Abstract
Notch (N) is a cell surface receptor that mediates an evolutionarily ancient signaling pathway to control an extraordinarily broad spectrum of cell fates and developmental processes. To gain insights into the functions of N signaling in the adult brain, we examined the involvement of N in Drosophila olfactory learning and memory. Long-term memory (LTM) was disrupted by blocking N signaling in conditional mutants or by acutely induced expression of a dominant-negative N transgene. In contrast, neither learning nor early memory were affected. Furthermore, induced overexpression of a wild-type (normal) N transgene specifically enhanced LTM formation. These experiments demonstrate that N signaling contributes to LTM formation in the Drosophila adult brain.
Activation of Notch (N) receptors has been linked to the specification of many cell types in both vertebrates and invertebrates (1). Binding of ligands such as Delta (2) causes cleavage of the intracellular domain of the N protein (3–5). The cleaved cytoplasmic domain of N (Nintra) enters the nucleus, in which it regulates expression of target genes (1). Intercellular communication mediated by N signaling consists of two different modes: lateral inhibitory signaling and inductive signaling. A prototypic example of lateral inhibitory signaling is that loss of N function causes a cluster of equivalent proneural cells to all assume the default neuronal fate rather than an epidermal fate (6). Inductive N signaling, on the other hand, mediates interactions between cells that are nonequivalent before the signal initiates, such as induction of cone cells by photoreceptor cells and specification of midline cells in the embryo (7–9).
In contrast to the extensive understanding of the vital role of N in development, the significance of N signaling in adult brains has yet to be revealed, although there is continuous presence of N protein and its ligands in the adult vertebrate nervous system (10). It has been reported that N signaling regulates neurite outgrowth in mammals (11), and chronic reduction of N activity in adult fruit flies also leads to progressive neurological syndromes (12). Processing of N requires γ-secretase activity, whereas presenilin (PS) is a critical component of the γ-secretase complex (13). Mutations in the PS genes are associated with the early onset of Alzheimer's disease (14). Conditional knockout of the PS1 gene in mice is also associated with reduced clearance of hippocampal memory traces (15). Although involvement of N in these PS mutant phenotypes remains to be determined, it has been shown that N plays a role in PS-mediated formation of neural projections in postmitotic neurons necessary for learned thermotaxis in Caenorhabditis elegans (16). In the present study, we examined the role of N signaling in learning and memory in Drosophila.
A Pavlovian procedure that pairs odors with foot-shock (17) was used in this study to assess the effects of N signaling on adult behavioral plasticity. Genetic analyses have demonstrated that memory formation after such Pavlovian training occurs in functionally distinct temporal phases (18). Two of these memory phases, short-term memory and middle-term memory, are labile and short-lived, whereas another two phases, anesthesia-resistant memory (ARM) and long-term memory (LTM), are resistant to various disruptive treatments and persist for several days. LTM and ARM have been dissected genetically. Disruptions of two transcription factors, cAMP-response element-binding protein (CREB) or alcohol dehydrogenase factor-1, abolish LTM without affecting ARM (19, 20). Conversely, ARM is disrupted but LTM is normal in radish mutants and in transgenic flies expressing a dominant-negative form of atypical protein kinase C designated PKM (18, 21, 22). Given these observations and the established role for N signaling in gene regulation (1), we focused our experiments on LTM formation in adult flies carrying a conditional mutation of N or expressing N transgenes.
Materials and Methods
Fly Stocks. Nts2 were obtained from the Bloomington Stock Center (Bloomington, IN) and were outcrossed with FM7 balancer flies for five generations. The presence of the Nts2 mutation was confirmed by embryonic lethality at restrictive temperature (30°C). Transgenic flies, heat-shock N+ (hs-N+) and hs-NΔcdc10rpts were gifts from M. Young's laboratory at The Rockefeller University (New York) (4, 23) and were outcrossed for five generations with our standard wild-type strain, w1118(isoCJ1) (19). For all behavioral analyses, w1118(isoCJ1) serves as the control.
