Background: Oxidized substrates such as Tim13 acquire two disulfide bonds simultaneously, but Mia40 has one active redox center that accepts two electrons.
Results: Mia40 can acquire up to six electrons when oxidizing substrates.
Conclusion: Mia40 has the flexibility to accept several electrons.
Significance: Mechanistic properties of the MIA pathway are unique compared with redox pathways in the endoplasmic reticulum and bacterial periplasm.
Keywords: disulfide, mitochondria, oxidation-reduction (redox), protein import, redox regulation, thiol, redox
Abstract
A redox-regulated import pathway consisting of Mia40 and Erv1 mediates the import of cysteine-rich proteins into the mitochondrial intermembrane space. Mia40 is the oxidoreductase that inserts two disulfide bonds into the substrate simultaneously. However, Mia40 has one redox-active cysteine pair, resulting in ambiguity about how Mia40 accepts numerous electrons during substrate oxidation. In this study, we have addressed the oxidation of Tim13 in vitro and in organello. Reductants such as glutathione and ascorbate inhibited both the oxidation of the substrate Tim13 in vitro and the import of Tim13 and Cmc1 into isolated mitochondria. In addition, a ternary complex consisting of Erv1, Mia40, and substrate, linked by disulfide bonds, was not detected in vitro. Instead, Mia40 accepted six electrons from substrates, and this fully reduced Mia40 was sensitive to protease, indicative of conformational changes in the structure. Mia40 in mitochondria from the erv1–101 mutant was also trapped in a completely reduced state, demonstrating that Mia40 can accept up to six electrons as substrates are imported. Therefore, these studies support that Mia40 functions as an electron sink to facilitate the insertion of two disulfide bonds into substrates.
Introduction
The mitochondrion has diverse pathways for translocation of proteins synthesized in the cytosol (1). The Mia40/Erv1 import pathway in the mitochondrial intermembrane space mediates the translocation of proteins that contain disulfide bonds (2–4). Substrates of this import pathway include the small Tim proteins (Tim8, Tim9, Tim10, Tim12, and Tim13) with a conserved twin CX3C motif (5) and proteins with a conserved twin CX9C motif (6). In addition, unconventional substrates include the copper chaperone for superoxide dismutase Ccs1, the iron-sulfur cluster protein Dre2, and Erv1 (7–9). The substrates are imported in a reduced state and acquire intramolecular bonds in the intermembrane space (10, 11).
The oxidoreductase Mia40 is the receptor in the intermembrane space (12). Mia40 has a twin CX9C motif that forms two intramolecular disulfide bonds (13); structural studies reveal that these disulfide linkages play a structural role (14, 15). The N terminus contains a redox-active CPC motif that is typically oxidized and specifically forms a transient disulfide bond with the substrate as the substrate enters the intermembrane space (16, 17). Formation of the intermolecular disulfide bond is required to trap the substrate (12). The substrate is subsequently released in an oxidized form, leaving reduced Mia40. For additional rounds of import, Mia40 is reoxidized by Erv1 (11).
The sulfhydryl oxidase Erv1 contains an N-terminal CX2C motif that shuttles electrons from the CPC motif in Mia40 to a second CX2C pair at the Erv1 active site (18–20). Electrons are transferred to a noncovalently bound FAD and subsequently donated to a variety of terminal electron acceptors, including oxygen and cytochrome c (21–24). Finally, electrons from cytochrome c are donated to Ccp1 or the respiratory chain, and electrons from oxygen yield hydrogen peroxide (22, 23).
The midpoint potentials of the reactive cysteine centers in proteins of the disulfide relay are poised to favor the transfer of electrons from the substrate to the terminal electron acceptors (19). The twin CX3C protein Tim13 acquires two disulfide bonds simultaneously when equilibrated in buffers with different redox potentials and has a single midpoint potential of −310 mV (19). Coordinated insertion of two disulfide bonds requires four electrons to be transferred at one time. However, the mechanism by which Mia40 with its single CPC active site can simultaneously transfer four electrons is not evident; several mechanisms have been suggested. A small molecule such as oxygen or glutathione may insert the second disulfide bond. Oxygen, however, is likely slow and cannot explain how substrate oxidation proceeds under anaerobic conditions (25). Glutathione has been shown to increase the efficiency of import into isolated mitochondria and to function in quality control by preventing the formation of partially oxidized intermediates (18). Alternatively, a ternary complex of the substrate, Mia40, and Erv1 may facilitate substrate oxidation by channeling electrons (26, 27). However, as the substrate and Erv1 both transfer electrons through the CPC site of Mia40, the logistics of moving four electrons simultaneously has not been resolved.
One potential mechanism is that Mia40 may serve as an electron sink by transferring electrons to the twin CX9C motif, resulting in oxidation of the substrate in one step. Excess Mia40 can oxidize substrates in vitro (18, 19, 26), and recent studies in vivo support that Mia40 is present in a partially reduced state (28, 29). Whereas these studies show that Mia40 is reduced, the specific redox state of Mia40 has not been determined. Here, we report that Mia40 can be fully reduced both in vitro and in mitochondria, indicating that Mia40 has the capacity to serve as an electron sink to collect electrons during the oxidation of imported substrates.
Experimental Procedures
Plasmids and Strains
Recombinant wild-type and cysteine point mutants of Erv1 and Mia40 were expressed and purified under native conditions as described previously (19, 22). Recombinant Tim13 was purified under denaturing conditions as described previously (30). Cysteine point mutants in Tim13 and Mia40 were generated by site-directed mutagenesis using the QuikChange II site-directed mutagenesis kit (Stratagene).
Reconstitution Studies
In the reconstitution assays, reduced Tim13 and Tim13 cysteine mutants were incubated with Mia40 or Mia40 mutants and/or Erv1 or Erv1 mutants as established previously (19). The redox state of Mia40 and Tim13 was determined by thiol trapping with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS)2 as described previously (19). All proteins were separated by nonreducing SDS-PAGE followed by immunoblot analysis with antibodies against Tim13, Erv1, and Mia40.
Import Studies
Mitochondria were purified from yeast cells grown in rich ethanol-glycerol media (YPEG) as described in previous studies (31). Yeast cultures were grown at 30 °C with vigorous shaking. Mitochondrial concentration was measured by the bicinchoninic acid assay, according to the vendor's protocol (Pierce). Mitochondrial (25 mg/ml) aliquots were stored at −80 °C. [35S]Methionine and cysteine-labeled precursor proteins were generated with the TnT® quick coupled transcription/translation kit (Promega) and plasmids carrying the gene of interest. Transcription of genes was driven by either the T7 or SP6 promoter. Import reactions were conducted as described previously (32, 33). For import assays in the presence of reductants, mitochondria were preincubated for 15 min in the indicated concentration of glutathione (GSH) or ascorbate.
