Abstract
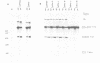
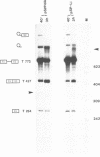
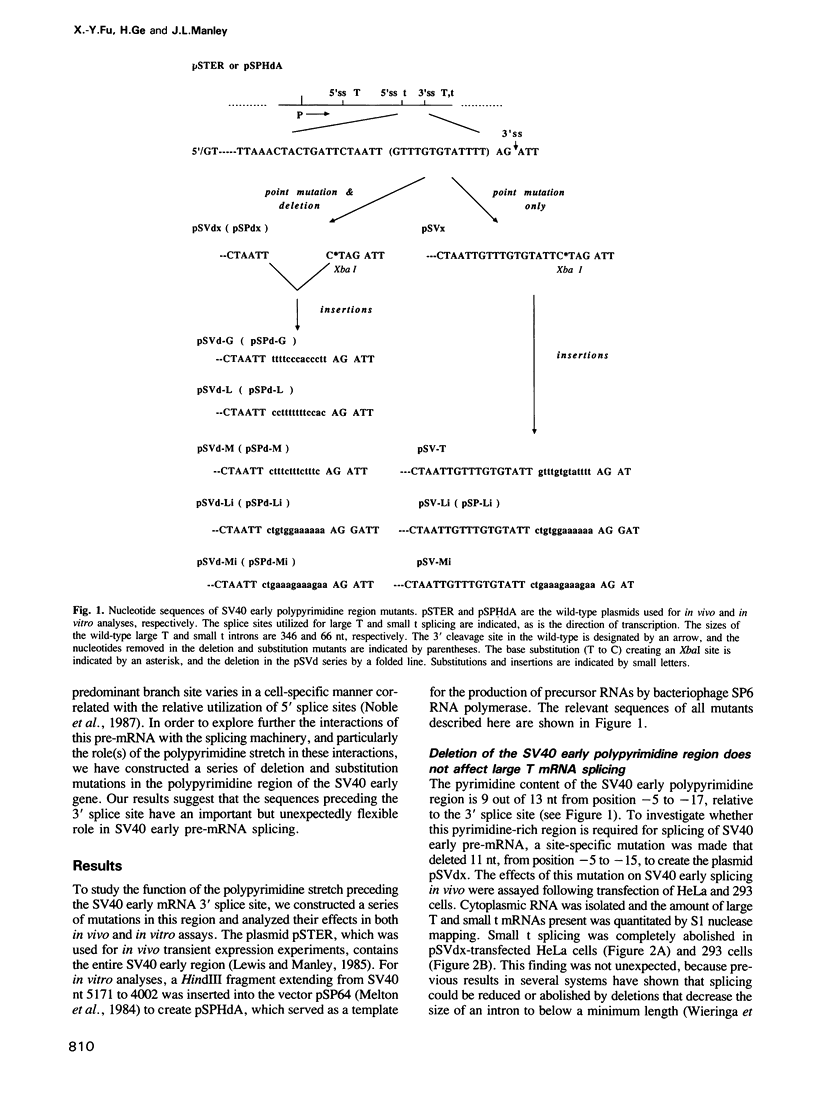
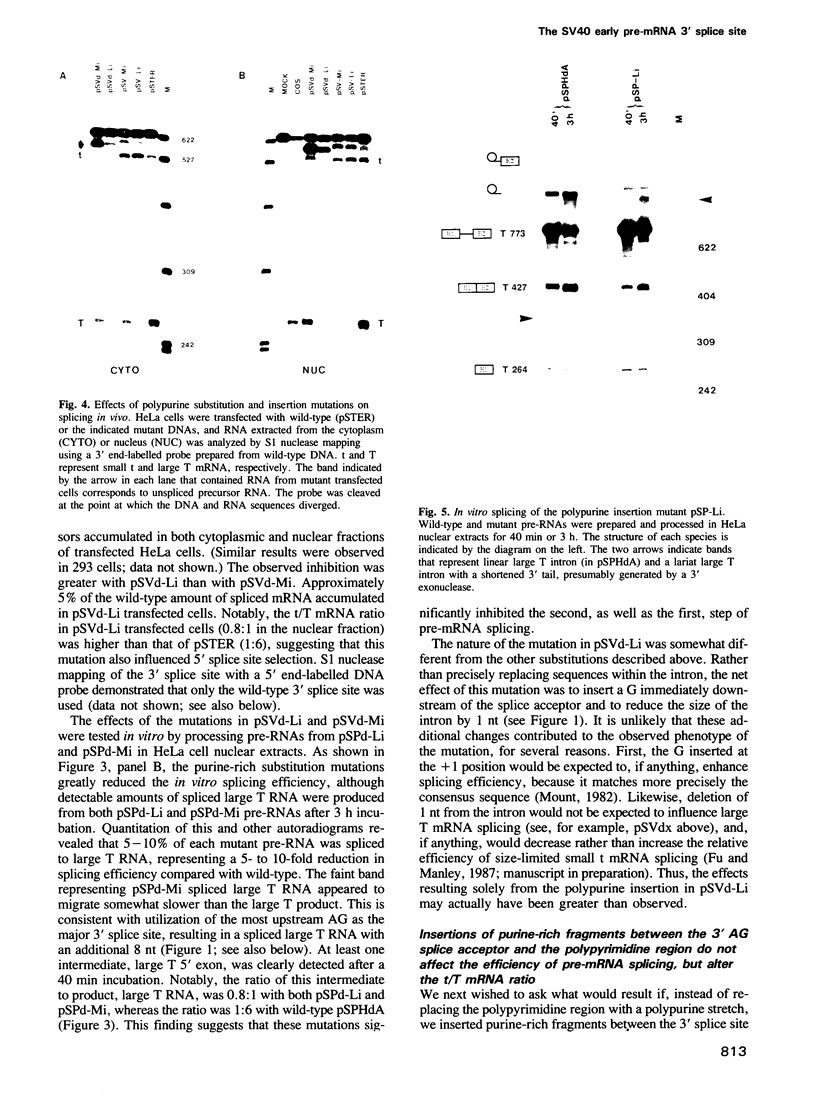
We have studied the role in pre-mRNA splicing of the nucleotide sequence preceding the SV40 early region 3' splice site. Somewhat surprisingly, neither the pyrimidine at the highly conserved -3 position, nor the polypyrimidine stretch that extends from -5 to -15, relative to the 3' splice site, were found to be required for efficient splicing. Mutations that delete this region or create polypurine insertions at position -2 had no significant effects on the efficiency of SV40 early pre-mRNA splicing in vivo or in vitro. Interestingly, however, the pyrimidine content of this region had substantial effects on the alternative splicing pattern of this pre-mRNA in vivo. Mutations that increased the number of pyrimidine residues resulted in more efficient utilization of the large T antigen mRNA 5' splice site relative to the small t 5' splice site, while mutations that increased the purine content enhanced small t mRNA splicing. A possible molecular mechanism for these findings, as well as a model that proposes a role for the polypyrimidine stretch in alternative splicing, are discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Berget S. M., Robberson B. L. U1, U2, and U4/U6 small nuclear ribonucleoproteins are required for in vitro splicing but not polyadenylation. Cell. 1986 Aug 29;46(5):691–696. doi: 10.1016/0092-8674(86)90344-2. [DOI] [PubMed] [Google Scholar]
- Bernstein S. I., Hansen C. J., Becker K. D., Wassenberg D. R., 2nd, Roche E. S., Donady J. J., Emerson C. P., Jr Alternative RNA splicing generates transcripts encoding a thorax-specific isoform of Drosophila melanogaster myosin heavy chain. Mol Cell Biol. 1986 Jul;6(7):2511–2519. doi: 10.1128/mcb.6.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Steitz J. A. Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein. Cell. 1986 Aug 29;46(5):697–704. doi: 10.1016/0092-8674(86)90345-4. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Chabot B., Black D. L., LeMaster D. M., Steitz J. A. The 3' splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science. 