Pavlovian Learning and Memory. The training and testing procedures were the same as described in refs. 17–19. Briefly, a group of ≈100 flies was sequentially exposed to two odors (60 s for each with 45 s of rest in between). During exposure to the first odor, flies were simultaneously subjected to electric shock (twelve 1.5-s pulses with 3.5-s intervals, 60 V); this constitutes a single training cycle. To measure “learning,” flies were transferred immediately after training to the choice point of a T maze and forced to choose between the two odors. For 24-h memory, flies were subjected to multiple spaced training sessions (1, 2, or 10 training sessions with a 15-min rest between each) or to massed training (10 sessions with no rest interval). After training, flies were transferred to food vials and stored at 18°C for 24 h before testing for their distribution in the T-maze arms. The performance index (PI) was calculated on the basis of the distribution of flies in each arm (17). A PI of 0 represented a 50:50 distribution, whereas a PI of 100 represented 100% avoidance of the shock-paired odor. For determining PIs in Nts2 at restrictive temperatures, flies were incubated at 30°C for 2 days, trained, stored during the retention interval, and tested at 30°C.
Sensorimotor Responses. Odor-avoidance responses were quantified by exposing naive flies to each odor (octanol or methyl-cyclohexanol) versus air in the T maze. After 120 s, the number of flies in each arm of the T maze was counted, and the PI was calculated for each odor individually as reported (17).
The ability to sense and escape from electric shock was quantified by inserting electrifiable grids into both arms of the T maze; shock pulses were delivered to one of the arms. Flies were transported to the choice point of the T maze, allowing them to choose between the two arms. After 60 s, the center compartment was closed, trapping flies in their respective arms. Individual PIs were calculated as for odor acuity. For determining PIs in Nts2, flies were incubated at 30°C for 2 days, and the measurements were also carried out at 30°C.
Drug Feeding. The drug-feeding protocol is the same as described in ref. 18. Flies were fed 35 mM cycloheximide in 5% glucose dissolved in 3% ethanol at 25°C for 12 h before heat-shock treatment. After heat-shock treatment, drug was fed for 3 h before training and for 24 h at 18°C after training during the retention period.
Heat-Shock Treatment. Flies subjected to heat shock were placed in empty vials in a water bath that was maintained at 37°C for 30 min. Flies then were transferred back to bottles with food and rested for 3 h at room temperature (20–24°C) before the training sessions. To minimize leaky expression for the groups not subjected to heat shock, flies were incubated at 18°C overnight before training, which was performed at room temperature.
RT-PCR of Induced N Expression. Flies were raised at either 18 or 25°C. One group of flies was shifted from 18 to 37°C for 30 min and returned to 18°C for 3 h before freezing flies at -70°C. Total RNA was prepared from 200 mg of frozen flies by using the RNeasy minikit (Qiagen, Valencia, CA), and mRNA was isolated by using the Dynabeads mRNA Direct microkit (Dynal, Madison, WI). Three independent RNA preparations were made for each temperature condition. First-strand cDNA was synthesized directly from the mRNA by using the Superscript first-strand synthesis system (Invitrogen). PCR amplification with a 25:1 mixture of Taq DNA polymerase (Invitrogen) and Pfu DNA polymerase (Stratagene) was carried out in Invitrogen 10× PCR buffer using 1.5 mM MgCl2 with 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min followed by extension at 72°C for 10 min. Three pairs of N-specific primers were designed against regions of the Drosophila N cDNA encoding the intracellular domain (GenBank accession no. NM_057511) (24). Primers N1 (5′-gatgccaattgtcaggataac-3′) and N2 (5′-gaatgttcaccgcttcggtat-3′) amplify a 285-bp fragment between bases 6722 and 7004; primers N3 (5′-agcgaaatggagtcggtcccg-3′) and N4 (5′-gaattgcttctgctgtgtggca-3′) amplify a 268-bp fragment between bases 7932 and 8197; primers N5 (5′-ctcggaggcctggagttcggttc-3′) and N6 (5′-ggatagctatccaacgtttggac-3′) amplify a 256-bp fragment between bases 8501 and 8754. The N3–N4 and N5–N6 primer pairs span regions of genomic DNA that contain introns, thus allowing distinction between cDNA products and potential genomic contaminants. Lack of genomic DNA contamination was confirmed by the absence of any PCR products when RT-PCR was carried out in control reactions without reverse transcriptase. Ribosomal protein 49 (rp49)-specific primers (5′-atgaccatccgcccagcatac-3′ and 5′-gagaacgcaggcgaccgttgg-3′) were designed to amplify a 391-bp fragment between bases 1 and 391 of the rp49-coding region (GenBank accession no. Y13939). The control rp49 mRNA should be expressed at equal levels in all cells at all stages (25).