Mixed Disulfide Trapping
Reduced Tim13 was incubated with equimolar amounts of Mia40 and Erv1. In a control reaction, Tim13 was pretreated with 60 mm iodoacetamide (IAA). The reaction was quenched with the addition of IAA as described previously (19). Proteins were resolved on reducing and nonreducing SDS-polyacrylamide gels and detected by immunoblot analysis.
In Organello Thiol Trapping
Mitochondria were isolated from the WT and erv1–101 yeast strains, as described (Table 1) (22). Mitochondria (2.5 mg/ml) were solubilized in buffer containing 1% digitonin, 50 mm Tris, pH 7.0, 150 mm KCl, and 1 mm EDTA supplemented with protease inhibitors: 200 mm PMSF, 10 mm leupeptin, 1 mm chymostatin, and 1 mm pepstatin for 15 min at 4 °C followed by centrifugation at 14,000 × g for 10 min at 4 °C. In control reactions, the mitochondrial lysate was reduced with 20 mm DTT and incubated at 25 or 95 °C. The mitochondrial lysate was precipitated with 20% TCA on ice for 15 min, and precipitated proteins were pelleted by centrifugation (14,000 × g for 30 min at 4 °C). The pellet was resuspended in 20 μl of 20 mm AMS and incubated for 1 h at 37 °C. Samples were separated by nonreducing SDS-PAGE, and Mia40 was detected by immunoblot analysis.
TABLE 1.
Strains used in this study
Protease Sensitivity Assay
1 μm Mia40 was incubated in the presence or absence of 15 μm reduced Tim13 in a buffer containing 20 mm Tris, pH 7.0, 150 mm KCl, and 1 mm EDTA. 5 μg/ml trypsin was added to the samples at 37 °C. At the indicated time points, 5-μl samples were removed, and the reaction was halted with the addition of 200 μm soybean trypsin inhibitor. For controls, 1 μm Mia40 was heated at 95 °C in the presence of 20 mm DTT or left untreated and 15 μm bovine serum albumin (BSA) was substituted for Tim13. Proteins were resolved on nonreducing SDS-PAGE followed by immunoblot analysis with antibodies against Mia40.
Results
Reductants Inhibit Substrate Oxidation and Import
Previous studies showed that Tim13 has a single midpoint potential and that two disulfide bonds are acquired simultaneously (19); a partially oxidized intermediate has not been detected. Because Mia40 only has one redox-active CPC motif, we investigated how four electrons might be handled during substrate oxidation in vitro. As a first approach, we considered whether the reductant glutathione (GSH) was required, based on the studies by Bien et al. (18). The oxidation of reduced Tim13 (15 μm) was tested in the presence of 5 and 10 mm GSH, 1 μm Erv1, and 1 μm Mia40 (Fig. 1, B, C, and E), using a reconstitution assay with purified recombinant proteins (19). The oxidation state of Tim13 was monitored by the addition of AMS, which covalently binds to reduced thiols and causes an increase in molecular mass of 500 daltons per addition; up to four AMS molecules can add to reduced Tim13.
FIGURE 1.
Treatment with reductant decreases the rate of Tim13 oxidation by Mia40 and Erv1. A, gel system was calibrated with Tim13 mutants in which the 1st and 4th cysteines (C1,4S) and the 2nd and 3rd cysteines (C2,3S) have been mutated to serine residues. Reduced Tim13 and mutants were left untreated (lanes 1–3) or treated with AMS (lanes 4–6). B, reduced Tim13 (15 μm) was incubated with combinations of Mia40 (1 μm) and Erv1 (1 μm) in a time course assay as indicated (lanes 5–13). Aliquots were removed, and the free thiols were blocked with 20 mm AMS treatment. Oxidized (Tim13) and reduced (Tim13-AMS4) Tim13 were detected by nonreducing SDS-PAGE followed by immunoblotting with antibodies against Tim13. To calibrate the gel system, WT Tim13 and Tim13 with the 2nd and 4th cysteines mutated to serine residues (Tim13 C2,4S) were included. Reduced Tim13 samples were left untreated (lanes 1 and 3) or treated with AMS (lanes 2 and 4). The asterisk denotes nonspecific bands (lanes 2 and 4). C, as indicated in B with the addition of 5 mm (lanes 7–10) and 10 mm (lanes 11–14) GSH. D, as indicated in B with the addition of 5 mm ascorbate in the oxidation reaction. E, reactions in the presence of 5 mm GSH and 5 mm ascorbate were quantitated using a Bio-Rad FX Molecular Imager and the affiliated Quantity 1 software; 100% was set as the amount of Tim13 that was oxidized at the end point in the absence of reductant (n = 3). Results with 10 mm GSH were identical to those of ascorbate and are not shown to simplify the figure.
A set of Tim13 standards was included to calibrate the gel system (Fig. 1, A, lanes 1–6, and B, lanes 1–4). Reduced wild-type (WT) Tim13 was left untreated (−AMS) or treated with AMS (+AMS), resulting in the addition of four AMS molecules (Fig. 1, A, lanes 1 and 4, and B, lanes 1 and 2). Two reduced mutant versions of Tim13 with the 1st and 4th or 2nd and 3rd cysteines changed to serines (Tim13 C1,4S and Tim13 C2,3S, respectively) were left untreated (−AMS) or treated with AMS (+AMS), resulting in the addition of two AMS molecules (Fig. 1A, lanes 2, 3, 5, and 6). Similarly, a reduced mutant version of Tim13 with the 2nd and 4th cysteine residues mutated to serine residues (Tim13 C2,4S) was left untreated (−AMS) or treated with AMS (+AMS), resulting in the addition of two AMS molecules (Fig. 1B, lanes 3 and 4). Mutant Tim13 that had only two AMS additions migrated at an intermediate molecular mass, between Tim13 with zero and four AMS additions, which can be detected in this gel system. In our experimental conditions, we have not identified a state in which Tim13 shows partial oxidation, which is in agreement with a single midpoint potential of −310 mV. However, nonspecific bands that migrated just above the Tim13 band (highlighted by an asterisk in lanes 2 and 4 of Fig. 1B) were sometimes detected; we do not know the specific reason for this band, but it may be an artifact from the polyclonal antibody detecting additional proteins in this molecular mass region.
Tim13 was oxidized in ∼1–2 h in the presence of Mia40, Erv1, and oxygen (Fig. 1, B, lanes 10–13, C, lanes 3–6, and E). Analysis of the migration of Tim13 in the 1st h of the time course (i.e. Fig. 1B, lanes 10 and 11) in comparison with the aforementioned Tim13 standards indicated that four AMS molecules added to the reduced Tim13 and an intermediate oxidation state in which Tim13 had only one disulfide bond were not detected. Whereas this time course is long, Tim13 does not spontaneously oxidize (Fig. 1B, lanes 5–7) nor does Mia40 or Erv1 alone (Fig. 1B, lanes 8 and 9) oxidize Tim13. Based on the addition of four AMS molecules simultaneously, Tim13 was oxidized in one step by Mia40 and Erv1; a partially oxidized intermediate was not observed.
