decreased by sh-MCPIP1#1 and sh-MCPIP4#1, respectively. Next, we transfected sh-Control, sh-MCPIP1#1, sh-MCPIP4#1, or sh-MCPIP1#1 plus sh-MCPIP4#1 into RAW264.7 cells, and the transfected cells were then treated with Pam3CSK4 (20 ng/ml) for 6 h. The mRNA level of IL-6 was examined by QPCR. As shown in Fig. 6C, knocking down of either MCPIP1 or MCPIP4 alone increased the level of IL-6 mRNA, whereas knocking down both MCPIP1 and MCPIP4 showed a more enhanced effect on the IL-6 mRNA in activated macrophages. To exclude any off-target effects, we also transfected another pairs of shRNAs into RAW264.7 cells and showed similar effects (Fig. 6C, right). The knocking-down efficiency of these shRNAs in the experiments described above was also confirmed by QPCR (data not shown). In contrast, overexpression of either MCPIP1 or MCPIP4 alone decreased IL-6 mRNA levels. Co-expression of MCPIP1 and MCPIP4 enhanced the repression on IL-6 mRNA level (Fig. 6D). The mutants of MCPIP1(D141N) and MCPIP4(D94N) failed to repress IL-6 mRNA expression. Neither overexpression of MCPIP1(D141N) with wild-type of MCPIP4 nor overexpression MCPIP4(D94N) with wild-type of MCPIP1 further enhanced their effects on IL-6 mRNA (Fig. 6D). These results suggest that the interaction of MCPIP1 and MCPIP4 is not required for their action in the regulation of IL-6 mRNA degradation. MCPIP1 and MCPIP4 may act independently in the regulation of IL-6 mRNA level.
FIGURE 6.
MCPIP1 and MCPIP4 additively contribute to control the IL-6 mRNA levels in activated macrophages. A, plasmids encoding four sets of shRNA targeting different positions of mouse MCPIP1 mRNA or MCPIP4 mRNA and control shRNA were transfected into RAW264.7 cells, a murine macrophage cell line. After 48 h of transfection, the transfected cells were treated with 20 ng/ml of Pam3CSK4 for 6 h. The mRNA levels of MCPIP1 and MCPIP4 were examined by QPCR. Data are presented as mean ± S.D., n = 4, *, p < 0.05; **, p < 0.01 versus control group. B, MCPIP1 and MCPIP4 protein levels in shRNA-transfected RAW264.7 cells were detected by Western blot with anti-MCPIP1 (Genetex) or anti-MCPIP4 (Proteintech). Actin was served as loading control. C, RAW264.7 cells were transfected with the plasmids encoding two-sets of different shRNAs as indicated and then treated with Pam3CSK4 (20 ng/ml) for 6 h. The mRNA level of IL-6 was examined by QPCR. Data are presented as mean ± S.D., n = 4, *, p < 0.05; **, p < 0.01 versus sh-control group. D, RAW264.7 cells were transfected with the expression vector for MCPIP1, MCPIP4, MCPIP1(D141N) (M1D141N), MCPIP4(D94N) (M4D94N), or their combinations as indicated. After 24 h, the transfected cells were treated with Pam3CSK4 (20 ng/ml) for 6 h. The mRNA level of IL-6 was examined by QPCR. Data are presented as mean ± S.D., n = 4, *, p < 0.05; **, p < 0.01.
Mapping the Functional Domains of MCPIP1 and MCPIP4 in Regulation of IL-6 3′-UTR
To further understand how MCPIP1 and MCPIP4 regulate IL-6 3′-UTR, we co-transfected the vectors containing serial deletions of MCPIP1 or MCPIP4 with the reporter of IL-6 3′-UTR into HEK293 cells. As shown in Fig. 7A, the region 81–457 of MCPIP1, containing the RNase domain and MCPIP4-interaction domain, is critical in the regulation of IL-6 3′-UTR. In addition, the region of 1–356 of MCPIP4, containing both RNase domain and MCPIP1-interaction domain, is required for suppressing the reporter of IL-6 3′-UTR. To further confirm that their RNase domain is critical in repression of IL-6 3′-UTR, we transfected the point mutations of both MCPIP1(D141N) and MCPIP4(D94N), which was previously demonstrated to be the active site for its RNase activity (2). The results showed that these point mutations also diminished their repressing activity on IL-6 3′-UTR. Taken together, these results suggest that the RNase activity of MCPIP1 and MCPIP4 is required for the regulation of mRNA destabilization.