Results
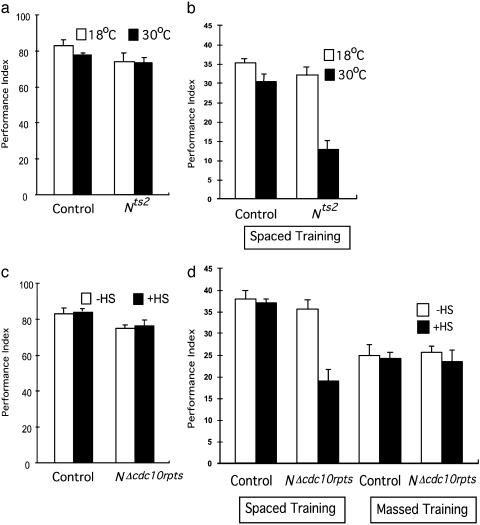
Consistent with the idea that N may affect LTM, we found that alteration of N activity did not affect learning or short-term memory. A temperature-sensitive N allele, Nts2, is viable at permissive temperature but embryonically lethal at restrictive temperature (26). As adults, these flies were behaviorally normal (shock reactivity and olfactory acuity) (Table 1) and showed normal learning scores (Fig. 1a) after 2 days of incubation at the restrictive temperature (30°C). Furthermore, learning was also not affected in transgenic flies that express dominant-negative N, hs-NΔcdc10rpts (Fig. 1c). Expression of hs-NΔcdc10rpts was induced acutely via heat-shock treatment (30 min at 37°C, with 3 h of recovery before training; see Materials and Methods). The NΔcdc10rpts protein lacks the intracellular domain that diffuses into the nucleus after N activation (4). Without the N intracellular domain, the NΔcdc10rpts protein binds ligands normally but is unable to regulate gene expression (27). All mutants tested were outcrossed to control lines to remove potential modifiers.
Table 1. Sensorimotor responses of different genotypes.
| Genotype (temp) | OA (MCH) | OA (OCT) | SR |
|---|---|---|---|
| w1118(isoCJ1) | 70 ± 3 | 69 ± 1 | 85 ± 2 |
| HS-w1118(isoCJ1) | 73 ± 4 | 67 ± 3 | 84 ± 2 |
| NΔcdc10rpts | 76 ± 4 | 74 ± 4 | 83 ± 1 |
| HS-NΔcdc10rpts | 75 ± 2 | 76 ± 2 | 83 ± 2 |
| hsN+ | 74 ± 5 | 73 ± 4 | 83 ± 1 |
| HS-hsN+ | 70 ± 5 | 71 ± 5 | 83 ± 2 |
| w1118(isoCJ1) (30°C) | 58 ± 6 | 42 ± 9 | 52 ± 7 |
| Nts2 (30°C) | 50 ± 9 | 35 ± 8 | 39 ± 11 |
There is no significant difference for “task-relevant” sensorimotor responses among all comparisons between the control and experimental groups except olfactory acuity for octanol between HS-w1118(isoCJ1) and HS-NΔcdc10rpts. All experiments were conducted at room temperature except those indicated as (30°C). OA, olfactory acuity; MCH, methylcyclohexanol; OCT, octanol; SR, electric-shock reactivity; HS, 37°C heat shock.
Fig. 1.