However, 5 mm GSH addition slowed Tim13 oxidation, with ∼50% of Tim13 oxidized over the 4-h time course (Fig. 1, C, lanes 7–10, and E). Moreover, the addition of 10 mm GSH inhibited Tim13 oxidation, with ∼95% remaining reduced after 4 h (Fig. 1C, lanes 17–20). Based on calibration of the gel system, a Tim13 intermediate that acquired one disulfide bond was not detected (Fig. 1A).
We tested whether the general reductant ascorbate acted similarly. Addition of 5 mm ascorbate inhibited Tim13 oxidation by 95% (Fig. 1, D, lanes 6–9, and E). As shown previously, Tim13 was not oxidized spontaneously or by Mia40 or Erv1 (Fig. 1D, lanes 1–5). Under these in vitro conditions, the addition of reductants impaired Tim13 oxidation in the presence of Mia40, Erv1, and oxygen.
We subsequently investigated the effect of reductants on the import of Tim13 and the CX9C substrate Cmc1 into isolated mitochondria (34), as described previously (18). Mitochondria were incubated with 2 and 5 mm GSH or ascorbate for 10 min followed by the addition of the precursor (Fig. 2, A–D). Again, the addition of reductants strongly inhibited Tim13 and Cmc1 import (Fig. 2, A and B), differing from results published by Herrmann and co-workers (18). In contrast, GSH and ascorbate addition did not interfere with the import of the TIM23 substrate Su9-DHFR (Fig. 2, C and D). Thus, GSH and ascorbate impair both the in vitro oxidation and in organello import of Mia40 substrates.
FIGURE 2.

Reductant treatment decreases in organello import of Cmc1 and Tim13, but not TIM23 substrate, Su9-DHFR. A, mitochondria were preincubated with 2 or 5 mm GSH for 10 min followed by the import of Tim13 or Cmc1. Protease treatment of mitochondria removed the precursor that failed to import, and the imported precursors were analyzed by reducing SDS-PAGE and autoradiography. A 10% standard (Std) from the translation reaction was included. B, as indicated in A but with ascorbate. C, as in A, except Su9-DHFR was imported in the presence of GSH. D, as in B, except Su9-DHFR was imported in the presence of ascorbate. Import reactions were quantitated, and 100% was set as the amount of precursor imported into WT mitochondria at the end point in the time course. Representative gels are shown (n = 4).
Another mechanism for oxidation of Mia40-dependent substrates is through the formation of a ternary complex of substrate, Mia40, and Erv1 in which the components cooperatively work together to insert two disulfide bonds into Mia40 (26–28). Previous work from our laboratory failed to identify a ternary complex that was linked by disulfide bonds in the reconstitution assay with purified components, but instead it showed the formation of Tim13-Mia40 and Mia40-Erv1 thiol-linked intermediates during Tim13 oxidation (19).
We expanded this analysis by investigating interactions with all combinations of reduced Tim13 with Mia40 and Erv1 followed by immunoblotting with antibodies against Tim13, Mia40, and Erv1. Disulfide-linked intermediates were trapped by post-treatment with IAA to stabilize the mixed disulfides, followed by SDS-PAGE on nonreducing gels (Fig. 3A). In the absence of reductant, an intermediate between Tim13 and Mia40 was detected (Fig. 3A, lanes 2, 6, 10, and 12). This intermediate was formed by free thiols in Tim13 because pretreatment of Tim13 with IAA (prior to incubating with Mia40 and Erv1) prevented formation of the Tim13-Mia40 mixed disulfide (Fig. 3A, lanes 3, 7, 11, and 13). In the presence of Tim13, Mia40 showed an unexpectedly large shift upward after the post-treatment with IAA (Fig. 3A, lane 10), suggesting that IAA might be adding to several reduced thiols; this shift was subsequently eliminated when Erv1 was added (Fig. 3A, lane 12), indicating that Erv1 subsequently oxidized Mia40.
FIGURE 3.
Mixed disulfide analysis shows that Mia40 is reduced by Tim13 in vitro. A, reduced Tim13 was preincubated in the absence or presence (+ IAA pretreatment of Tim13, lanes 3, 5, 7, 11, 13, 17, and 19) of IAA as indicated. Subsequently, equimolar amounts of recombinant Mia40, Erv1, and Tim13 were incubated in combination as indicated for 10 min. All samples were incubated with IAA post-treatment to block free thiols. Samples were separated by nonreducing SDS-PAGE followed by immunoblot analysis with antibodies against Tim13 (lanes 1–7), Mia40 (lanes 8–13), and Erv1 (lanes 14–19). Asterisks denote multimers of Tim13; the diamond marks a Mia40 dimer, and the square marks an Erv1 dimer (19). B, as in A, samples were separated on a reducing gel. Representative gels are shown (n = 4).
An intermediate between Tim13-Erv1 (Fig. 3A, lanes 4–7 and 16–19 (obscured by an Erv1 dimer, indicated by ■)) was detected, even when reduced Tim13 was pretreated with IAA to block free thiols (Fig. 3A, lanes 5, 7, 17, and 19); the formation of this Tim13-Erv1 complex seemingly is not mediated through disulfide bonds and likely represents a nonspecific complex. A Mia40-Erv1 intermediate (Fig. 3A, lanes 9, 12, and 13 (obscured by a Mia40 dimer, indicated by♦) and lanes 15, 18 and 19) was also detected. Note that the Mia40-Erv1 complex was only detected upon longer exposure of the blot (Fig. 3A, lanes 15, 18, and 19). Nonspecific multimers of Tim13 (Fig. 3A, indicated by the asterisks) also formed. A stable ternary complex of Tim13, Mia40, and Erv1 linked by disulfide bonds above the molecular mass of 72 kDa was not detected (data not shown). As a control, resolving the intermediates on a reducing gel resulted in migration of the proteins at their native molecular mass (Fig. 3B).
The increased shift in molecular mass of Mia40 and the Tim13-Mia40 intermediate after IAA post-treatment (Fig. 3A, lane 10 versus lane 12) was unexpectedly large in this gel system. Based on the published literature (13), we predicted two IAA additions at the CPC motif (and not the core CX9C motif) to increase the molecular mass by 116 Da, which was not expected to markedly increase the migration of Mia40 and the Tim13-Mia40 intermediate in this gel system. In a previous study, we showed that Mia40 had a single midpoint potential of −290 mV, and at different redox potentials, one disulfide bond was reduced (19). However, recent studies suggested that Mia40 exists in intermediate oxidation states in mitochondria (28, 29). Based on earlier publications in the literature that supported that only the CPC motif of Mia40 was redox active and could be reduced, we may have misinterpreted Mia40 oxidation in the presence of excess Tim13 in our previous study such that more than two cysteines of Mia40 were reduced by excess Tim13 (19). Therefore, this increased molecular mass in Mia40 warranted further investigation.