FIGURE 7.
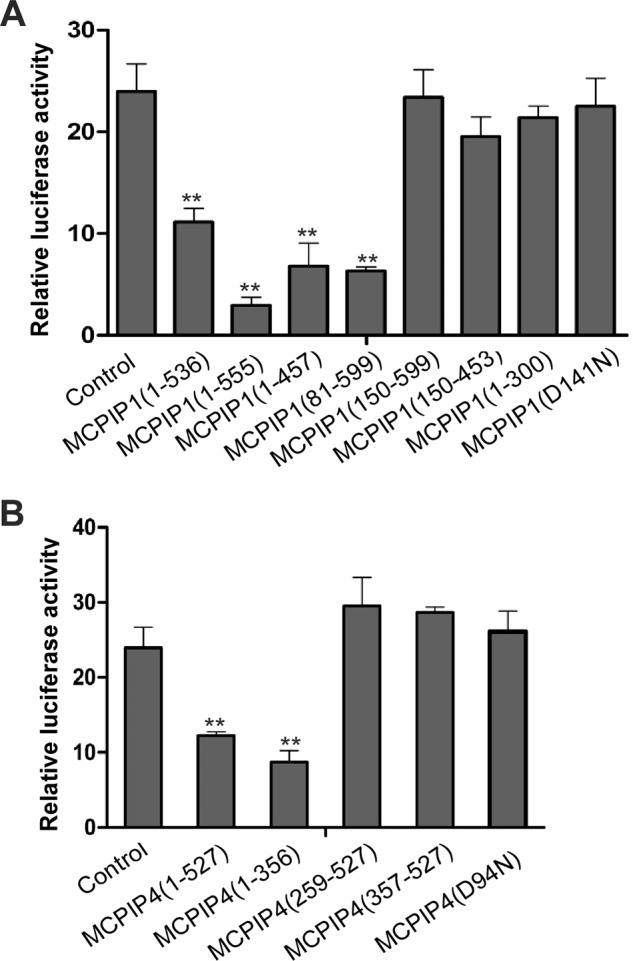
Mapping the functional domains of MCPIP1 and MCPIP4 in repression of IL-6 3′-UTR. The expression plasmids containing serial truncations or point mutation of MCPIP1 (A) or MCPIP4 (B) were co-transfected with the reporter of IL-6 3′-UTR and a control vector pRL-TK into HEK293 cells. After 24 h, the cell lysates were prepared, and the luciferase activity was measured by the dual luciferase assay system. Data are presented as mean ± S.D., n = 4, **, p < 0.01 versus control group.
Discussion
MCPIP1 is a newly identified CCCH-zinc finger-containing protein, which belongs to a subfamily including MCPIP1, -2, -3, and -4. Though it was recently demonstrated that MCPIP1 is a critical factor that controls inflammatory cell activation, immune homeostasis, and viral infection (30–34), the function of the other three members remains largely unknown. In addition, emerging evidence suggests that MCPIP1 is an endonuclease and selectively destabilizes mRNAs that encode certain inflammatory cytokines and immune molecules (24). However, the detailed molecular mechanisms by which MCPIP1 promotes mRNA degradation are obscure. To understand the molecular mechanisms, we searched MCPIP1-interacting proteins by Co-IP combined with proteomic analysis. Interestingly, MCPIP4 was co-purified with MCPIP1 in this approach. Further studies demonstrated that MCPIP4 is a MCPIP1-interacting protein, which may work together with MCPIP1 to control inflammatory cytokine production.
Both MCPIP1 and MCPIP4 contain a single CCCH-type zinc finger domain at the middle region, a highly conserved NYN-type RNase domain at the N terminus, and a proline-rich domain at the C terminus (Fig. 1C). Many studies have shown that MCPIP1 exerts the most potent RNase activity on certain mRNAs and pre-miRNAs among the MCPIP family (22, 33). In the gene knock-out mouse models, MCPIP1-deficient mice developed severe and complex inflammatory phenotypes including splenomegaly, lymphadenopathy, multi-organ inflammatory cell infiltration, and premature death (2, 11). However, MCPIP4-deficient mice did not show gross developmental defects (21). On the other hand, emerging evidence suggests that MCPIP1 and MCPIP4 have some similar effects on the regulation of IL-6 mRNA degradation and endothelial inflammation (21, 35, 36). Taken together, these studies suggest that both MCPIP1 and MCPIP4 may involve in the regulation of inflammatory cytokine production. In our present study, we have evidence to suggest that MCPIP1 interacts with MCPIP4 to form a protein complex, but they may act independently in the regulation of IL-6 mRNA degradation. Based current evidence, MCPIP1 and MCPIP4 may additively contribute to control IL-6 production in vivo.