Blockade of LTM formation by disruption of N signaling. (a) Learning is not affected in temperature-sensitive N mutants. Learning scores were obtained at permissive (18°C) and restrictive (30°C) temperatures (see Materials and Methods) for the temperature-sensitive N mutant, Nts2, and for control flies. In this and all following figures, the isogenic line, w1118(isoCj1) (19), was used as a control (n = 7, 2, 8, and 7 for data points from left to right). (b) Reduced 24-h memory at the restrictive temperature for Nts2. Twenty-four-hour memory was determined after spaced training (10 training sessions with a 15-min rest interval between each). Spaced training induces formation of persistent memory that consists of two distinct components: protein synthesis-dependent LTM and ARM (n = 4 for all data points). (c) Learning is not affected by acutely induced expression of a dominant-negative N transgene (NΔcdc10rpts). Expression of NΔcdc10rpts was induced by 37°C heat shock for 1 h followed by 3 h of rest before training (n = 6 for all data points). (d) Blockade of LTM but not ARM by acutely induced expression of dominant-negative NΔcdc10rpts. The LTM component of persistent memory induced by spaced training is blocked, but ARM induced by massed (or spaced) training is not affected (n = 8, 10, 8, 8, 15, 15, 17, and 17 for data points from left to right).
In contrast, reduction of N activity disrupted 1-day memory. One-day memory after spaced training (10 training sessions with a 15-min rest interval between each) normally is composed of roughly equal amounts of ARM and LTM, whereas 1-day memory after massed training (10 training sessions with no rest interval) is composed only of ARM (18). One-day memory was significantly reduced in Nts2 mutants at a restrictive temperature (30°C) when compared with the permissive temperature (18°C) (Fig. 1b). There was also a slight but statistically insignificant reduction in memory scores in the control group when comparing the higher temperature with the lower temperature (Fig. 1b). Induced expression of dominant-negative N in hs-NΔcdc10rpts flies also significantly reduced 1-day memory after spaced training (Fig. 1d). Flies were given a 30-min heat shock and subjected to spaced training 3 h afterward. To determine which components (i.e., ARM or LTM) were affected, we also assayed 1-day memory after massed training in hs-NΔcdc10rpts transgenic flies (Fig. 1d). There was no difference for 1-day memory after massed training, indicating disruption of LTM but not ARM. The heat-shock treatment did not exert any detectable effect on control flies (Fig. 1d). Thus, reduced N activity, resulting either from a conditional mutation or induced expression of a dominant-negative N, specifically blocked the formation of LTM without affecting early memory or ARM.
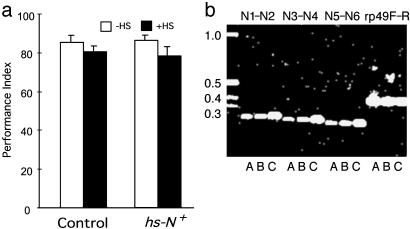
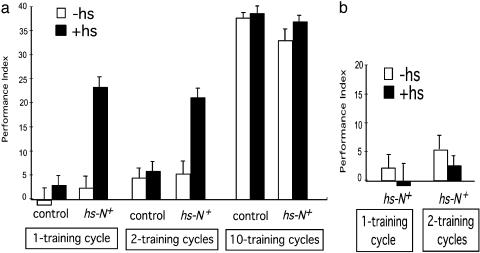
We then evaluated the effects of overexpression of wild-type N+ in hs-N+ transgenic flies. Flies were given a 30-min heat shock and trained and tested 3 h afterward. Overexpression of N+ had no significant effect on learning (Fig. 2a), consistent with our observations that disruption of N does not affect learning. Overexpression of N+ after heat-shock treatment was confirmed by RT-PCR of adult head mRNA (Fig. 2b). Acutely induced overexpression of N+ also did not affect 1-day memory for flies subjected to the normal spaced training procedure (10 training cycles) (Fig. 3a). However, 1-day memory was enhanced significantly for flies overexpressing N+ that had only one or two spaced training sessions (Fig. 3a). To characterize the nature of this enhancement in 1-day memory, we showed that enhanced memory elicited by one or two spaced training sessions in flies overexpressing N+ could be blocked by feeding of cycloheximide, a drug blocking protein synthesis (18), to flies (Fig. 3b). It has been shown that feeding cycloheximide to flies can block formation of protein-synthesis-dependent LTM (18). There was no significant difference for sensory modalities necessary for performing the learning task (Table 1). Thus, overexpression of the N+ gene facilitates LTM formation.