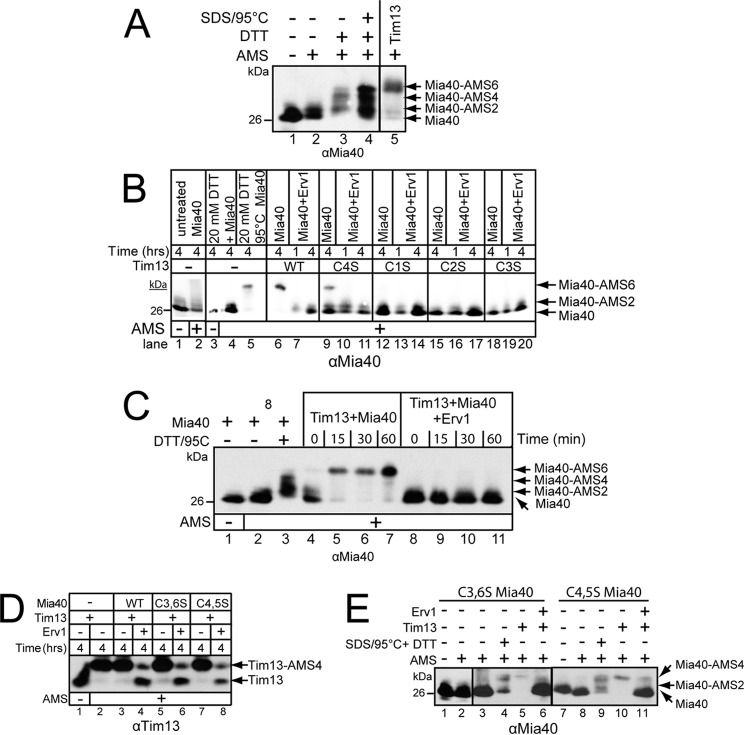
Tim13 Reduces Six Cysteines of Mia40
To determine the number of AMS additions to Mia40, the gel system was calibrated (Fig. 4A). Oxidized Mia40 was reduced using parameters that have been published previously (13). Mia40 treated with 20 mm AMS (Fig. 4A, lane 2) migrated at the same mass as the untreated Mia40 (Fig. 4A, lane 1), indicating that Mia40 was fully oxidized at the start of the analysis. When Mia40 was incubated in 20 mm DTT at 25 °C, two and four AMS additions were observed, demonstrating that Mia40 was partially reduced (Fig. 4A, lane 3); Grumbt et al. (13) similarly showed a partial reduction of Mia40. When Mia40 was incubated in 0.05% SDS, 20 mm DTT, and heated to 95 °C, Mia40 continued to unfold, and up to six AMS additions were detected (Fig. 4A, lane 4). However, it was typically difficult to completely reduce Mia40, and the extent of reduction with chemical treatment varied. Only Mia40 that was incubated with excess reduced Tim13 (15 μm) showed completed reduction, supported by the addition of six AMS molecules (Figs. 4, A, lane 5, and C, lanes 5–7, and 5A, lanes 13–18). Based on migration in the gel, the addition of two, four, and six AMS molecules to Mia40 caused distinct mass differences that can be separated in the gel system. Such a gel-based system can thus be used to assess the oxidation status of Mia40. Using the in vitro reconstitution system (19), we determined the specific redox state of Mia40 in the presence of excess reduced Tim13 or Tim13 mutants in which each cysteine was individually replaced with serine (Fig. 4B). The Tim13 mutants have been designated C1S (C57S), C2S (C61S), C3S (C73S), and C4S (C77S). Reduced Tim13 (15 μm) was incubated with Mia40 (1 μm) or both Mia40 (1 μm) and Erv1 (1 μm); the redox status of Mia40 was monitored at 1 and 4 h by the addition of AMS followed by nonreducing SDS-PAGE and immunoblot analysis with anti-Mia40. In a control reaction, Mia40 was treated with 20 mm DTT at 25 °C for 1 h and two AMS molecules were added to a fraction of the Mia40 (Fig. 4B, lanes 3 and 4) (13), in contrast to the untreated Mia40, in which no AMS addition was detected (Fig. 4B, lanes 1 and 2). In addition, Mia40 incubation at 95 °C in the presence of DTT resulted in the addition of six AMS moieties (13), confirming complete reduction of Mia40 (Fig. 4B, lane 5). When excess WT Tim13 was mixed with Mia40, six AMS molecules were added, demonstrating Mia40 was completely reduced (Fig. 4B, lane 6). Subsequent Erv1 addition oxidized Mia40 (Fig. 4B, lanes 7 and 8). With the Tim13 C4S mutant, two and six AMS molecules were added to a subset of the Mia40 pool (Fig. 4B, lane 9), which was mostly oxidized by the addition of Erv1 (Fig. 4B, lanes 10 and 11). Theoretically, the C4S Tim13 mutant donated two electrons to Mia40, yet Mia40 accepted six electrons; this implies that Mia40 likely accepts electrons from several Tim13 molecules. In contrast, the Tim13 mutants C1S, C2S, and C3S failed to oxidize Mia40 (Fig. 4B, lanes 12, 15, and 18). Thus, the first three cysteine residues of Tim13 are required to drive reduction of Mia40, which aligns with studies by Chacinska and co-workers (17) that the first cysteine is critical for import of the small Tim proteins into the intermembrane space. Our in vitro studies recapitulate the interactions that occur during import, signifying that the in vitro system has merit for dissecting potential reactions in vivo.
FIGURE 4.
Mia40 can accept up to six electrons from Tim13. A, control reactions to assess the Mia40 oxidation status. 1 μm Mia40 was left untreated (lane 1), treated with 20 mm AMS for 60 min (lane 2), incubated in 20 mm DTT for 60 min followed by 20 mm AMS addition (lane 3), or incubated at 95 °C with 0.05% SDS for 10 min followed by incubation in 20 mm DTT for 60 min and subsequent 20 mm AMS addition (lane 4). 15 μm reduced Tim13 was incubated with oxidized Mia40 at 25 °C for 60 min followed by addition of 20 mm AMS (lane 5). Proteins were separated by nonreducing SDS-PAGE and Mia40 was detected by immunoblot analysis. B, redox state of Mia40 was determined by incubation of 15 μm reduced Tim13 (WT, or C1S, C2S, C3S, and C4S mutants) with either 1 μm Mia40 or 1 μm Mia40 and 1 μm Erv1 for up to 4 h followed by 20 mm AMS addition (lanes 6–20). The samples were resolved by nonreducing SDS-PAGE and immunoblotted with anti-Mia40. In control reactions, Mia40 was left untreated for 4 h (lanes 1 and 2), reduced with 20 mm DTT (lanes 3 and 4), and heated to 95 °C with addition of 0.05% SDS (lane 5) followed by AMS treatment as indicated. C, as in B except reduced Tim13 (15 μm) was incubated with combinations of 1 μm Mia40 or 1 μm Mia40 and 1 μm Erv1 in a time course assay as indicated (lanes 4–11). Aliquots were removed at the indicated times and AMS was added. In control reactions, Mia40 was incubated in reaction buffer for 60 min (lanes 1 and 2) or in the presence of 20 mm DTT and 95 °C (lane 3) followed by 20 mm AMS addition (lanes 2 and 3). D, reduced Tim13 (15 μm) was incubated with a combination of WT Mia40 or Mia40 mutants C3,6S and C4,5S (1 μm) and Erv1 (1 μm) for 4 h. The reaction was stopped, and free thiols were blocked with AMS treatment. Oxidized (Tim13) and reduced (Tim13-AMS4) Tim13 were separated on nonreducing SDS-PAGE followed by immunoblotting with antibodies against Tim13. E, as in D, except that the redox status of mutant Mia40 (C3,6S and C4,5S) was investigated by treatment of AMS and subsequent nonreducing SDS-PAGE and immunoblot analysis with anti-Mia40 (n = 3).