We and others have previously observed that overexpressed exogenous or endogenous MCPIP1 protein formed granule-like structure in the cytoplasm (22, 27). In our previous work, we have observed that MCPIP1-granules were not overlapped with mitochondria, Golgi network, lysosome, endosome, and stress granules, etc. (27). The exact identity of MCPIP1-granules is still obscure. In the present study, we have found that both MCPIP1 and MCPIP4 formed similar granule-like structures in cytoplasm. Moreover, both MCPIP1 and MCPIP4 granules are partly overlapped with GW182 and Ago2, which are typical markers for the GW-body (28) and the major components of miRNA-induced silencing complex (miRISC) (37). Further studies suggest that MCPIP1 is associated with the C terminus of GW182, but not associated with Ago2. As it was known that Ago2 interacts with the N terminus of GW182 (38), we propose that GW182 may act as an adaptor protein to recruit both Ago2 and MCPIP1 into GW-body through its N terminus and C terminus, respectively. Nevertheless, the relationship between MCPIP1 and miRISC need to be further investigated both in vitro and in vivo.
In summary, we here report that MCPIP4 is an MCPIP1-interacting protein, and both MCPIP1 and MCPIP4 may be important in controlling IL-6 production. However, the interaction of MCPIP1 and MCPIP4 is not required for their regulation of IL-6 mRNA degradation. MCPIP1 and MCPIP4 act independently and additively contribute to controlling IL-6 production in activated macrophages.
Author Contributions
M. F. conceived and coordinated the study and wrote the paper. S. H. designed, performed, and analyzed the experiments shown in Figs. 1–7. S. L. and T. T. W. designed, performed, and analyzed the experiments shown in Fig. 1, A and B. J. J. F. provided technical assistance and contributed to the preparation of the figures. X. Y., A. K., and G. L. helped to revise the manuscript and provide consultation. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Hiroshi Suzuki, Dr. Keith L. Kirkwood, and Dr. Toshimitsu Matsui for kindly providing plasmids and anti-MCPIP4 antibody.
This work was supported by National Institutes of Health Grant AI103618 (to M. F.) and a UMKC SOM Dean's Support Fund (to M. F.). This work was also supported by National Institutes of Health Grants HL105473 and HL076206 (to G. L.), National Institutes of Health Grants DA025528, DA025011, and AA020806 (to A. K.), and National Institutes of Health Grant DK088787 (to T. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- MCPIP1
- MCP-induced protein 1
- MCPIP4
- MCP-induced protein 4
- IL-6
- interleukin-6
- UTR
- untranslated region
- Ago2
- Argonaute 2
- miRISC
- miRNA-induced silencing complex
- QPCR
- quantitative real-time PCR
- MS
- mass spectrometry.