Fig. 2.
Learning is not affected by overexpression of wild-type N+ (hs-N+). Flies were subjected to 30 min of heat shock at 37°C followed by 3 h of rest. (a) Learning scores were similar for controls and hs-N+ flies regardless of whether they were subjected to heat-shock treatment (n = 2, 4, 2, and 4 for data points from left to right). (b) Heat shock induced expression of hs-N+ cDNA in adult heads. Semiquantitative RT-PCR using N primer pairs N1–N2, N3–N4, and N5–N6 shows induction of the hs-N+ transgene by 30 min of heat shock at 37°C followed by 3 h of rest (lane C) when compared with PCR from control flies that were kept at 18°C (lane A) or 25°C (lane B). The rp49F-R control primers show no temperature-induced increase in expression of rp49. Details of PCR primer pairs and expected products are given in Materials and Methods. Three separate mRNA isolations showed the same pattern of increased expression of the hs-N+ transgene after 37°C heat shock.
Fig. 3.
LTM is enhanced by overexpression of wild-type N+ (hs-N+). (a) Enhanced LTM after acutely induced overexpression of wild-type N+. Flies were subjected to the same heat-shock treatment as described above. Twenty-four-hour memory was compared between hs-N+ flies with and without heat shock for 1, 2, and 10 spaced training cycles, respectively (n = 6, 6, 5, 7, 7, 9, 5, 5, 6, 6, 7, and 8 for data points from left to right). (b) Blockade of enhanced LTM by an inhibitor of protein synthesis (cycloheximide). Flies were subjected to the same heat-shock treatment as described for a. Twenty-four-hour memory was compared between hs-N+ flies with and without heat shock for one and two spaced training cycles, respectively (n = 7, 9, 5, and 5 for data points from left to right; see Materials and Methods for drug-feeding conditions.)
Discussion
In the present study, our data revealed that one function for N in the adult brain is to mediate the formation of LTM. Three different lines of evidence support this conclusion. First, 1-day memory was reduced in a temperature-sensitive N mutant at the restrictive temperature (30°C) at which N receptor function is supposedly defective. Second, 1-day memory was diminished in transgenic flies that express a dominant-negative NΔcdc10rpts. Third, 1-day memory formation was facilitated by overexpression of N+. Although known for its crucial role in development of the nervous system, the use of temperature-sensitive mutations and of transgenic flies carrying inducible genes through acute treatment have allowed us to disassociate the role of N in adult physiology from its role in development. For all experiments that led to the observations noted above, flies were allowed to develop under relatively normal conditions and N function was perturbed acutely at the adult stage before the training. Moreover, we also showed that temporary disruption of N function at the adult stage exerted no significant effects on sensorimotor responses as well as on learning. Thus, the observed effects can be attributed specifically to memory, confirming that 1-day memory can be affected by manipulation of N function. Our finding is corroborated by an independent observation in which 1-day memory is reduced by disturbing N function through the use of RNA interference and Nts1 loss of function in adult flies (28).
One-day memory consists of both ARM and LTM phases (18). Our observation suggests a specific effect of manipulation of N function on LTM. One-day memory elicited by massed training (ARM only) (18) was not affected by expression of dominant-negative NΔcdc10rpts, whereas 1-day memory after spaced training (ARM and LTM) was reduced significantly. Consistent with this observation, enhanced memory induced by one or two spaced training sessions was blocked by a drug that inhibits protein synthesis. Such drugs are known to block LTM specifically (18).