FIGURE 5.
Mia40 is reduced by Tim13 at an equimolar cysteine concentration. The redox state of WT Mia40 (A), C3,6S Mia40 (B), or C4,5S Mia40 (C) was determined by incubating increasing amounts of reduced Tim13 with 1 μm Mia40 for 2 h followed by addition of 20 mm AMS. The protein ratio (μm Tim13:μm Mia40) and cysteine ratio (Tim13 cysteines/Mia40 cysteines) are indicated. In control reactions, Mia40 was reduced with 20 mm DTT and incubated at 25 or 95 °C in the presence of 0.05% SDS along with 20 mm DTT. A representative gel is shown (n = 3).
We investigated Mia40 reduction in a time course assay (Fig. 4C). As in Fig. 4B, excess Tim13 was incubated with Mia40. During the first 15 min (Fig. 4C, lanes 4 and 5), a progression of two and six AMS additions on Mia40 was detected, until 60 min when the Mia40 pool was reduced (Fig. 4C, lane 7). The addition of Erv1 resulted in efficient oxidation of Mia40 (Fig. 4C, lanes 8–11). Thus, Mia40 can be fully reduced by Tim13.
The importance of the Mia40 cysteines in reduction by Tim13 was investigated in more detail. Previously, we have shown the mutations in the Mia40 redox center, denoted CPS and SPC, failed to oxidize Tim13 (19). As this mutant version of Mia40 did not oxidize Tim13, we also found that Mia40 was not reduced (data not shown). Two additional mutants in Mia40 that lacked one of the structural disulfide bonds in the core were constructed and designated Mia40 C3,6S (C307S and C340S) (lacks the disulfide bond proximal to the CPC motif) and Mia40 C4,5S (C317S and C329S) (lacks the disulfide bond distal to the CPC motif). Oxidation of reduced Tim13 (15 μm) by the Mia40 C3,6S or Mia40 C4,5S mutants (1 μm) alone or in the presence of Erv1 (1 μm) in a reconstitution assay was tested (Fig. 4D). After 4 h, Tim13 was partially oxidized by mutant Mia40 C3,6S and C4,5S. In reactions lacking Erv1, Tim13 was not oxidized efficiently, indicating that the redox relay was required. The subsequent redox status of the Mia40 mutants was investigated by thiol trapping (Fig. 4E). Both C3,6S and C4,5S Mia40 mutants were fully reduced by excess Tim13 (Fig. 4E, lanes 5 and 10) and oxidized by Erv1 (Fig. 4E, lanes 6 and 11). Note that reduced Mia40 was not quantitatively recovered, despite numerous repeats (Fig. 4E, lanes 4, 5, 9, and 10 versus lanes 6 and 11); the Mia40 mutants may be less stable when not oxidized. (Detailed analysis of Mia40 controls are shown in Fig. 5, B and C, lanes 1–4.) Thus, the C3,6S and C4,5S Mia40 mutants are functional in the in vitro reconstitution assay. Moreover, these in vitro results are in agreement with results from Terziyska et al. (35) that showed that these Mia40 mutants are functional in vivo.
We adjusted the ratio of Tim13 to Mia40 to estimate the amount of Tim13 that was required to reduce Mia40 (Fig. 5A). In a control reaction, we incubated the buffer, excluding Tim13, with Mia40 for 2 h; Mia40 remained oxidized (Fig. 5A, lanes 4–10). Addition of the reduced Tim13 with Mia40 resulted in partial Mia40 reduction when the ratio of thiols was Tim13 four Cys:Mia40 six Cys (Fig. 5A, lane 13). Reduction of Mia40 was complete at an equal ratio of cysteines (Tim13 six Cys:Mia40 six Cys) and at ratios with excess cysteines in Tim13 (Fig. 5A, lanes 14–18).
Reduction of mutant C3,6S and C4,5S Mia40 with Tim13 in a similar titration assay was also tested (Fig. 5, B and C). In control reactions, DTT treatment at 25 °C resulted in a Mia40 intermediate with two AMS additions (Fig. 5, B and C, lane 3). As in Fig. 5A, Mia40 mutants were not spontaneously reduced in buffer (Fig. 5, B and C, lanes 5–10). At 95 °C, Mia40 was completely reduced, and four AMS additions were detected (Fig. 5, B and C, lane 4). With mutant C3,6S Mia40, reduction by Tim13 was less efficient, as complete Mia40 reduction was observed when the cysteine ratio of Tim13:Mia40 was 24:4 (Fig. 5B, lanes 15 and 16). At ratios lower than 24:4 (Fig. 5B, lanes 11–14), C3,6S Mia40 was not efficiently reduced, but an intermediate in which C3,6S Mia40 was reduced at two cysteine residues (two AMS additions) was not readily detected. With the mutant C4,5S Mia40 (Fig. 5C), reduction by Tim13 was similar to the titration observed with WT Mia40 (Fig. 5A). At an equimolar ratio of cysteine residues (4:4), Mia40 was efficiently reduced (Fig. 5C, lane 12). Again, an intermediate in which Mia40 was partially reduced was not readily observed (Fig. 5C, lane 11). Thus, mutant Mia40 was reduced by Tim13, with reduction of the C4,5S mutant being more efficient than reduction of the C3,6S mutant.