References
- 1. Liang J., Wang J., Azfer A., Song W., Tromp G., Kolattukudy P. E., Fu M. (2008) A novel CCCH-Zinc finger protein family regulates proinflammatory activation of macrophages. J. Biol. Chem. 283, 6337–6346 [DOI] [PubMed] [Google Scholar]
- 2. Matsushita K., Takeuchi O., Standley D. M., Kumagai Y., Kawagoe T., Miyake T., Satoh T., Kato H., Tsujimura T., Nakamura H., Akira S. (2009) Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190 [DOI] [PubMed] [Google Scholar]
- 3. Liang J., Song W., Tromp G., Kolattukudy P. E., Fu M. (2008) Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS ONE. 3, e2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carballo E., Lai W. S., Blackshear P. J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281, 1001–1005 [DOI] [PubMed] [Google Scholar]
- 5. Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tan D., Zhou M., Kiledjian M., Tong L. (2014) The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nat. Struct. Mol. Biol. 21, 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leppek K., Schott J., Reitter S., Poetz F., Hammond M. C., Stoecklin G. (2013) Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 153, 869–881 [DOI] [PubMed] [Google Scholar]
- 8. Gao G., Guo X., Goff S. P. (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297, 1703–1706 [DOI] [PubMed] [Google Scholar]
- 9. Zhu Y., Chen G., Lv F., Wang X., Ji X., Xu Y., Sun J., Wu L., Zheng Y. T., Gao G. (2011) Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 15834–15839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mino T., Murakawa Y., Fukao A., Vandenbon A., Wessels H-H., Ori D., Uehata T., Tartey S., Akira S., Suzuki Y., Vinuesa C. G., Ohler U., Standley D. M., Landthaler M., Fujiwara T., Takeuchi O. (2015) Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161, 1058–1073 [DOI] [PubMed] [Google Scholar]
- 11. Liang J., Saad Y., Lei T., Wang J., Qi D., Yang Q., Kolattukudy P. E., Fu M. (2010) MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-κB signaling. J. Exp. Med. 207, 2959–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miao R., Huang S., Zhou Z., Quinn T., Van Treeck B., Nayyar T., Dim D., Jiang Z. S., Papasian C. J., Wang Y., Liu G., Eugene Chen Y., Fu M. (2013) Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol. Cell Biol. 91, 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uehata T., Iwasaki H., Vandenbon A., Matsushita K., Hernandez-Cuellar E., Kuniyoshi K., Satoh T., Mino T., Suzuki Y., Standley D. M., Tsujimura T., Rakugi H., Isaka Y., Takeuchi O., Akira S. (2013) Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 153, 1036–1049 [DOI] [PubMed] [Google Scholar]
- 14. Xu J., Peng W., Sun Y., Wang X., Xu Y., Li X., Gao G., Rao Z. (2012) Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase. Nucleic Acids Res. 40, 6957–6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mizgalska D., Wegrzyn P., Murzyn K., Kasza A., Koj A., Jura J., Jarzab B., Jura J. (2009) Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1β mRNA. FEBS J. 276, 7386–7399 [DOI] [PubMed] [Google Scholar]
- 16. Li M., Cao W., Liu H., Zhang W., Liu X., Cai Z., Guo J., Wang X., Hui Z., Zhang H., Wang J., Wang L. (2012) MCPIP1 down-regulates IL-2 expression through an ARE-independent pathway. PLoS ONE 7, e49841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Minagawa K., Yamamoto K., Nishikawa S., Ito M., Sada A., Yakushijin K., Okamura A., Shimoyama M., Katayama Y., Matsui T. (2007) Deregulation of a possible tumour suppressor gene, ZC3H12D, by translocation of IGKα in transformed follicular lymphoma with t(2;6)(p12;q25). Br. J. Haematol. 139, 161–163 [DOI] [PubMed] [Google Scholar]
- 18. Wang M., Vikis H. G., Wang Y., Jia D., Wang D., Bierut L. J., Bailey-Wilson J. E., Amos C. I., Pinney S. M., Petersen G. M., de Andrade M., Yang P., Wiest J. S., Fain P. R., Schwartz A. G., Gazdar A., Minna J., Gaba C., Rothschild H., Mandal D., Kupert E., Seminara D., Liu Y., Viswanathan A., Govindan R., Anderson M. W., You M. (2007) Identification of a novel tumor suppressor gene p34 on human chromosome 6q25.1. Cancer Res. 67, 93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Owerbach D., Piña L., Gabbay K. H. (2004) A 212-kb region on chromosome 6q25 containing the TAB2 gene is associated with susceptibility to type 1 diabetes. Diabetes 53, 1890–1893 [DOI] [PubMed] [Google Scholar]