Over the last several decades, a large number of genes in Drosophila have been identified to affect learning and memory through both forward and reverse genetic approaches (for review see ref. 29). Many signal transduction pathways have been implicated, such as cAMP, Ca2+/CaM-dependent kinase, protein kinase C, and mitogen-activated protein kinase pathways. However, none of the components of the N signaling pathway have been implicated by previous studies, which may reflect a crucial role of N signaling in development for which most mutations in this pathway are lethal and therefore unsuitable for behavioral studies.
It is notable that in flies with overexpression of N+, only one training session was required to elicit LTM instead of the usual 10 spaced training cycles. This enhancement is very similar to that reported for CREB (30). Induced expression of hs-dCREb2-activator also reduced the number of spaced training sessions, required to yield the maximal level of LTM in normal flies, from 10 sessions to 1 session (30). Future experiments may examine specific roles for the CREB and N pathways in the formation of LTM. Involvement of the CREB pathway in LTM formation is a mechanism observed in a wide range of organisms including Drosophila, Aplysia, and vertebrates (19, 30–32). The N signaling pathway is also highly conserved evolutionarily (1). In fact, a recent report showed that learning and memory are defective in a heterozygous Notch1+/- mouse knockout (33), although a developmental etiology could not be ruled out for such knockout mice. Our results extend this observation and the evolutionary implications thereof by revealing an acute role for N signaling during memory formation.
Acknowledgments
We thank A. Andres and S. deBelle for sharing unpublished data, Dr. M. Young for providing fly stocks, and Dr. J. Dubnau (Cold Spring Harbor Laboratory) for comments on the manuscript. This work was supported by National Institutes of Health grants (to T.T. and Y.Z.) and the Dart Foundation, as well as a grant from the National Natural Sciences Foundation of China (to Y.Z. and Z.X.). T.T. is a St. Giles Foundation Professor of Neuroscience.
Abbreviations: N, Notch; PS, presenilin; ARM, anesthesia-resistant memory; LTM, long-term memory; CREB, cAMP-response element-binding protein; hs-N, heat-shock N; PI, performance index; rp, ribosomal protein.
References
- 1.Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. (1999) Science 284, 770-776. [DOI] [PubMed] [Google Scholar]
- 2.Parks, A. L., Turner, F. R. & Muskavitch, M. A. (1995) Mech. Dev. 50, 201-216. [DOI] [PubMed] [Google Scholar]
- 3.Kidd, S., Lieber, T. & Young, M. W. (1998) Genes Dev. 12, 3728-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroeter, E. H., Kisslinger, J. A. & Kopan, R. (1998) Nature 393, 382-386. [DOI] [PubMed] [Google Scholar]
- 5.Struhl, G. & Adachi, A. (1998) Cell 93, 649-660. [DOI] [PubMed] [Google Scholar]
- 6.Artavanis-Tsakonas, S., Matsuno, K. & Fortini, M. E. (1995) Science 268, 225-232. [DOI] [PubMed] [Google Scholar]
- 7.Flores, G. V., Duan, H., Yan, H., Nagaraj, R., Fu, W., Zou, Y., Noll, M. & Banerjee, U. (2000) Cell 103, 75-85. [DOI] [PubMed] [Google Scholar]
- 8.Lecourtois, M. & Schweisguth, F. (1995) Genes Dev. 9, 2598-2608. [DOI] [PubMed] [Google Scholar]