Mia40 Undergoes Conformational Changes When Reduced by Tim13
Because Mia40 can accept up to six electrons from Tim13 in vitro, we tested the stability of Mia40 in limited proteolysis experiments to determine whether the transfer of electrons to Mia40 was accompanied by conformational changes (Fig. 6). Reduced Tim13 and oxidized Mia40 were incubated together for 2 h at 25 °C and then treated with 5 μg/ml trypsin at 37 °C. Aliquots were removed at the indicated time points, stopped with the addition of soybean trypsin inhibitor, and analyzed by SDS-PAGE and immunoblot analysis with anti-Mia40. As a control, Mia40 was reduced with 20 mm DTT and incubated at 95 °C for 5 min to induce unfolding followed by treatment with trypsin. Untreated Mia40 was less sensitive to trypsin, because fragments were resistant to trypsin after 20 min (Fig. 6A). In the presence of 15 μm Tim13 after 2 h, 1 μm Mia40 was degraded in 2–5 min and resistant products were not detected (Fig. 6A). Bovine serum albumin (BSA) was swapped for Tim13, and under similar conditions, Mia40 was resistant to cleavage (Fig. 6B). Thus, the protease sensitivity of Mia40 in the presence of excess Tim13 is similar to that of reduced/heated Mia40.
FIGURE 6.

Tim13 induces conformational changes in Mia40. A, reduced Tim13 (15 μm) was incubated with Mia40 (1 μm) followed by addition of 5 μg/ml trypsin at 25 °C. At the indicated times, aliquots were removed, and soybean trypsin inhibitor was added. Control reactions include untreated Mia40 and Mia40 incubated in 20 mm DTT at 95 °C. Samples were separated by SDS-PAGE followed by immunoblot analysis with antibodies against Mia40. A representative gel is shown. B, as in A, in the presence of 15 μm BSA (n = 4).
Mia40 Is Completely Reduced in the Erv1–101 Mutant
The requirement for functional Erv1 in the oxidation of Mia40 was tested. Recombinant Tim13 and Mia40 were incubated with WT Erv1 or Erv1 mutants C30S or C133S (Fig. 7A) (19). Aliquots were removed at 4 h, and the redox state of Mia40 was analyzed by addition of AMS. The WT Erv1 oxidized Mia40 (Figs. 4C, lanes 8–11, and 7A, lane 6). In contrast, Mia40 remained reduced in the control reaction of Tim13 and Mia40 lacking Erv1 (Fig. 7A, lane 5). The Erv1 mutants C30S or C133S failed to oxidize Mia40 (Fig. 7A, lanes 7 and 8). Excess Cmc1 also fully reduced Mia40 resulting in six AMS additions (Fig. 7B, lane 5), and Mia40 was subsequently oxidized by Erv1 (Fig. 7B, lane 6).
FIGURE 7.
Mia40 is reduced in the erv1–101 mutant at the restrictive temperature 37 °C. A, redox status of Mia40 was analyzed in the presence of WT and Erv1 mutants (C30S or C133S) (lanes 6–8). Reduced Tim13 (15 μm) was incubated with combinations of Mia40 (1 μm), Erv1 (1 μm), and mutant Erv1 (C133S or C30S, 1 μm). Aliquots were removed at 4 h, and free thiols were blocked with AMS. Mia40 was separated by nonreducing SDS-PAGE followed by immunoblotting with antibodies against Mia40. Control reactions for Mia40 were included as described previously in Fig. 4A (lanes 1–4). B, reduced Cmc1 (15 μm) was incubated in combination with Mia40 (1 μm) and/or Erv1 (1 μm) for 4 h, followed by AMS treatment (lanes 5 and 6). Samples were separated by nonreducing SDS-PAGE followed by immunoblot analysis with antibodies against Mia40. Arrows depict oxidized Mia40, reduced Mia40 (Mia40-AMS6), a Cmc1-Mia40 complex, and the Mia40 dimer. Control reactions for Mia40 (lanes 1–4) as described in A. C, radiolabeled WT Mia40 and Mia40 CPC-SX9S-SX9S (mutant that lacks the four cysteines that form the structural disulfide bonds) were synthesized in vitro in a reticulocyte transcription/translation system. Aliquots were removed at 90 min, and free thiols on Mia40 were blocked with AMS and separated by nonreducing SDS-PAGE. D, redox state of Mia40 was investigated in mitochondria isolated from WT and erv1-101 yeast strains grown at the permissive temperature (25 °C) or nonpermissive temperature (37 °C). Isolated mitochondria were precipitated in 20% TCA in the presence of AMS. In control reactions, WT mitochondria were left untreated or incubated with 20 mm DTT at 25 or 95 °C prior to TCA precipitation and AMS treatment. Proteins were resolved on nonreducing SDS-PAGE and immunoblotted with anti-Mia40. Representative gels are shown (n = 4).
We also performed in organello thiol trapping to assess the redox state of Mia40 in mitochondria from the erv1–101 temperature-sensitive yeast strain (22). To confirm that differences in Mia40 migration corresponding with the addition of two and six AMS molecules could be separated in the gel system, we synthesized full-length Mia40 or mutant Mia40 that lacked the four cysteine residues (replaced by serine residues) in the core region (Mia40-CPC-SX9S-SX9S) in the reticulocyte lysate system, followed by AMS treatment and separation by SDS-PAGE (Fig. 7C). Whereas the addition of two AMS moieties resulted in a small shift in migration on the SDS gel, six AMS additions markedly slowed Mia40 migration in the gel. Thus, the gel system is adequate for detecting differences in the addition of two and six AMS molecules to Mia40.
We purified mitochondria from WT and erv1–101 yeast strains and TCA-precipitated mitochondrial proteins followed by treatment with AMS and SDS-PAGE analysis with anti-Mia40 (Fig. 7D). In WT mitochondria, AMS did not modify untreated Mia40, indicating that Mia40 was oxidized (Fig. 7D, lanes 1, 2, and 11–13). As a control to verify that Mia40 could be reduced, mitochondria from the WT strain were treated with 20 mm DTT and incubated at 25 or 95 °C, respectively, followed by AMS treatment. Under these conditions, six AMS molecules were added to Mia40 (based on calibration from the gel in Fig. 7C), supporting that Mia40 could be reduced in mitochondria (Fig. 7D, lanes 3–6). In purified mitochondria in which the erv1–101 strain was grown at the permissive temperature of 25 °C, the majority of Mia40 remained oxidized (Fig. 7D, lanes 7, 8, and 14). In contrast, when the erv1–101 strain was shifted to the nonpermissive temperature (37 °C), Mia40 was reduced (Fig. 7D, lanes 9, 10, 15, and 18). In an additional control reaction, Mia40 remained oxidized when the WT strain was grown at 37 °C (Fig. 7D, lanes 13 and 16), indicating that a temperature shift does not alter the redox state of Mia40. Note that based on the gel calibration (Fig. 7C), an addition of six AMS and not two AMS was detected. Taken together, these studies provide evidence that Mia40 can serve as an electron sink to accept multiple electrons from reduced substrates.