- 20. Huang S., Qi D., Liang J., Miao R., Minagawa K., Quinn T., Matsui T., Fan D., Liu J., Fu M. (2012) The putative tumor suppressor Zc3h12d modulates toll-like receptor signaling in macrophages. Cell. Signal. 24, 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minagawa K., Wakahashi K., Kawano H., Nishikawa S., Fukui C., Kawano Y., Asada N., Sato M., Sada A., Katayama Y., Matsui T. (2014) Posttranscriptional modulation of cytokine production in T cells for the regulation of excessive inflammation by TFL. J. Immunol. 192, 1512–1524 [DOI] [PubMed] [Google Scholar]
- 22. Suzuki H. I., Arase M., Matsuyama H., Choi Y. L., Ueno T., Mano H., Sugimoto K., Miyazono K. (2011) MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell 44, 424–436 [DOI] [PubMed] [Google Scholar]
- 23. Zhao W., Liu M., Kirkwood K. L. (2008) p38α stabilizes interleukin-6 mRNA via multiple AU-rich elements. J. Biol. Chem. 283, 1778–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uehata T., Akira S. (2013) mRNA degradation by the endoribonuclease Regnase-1/ZC3H12a/MCPIP-1. Biochim. Biophys. Acta. 1829, 708–713 [DOI] [PubMed] [Google Scholar]
- 25. Jura J., Skalniak L., Koj A. (2012) Monocyte chemotactic protein-1-induced protein-1 (MCPIP1) is a novel multifunctional modulator of inflammatory reactions. Biochim. Biophys. Acta. 1823, 1905–1913 [DOI] [PubMed] [Google Scholar]
- 26. Akira S. (2013) Regnase-1, a ribonuclease involved in the regulation of immune responses. Cold Spring Harb. Symp. Quant. Biol. 78, 51–60 [DOI] [PubMed] [Google Scholar]
- 27. Qi D., Huang S., Miao R., She Z. G., Quinn T., Chang Y., Liu J., Fan D., Chen Y. E., Fu M. (2011) Monocyte chemotactic protein-induced protein 1 (MCPIP1) suppresses stress granule formation and determines apoptosis under stress. J. Biol. Chem. 286, 41692–41700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chan E. K., Yao B., Fritzler M. J. (2013) Reflections on ten years of history of, and future prospects for, GW182 and GW/P body research. Adv. Exp. Med. Biol. 768, 261–270 [DOI] [PubMed] [Google Scholar]
- 29. Huang S., Miao R., Zhou Z., Wang T., Liu J., Liu G., Chen Y. E., Xin H. B., Zhang J., Fu M. (2013) MCPIP1 negatively regulates Toll-like receptor 4 signaling and protects mice from LPS-induced septic shock. Cell. Signal. 25, 1228–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwasaki H., Takeuchi O., Teraguchi S., Matsushita K., Uehata T., Kuniyoshi K., Satoh T., Saitoh T., Matsushita M., Standley D. M., Akira S. (2011) The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat. Immunol. 12, 1167–1175 [DOI] [PubMed] [Google Scholar]
- 31. Niu J., Shi Y., Xue J., Xu M., Miao R., Huang S., Wang T., Wu J., Fu M., Wu Z-H. (2013) USP10 inhibits genotoxic NF-κB activation by MCPIP1-facilitated deubiquitination of NEMO. EMBO J. 32, 3206–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu S., Qiu C., Miao R., Zhou J., Fu W., Zhu L., Zhang L., Xu J., Fan D., Li K., Fu M., Wang T. (2013) MCPIP1 restricts HIV infection and is rapidly degraded in activated CD4+ T cells. Proc. Natl. Acad. Sci. U.S.A., 110, 19083–19088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin R. J., Chien H. L., Lin S. Y., Chang B. L., Yu H. P., Tang W. C., Lin Y. L. (2013) MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 41, 3314–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin R. J., Chu J. S., Chien H. L., Tseng C. H., Ko P. C., Mei Y. Y., Tang W. C., Kao Y. T., Cheng H. Y., Liang Y. C., Lin S. Y. (2014) MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J. Immunol. 193, 4159–4168 [DOI] [PubMed] [Google Scholar]
- 35. Liu L., Zhou Z., Huang S., Guo Y., Fan Y., Zhang J., Zhang J., Fu M., Chen Y. E. (2013) Zc3h12c inhibits vascular inflammation by repressing NF-κB activation and pro-inflammatory gene expression in endothelial cells. Biochemistry J. 451, 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qi Y., Liang J., She Z. G., Cai Y., Wang J., Lei T., Stallcup W. B., Fu M. (2010) MCP-induced protein 1 suppresses TNFα-induced VCAM-1 expression in human endothelial cells. FEBS Lett. 584, 3065–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ding L., Han M. (2007) GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 17, 411–416 [DOI] [PubMed] [Google Scholar]
- 38. Eulalio A., Huntzinger E., Izaurralde E. (2008) GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15, 346–353 [DOI] [PubMed] [Google Scholar]