- 9.Morel, V., Lecourtois, M., Massiani, O., Maier, D., Preiss, A. & Schweisguth, F. (2001) Curr. Biol. 11, 789-792. [DOI] [PubMed] [Google Scholar]
- 10.Stump, G., Durrer, A., Klein, A. L., Lutolf, S., Suter, U. & Taylor, V. (2002) Mech. Dev. 114, 153-159. [DOI] [PubMed] [Google Scholar]
- 11.Sestan, N., Artavanis-Tsakonas, S. & Rakic, P. (1999) Science 286, 741-746. [DOI] [PubMed] [Google Scholar]
- 12.Presente, A., Andres, A. & Nye, J. S. (2001) NeuroReport 12, 3321-3325. [DOI] [PubMed] [Google Scholar]
- 13.Sisodia, S. S. & St. George-Hyslop, P. H. (2002) Nat. Rev. Neurosci. 3, 281-290. [DOI] [PubMed] [Google Scholar]
- 14.Price, D. L., Tanzi, R. E., Borchelt, D. R. & Sisodia, S. S. (1998) Annu. Rev. Genet. 32, 461-493. [DOI] [PubMed] [Google Scholar]
- 15.Feng, R., Rampon, C., Tang, Y.-P., Shrom, D., Jin, J., Kyin, M., Sopher, B., Martin, G. M., Kim, S.-H., Langdon, R. B., Sisodia, S. S. & Tsien, J. Z. (2001) Neuron 32, 911-926. [DOI] [PubMed] [Google Scholar]
- 16.Wittenburg, N., Eimer, S., Lakowski, B., Rohrig, S., Rudolph, C. & Baumeister, R. (2000) Nature 406, 306-309. [DOI] [PubMed] [Google Scholar]
- 17.Tully, T. & Quinn, W. G. (1985) J. Comp. Physiol. A 157, 263-277. [DOI] [PubMed] [Google Scholar]
- 18.Tully, T., Preat, T., Boynton, S. C. & Del Vecchio, M. (1994) Cell 79, 35-47. [DOI] [PubMed] [Google Scholar]
- 19.Yin, J. C., Wallach, J. S., Del Vecchio, M., Wilder, E. L., Zhou, H., Quinn, W. G. & Tully, T. (1994) Cell 79, 49-58. [DOI] [PubMed] [Google Scholar]
- 20.DeZazzo, J., Sandstrom, D., de Belle, S., Velinzon, K., Smith, P., Grady, L., Del Vecchio, M., Ramaswami, M. & Tully T. (2000) Neuron 27, 145-158. [DOI] [PubMed] [Google Scholar]
- 21.Drier, E. A., Tello, M. K., Cowan, M., Wu, P., Blace, N., Sacktor, T. C. & Yin, J. C. (2002) Nat. Neurosci. 5, 316-324. [DOI] [PubMed] [Google Scholar]
- 22.Chiang, A.-S., Blum, A., Barditch, J., Chen, Y.-H., Chiu, S.-L., Regulski, M., Armstrong, J. D., Tully T. & Dubnau, J. (2004) Curr. Biol. 14, 263-272. [DOI] [PubMed] [Google Scholar]
- 23.Shellenbarger, D. L. & Mohler, J. D. (1975) Genetics 81, 143-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- 25.O'Connell, P. & Rosbash, M. (1984) Nucleic Acids Res. 12, 5495-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsley, D. L. & Grell, E. H., eds. (1968) Genetic Variations of Drosophila melanogaster (Carnegie Institution, Washington, DC).
- 27.Lieber, T., Kidd, S., Alcamo, E., Corbin, V. & Young, M. W. (1993) Genes Dev. 7, 1949-1965. [DOI] [PubMed] [Google Scholar]
- 28.Presente, A., Boyles, R. S., Serway, C. N., de Belle, S. & Andres, A. J. (2004) Proc. Natl. Acad. Sci. USA 101, 1764-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubnau, J. & Tully, T. (1998) Annu. Rev. Neurosci. 21, 407-444. [DOI] [PubMed] [Google Scholar]
- 30.Yin, J. C., Del Vecchio, M., Zhou, H. & Tully, T. (1995) Cell 81, 107-115. [DOI] [PubMed] [Google Scholar]
- 31.Bartsch, D., Ghirardi, M., Skehel, P. A., Kart, K. A., Herder, S. P., Chen, M., Bailey, C. H. & Kandel, E. R. (1995) Cell 83, 979-992. [DOI] [PubMed] [Google Scholar]
- 32.Bourtchuladze, R., Frenguelli, B., Blendy, J., Cioffi, D., Schutz, G. & Silva, A. J. (1994) Cell 79, 59-68. [DOI] [PubMed] [Google Scholar]
- 33.Costa, R. M., Honjo, T. & Silva, A. J. (2003) Curr. Biol. 13, 1348-1354. [DOI] [PubMed] [Google Scholar]