Discussion
Based on our previous studies, the small Tim proteins acquire two disulfide bonds simultaneously, inferring that four electrons are rapidly removed from the substrate. Indeed, reduced Tim13 quickly folds at a single midpoint potential and acquired two disulfide bonds (19). An efficient system must be in place to accept numerous electrons as the substrate is oxidized. However, the specific mechanism by which the substrate is oxidized by Mia40 has been difficult to reconcile, because Mia40 contains a single redox-active CPC motif that interacts with the substrate and can theoretically only accept two electrons (11, 36). Studies by Bien et al. (18) proposed that GSH functions in this pathway to facilitate substrate oxidation, potentially in a quality control pathway. Here, GSH and the reductant ascorbate inhibited the oxidation and import of Tim13 and Cmc1. It is not clear how a reductant might stimulate protein import and substrate oxidation, because in an in vitro assay that we developed to identify small molecule inhibitors of Erv1 (32), the nonphysiologic substrate DTT was used, and DTT inhibited the ability of Erv1 to oxidize substrates. Thus, the reductants seem to inhibit oxidation of substrates.
In this study, we developed in vitro studies to characterize the redox state of Mia40 using thiol trapping with AMS, which increases the molecular mass by 0.5 kDa per addition. With our gel systems, we can distinguish between two, four, and six AMS additions. Previous studies have relied on assays in which free thiols of Mia40 are blocked with IAA or N-ethylmaleimide to address the redox state of Mia40 (11, 18, 37); because addition of IAA and N-ethylmaleimide does not yield appreciable mass differences, the specific redox status, other than oxidized or reduced, is difficult to interpret.
A second molecule of Mia40 binding to substrate is another potential mechanism to allow the substrate to acquire two disulfide bonds (18). However, given that interactions between Mia40 and the substrate are driven by specific sequences in the substrate (16, 17), a second Mia40 molecule mediating insertion of the second disulfide bond does not seem likely. Indeed, our in vitro studies indicate that Tim13 oxidation by Mia40 requires all four cysteine residues in the Tim13 substrate (Fig. 4B), although partial oxidation of Tim13 proceeds when the fourth cysteine of Tim13 was mutated.
Another mechanism to facilitate the transfer of four electrons from the substrate is the formation of a ternary complex with substrate, Mia40, and Erv1 (27, 28). However, we and others could not isolate a ternary complex that was stabilized by disulfide linkages (19, 28). Instead, the ternary complex is proposed to be stabilized by noncovalent interactions (27).
The mechanism by which electrons are transferred through this ternary complex has not been elucidated. Electron shuttling is complicated by structural restrictions. The reduced substrate docks on the redox-active CPC center of Mia40 (25). However, reoxidizing the CPC motif by Erv1 requires that Erv1 binds to the same region in Mia40 (20, 36). As both the substrate and Erv1 cannot bind at the same time, this mechanism would only allow the transfer of two electrons and subsequent release of a partially oxidized substrate, which has not been observed for Tim13. Our in vitro studies, however, support the formation of a ternary complex, in which Mia40 acts as an electron sink to acquire multiple electrons from the substrate. We show that Mia40 has the ability to accept up to six electrons (Figs. 4 and 5), which is accompanied by structural changes (Fig. 6). In addition, Mia40 mutants that lack one of the two structural disulfide bonds are also functional in the reconstitution assays (Fig. 4, D and E). Our studies support that Mia40 may first interact with the reduced substrate to accept four electrons, leaving the substrate oxidized. Subsequently, an interaction of reduced Mia40 with Erv1 then shuttles four electrons, leaving an oxidized Mia40 for another round of import. A ternary complex stabilized by noncovalent interactions would facilitate electron shuttling. In corroboration of this model, Chacinska and co-workers (28) use mutational analysis in Mia40 to show that the Cys-3(Cys-307)–Cys-6(Cys-340) couple (near the CPC motif) may be accessible to Erv1 binding. Thus, C3 and C6 residues may have a redox role in addition to a structural role. In addition, Mia40 is present in various redox states in vivo and is fully reduced in the erv1 mutant (Fig. 7) (29, 37).
In sum, our in vitro studies support that Mia40 has the flexibility to accept multiple electrons, and Erv1 can subsequently oxidize Mia40. Our analysis suggests that Mia40 can function as an electron sink to accommodate substrates that acquire more than one disulfide bond. As a result, linking in vitro and in vivo studies begins to provide a kinetic analysis of how multiple electrons are simultaneously shuttled from substrate through Mia40 to Erv1, resulting in an oxidized substrate. This is likely important as an editing pathway to repair incorrectly placed disulfide bonds similar to the endoplasmic reticulum and bacterial periplasm has not been identified in the mitochondrial intermembrane space (38).
Author Contributions
S. E. N., D. V. D., and C. M. K. designed the study and wrote the paper. S. E. N., D. V. D., and R. O. L. designed, performed, and analyzed experiments. H. L. T., D. M. H., and K. G. provided technical assistance and contributed reagents and vectors. A. B. and D. M. H. designed experiments in Fig. 1D, and A. B. analyzed experiments. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Janos Steffen and Juwina Wijaya for helpful comments and for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Ruth L. Kirschstein National Research Service Awards GM871082 (to S. E. N.) and GM007185 (to H. L. T.) and National Institutes of Health Grants R01 GM061721 and R01 GM071775 (to A. B.). This work was also supported by the UCLA Cota-Robles Fellowship, American Heart Association Grant 0640076N (to C. M. K.), and Muscular Dystrophy Association Grant 022398. The authors declare that they have no conflicts of interest with the contents of this article.
- AMS
- 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- IAA
- iodoacetamide.
References
- 1. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Herrmann J. M., Riemer J. (2012) Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J. Biol. Chem. 287, 4426–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koehler C. M., Tienson H. L. (2009) Redox regulation of protein folding in the mitochondrial intermembrane space. Biochim. Biophys. Acta 1793, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stojanovski D., Bragoszewski P., Chacinska A. (2012) The MIA pathway: a tight bond between protein transport and oxidative folding in mitochondria. Biochim. Biophys. Acta 1823, 1142–1150 [DOI] [PubMed] [Google Scholar]
- 5. Koehler C. M., Merchant S., Schatz G. (1999) How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci. 24, 428–432 [DOI] [PubMed] [Google Scholar]
- 6. Longen S., Bien M., Bihlmaier K., Kloeppel C., Kauff F., Hammermeister M., Westermann B., Herrmann J. M., Riemer J. (2009) Systematic analysis of the twin cx(9)c protein family. J. Mol. Biol. 393, 356–368 [DOI] [PubMed] [Google Scholar]
- 7. Banci L., Bertini I., Ciofi-Baffoni S., Boscaro F., Chatzi A., Mikolajczyk M., Tokatlidis K., Winkelmann J. (2011) Anamorsin is a [2Fe-2S] cluster-containing substrate of the Mia40-dependent mitochondrial protein trapping machinery. Chem. Biol. 18, 794–804 [DOI] [PubMed] [Google Scholar]
- 8. Terziyska N., Grumbt B., Bien M., Neupert W., Herrmann J. M., Hell K. (2007) The sulfhydryl oxidase Erv1 is a substrate of the Mia40-dependent protein translocation pathway. FEBS Lett. 581, 1098–1102 [DOI] [PubMed] [Google Scholar]
- 9. Gross D. P., Burgard C. A., Reddehase S., Leitch J. M., Culotta V. C., Hell K. (2011) Mitochondrial Ccs1 contains a structural disulfide bond crucial for the import of this unconventional substrate by the disulfide relay system. Mol. Biol. Cell 22, 3758–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Curran S. P., Leuenberger D., Schmidt E., Koehler C. M. (2002) The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 158, 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. (2005) A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 12. Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuán Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., Pfanner N. (2004) Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grumbt B., Stroobant V., Terziyska N., Israel L., Hell K. (2007) Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J. Biol. Chem. 282, 37461–37470 [DOI] [PubMed] [Google Scholar]
- 14. Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., Sideris D. P., Katrakili N., Tokatlidis K. (2009) MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 16, 198–206 [DOI] [PubMed] [Google Scholar]
- 15. Kawano S., Yamano K., Naoé M., Momose T., Terao K., Nishikawa S., Watanabe N., Endo T. (2009) Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc. Natl. Acad. Sci. U.S.A. 106, 14403–14407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sideris D. P., Petrakis N., Katrakili N., Mikropoulou D., Gallo A., Ciofi-Baffoni S., Banci L., Bertini I., Tokatlidis K. (2009) A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol. 187, 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Milenkovic D., Ramming T., Müller J. M., Wenz L. S., Gebert N., Schulze-Specking A., Stojanovski D., Rospert S., Chacinska A. (2009) Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell 20, 2530–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bien M., Longen S., Wagener N., Chwalla I., Herrmann J. M., Riemer J. (2010) Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell 37, 516–528 [DOI] [PubMed] [Google Scholar]
- 19. Tienson H. L., Dabir D. V., Neal S. E., Loo R., Hasson S. A., Boontheung P., Kim S. K., Loo J. A., Koehler C. M. (2009) Reconstitution of the Mia40-Erv1 oxidative folding pathway for the small tim proteins. Mol. Biol. Cell 20, 3481–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lionaki E., Aivaliotis M., Pozidis C., Tokatlidis K. (2010) The N-terminal shuttle domain of Erv1 determines the affinity for Mia40 and mediates electron transfer to the catalytic Erv1 core in yeast mitochondria. Antioxid. Redox Signal. 13, 1327–1339 [DOI] [PubMed] [Google Scholar]
- 21. Farrell S. R., Thorpe C. (2005) Augmenter of liver regeneration: a flavin-dependent sulfhydryl oxidase with cytochrome c reductase activity. Biochemistry 44, 1532–1541 [DOI] [PubMed] [Google Scholar]
- 22. Dabir D. V., Leverich E. P., Kim S. K., Tsai F. D., Hirasawa M., Knaff D. B., Koehler C. M. (2007) A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 26, 4801–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bihlmaier K., Mesecke N., Terziyska N., Bien M., Hell K., Herrmann J. M. (2007) The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 179, 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allen S., Balabanidou V., Sideris D. P., Lisowsky T., Tokatlidis K. (2005) Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 353, 937–944 [DOI] [PubMed] [Google Scholar]
- 25. Banci L., Bertini I., Cefaro C., Cenacchi L., Ciofi-Baffoni S., Felli I. C., Gallo A., Gonnelli L., Luchinat E., Sideris D., Tokatlidis K. (2010) Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc. Natl. Acad. Sci. U.S.A. 107, 20190–20195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bourens M., Dabir D. V., Tienson H. L., Sorokina I., Koehler C. M., Barrientos A. (2012) Role of twin Cys-Xaa9-Cys motif cysteines in mitochondrial import of the cytochrome C oxidase biogenesis factor Cmc1. J. Biol. Chem. 287, 31258–31269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stojanovski D., Milenkovic D., Müller J. M., Gabriel K., Schulze-Specking A., Baker M. J., Ryan M. T., Guiard B., Pfanner N., Chacinska A. (2008) Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J. Cell Biol. 183, 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Böttinger L., Gornicka A., Czerwik T., Bragoszewski P., Loniewska-Lwowska A., Schulze-Specking A., Truscott K. N., Guiard B., Milenkovic D., Chacinska A. (2012) In vivo evidence for cooperation of Mia40 and Erv1 in the oxidation of mitochondrial proteins. Mol. Biol. Cell 23, 3957–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kojer K., Bien M., Gangel H., Morgan B., Dick T. P., Riemer J. (2012) Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 31, 3169–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beverly K. N., Sawaya M. R., Schmid E., Koehler C. M. (2008) The Tim8-Tim13 complex has multiple substrate binding sites and binds cooperatively to Tim23. J. Mol. Biol. 382, 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glick B. S., Pon L. A. (1995) Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213–223 [DOI] [PubMed] [Google Scholar]
- 32. Dabir D. V., Hasson S. A., Setoguchi K., Johnson M. E., Wongkongkathep P., Douglas C. J., Zimmerman J., Damoiseaux R., Teitell M. A., Koehler C. M. (2013) A small molecule inhibitor of redox-regulated protein translocation into mitochondria. Dev. Cell 25, 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hasson S. A., Damoiseaux R., Glavin J. D., Dabir D. V., Walker S. S., Koehler C. M. (2010) Substrate specificity of the TIM22 mitochondrial import pathway revealed with small molecule inhibitor of protein translocation. Proc. Natl. Acad. Sci. U.S.A. 107, 9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horn D., Al-Ali H., Barrientos A. (2008) Cmc1p is a conserved mitochondrial twin CX9C protein involved in cytochrome c oxidase biogenesis. Mol. Cell. Biol. 28, 4354–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Terziyska N., Grumbt B., Kozany C., Hell K. (2009) Structural and functional roles of the conserved cysteine residues of the redox-regulated import receptor Mia40 in the intermembrane space of mitochondria. J. Biol. Chem. 284, 1353–1363 [DOI] [PubMed] [Google Scholar]
- 36. Banci L., Bertini I., Calderone V., Cefaro C., Ciofi-Baffoni S., Gallo A., Kallergi E., Lionaki E., Pozidis C., Tokatlidis K. (2011) Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proc. Natl. Acad. Sci. U.S.A. 108, 4811–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spiller M. P., Ang S. K., Ceh-Pavia E., Fisher K., Wang Q., Rigby S. E., Lu H. (2013) Identification and characterization of mitochondrial Mia40 as an iron-sulfur protein. Biochem. J. 455, 27–35 [DOI] [PubMed] [Google Scholar]
- 38. Depuydt M., Messens J., Collet J. F. (2011) How proteins form disulfide bonds. Antioxid. Redox Signal. 15, 49–66 [DOI] [PubMed] [Google Scholar]
- 39. Koehler C. M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R. J., Schatz G. (1998) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279, 369–373 [DOI] [PubMed] [Google Scholar]