1985 Dec 20;230(4732):1344–1349. doi: 10.1126/science.2933810. [DOI] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveleth D. D., Gietz R. D., Spencer C. A., Nargang F. E., Hodgetts R. B., Marsh J. L. Sequence and structure of the dopa decarboxylase gene of Drosophila: evidence for novel RNA splicing variants. EMBO J. 1986 Oct;5(10):2663–2672. doi: 10.1002/j.1460-2075.1986.tb04549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenthal S., Parker V. P., Davidson N. Developmental variations in the splicing pattern of transcripts from the Drosophila gene encoding myosin alkali light chain result in different carboxyl-terminal amino acid sequences. Proc Natl Acad Sci U S A. 1985 Jan;82(2):449–453. doi: 10.1073/pnas.82.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin A., Jove R., Hemenway C., Keiser H. D., Manley J. L., Prives C. Splicing pathways of SV40 mRNAs in X. laevis oocytes differ in their requirements for snRNPs. Cell. 1984 Jul;37(3):927–936. doi: 10.1016/0092-8674(84)90427-6. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Glenney J. R., Jr, Glenney P. The microvillus 110K cytoskeletal protein is an integral membrane protein. Cell. 1984 Jul;37(3):743–751. doi: 10.1016/0092-8674(84)90410-0. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner M. B., Martin G. A., Noble J. A., Nikoloff D. M., Tal R., Kawasaki E. S., White T. J. Human CSF-1: gene structure and alternative splicing of mRNA precursors. EMBO J. 1987 Sep;6(9):2693–2698. doi: 10.1002/j.1460-2075.1987.tb02561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Lewis E. D., Manley J. L. Repression of simian virus 40 early transcription by viral DNA replication in human 293 cells. Nature. 1985 Sep 12;317(6033):172–175. doi: 10.1038/317172a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B. A., Johnson W. A., Hirsh J. Regulated splicing produces different forms of dopa decarboxylase in the central nervous system and hypoderm of Drosophila melanogaster. EMBO J. 1986 Dec 1;5(12):3335–3342. doi: 10.1002/j.1460-2075.1986.tb04648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawa H., Kotani H., Nakanishi S. Tissue-specific generation of two preprotachykinin mRNAs from one gene by alternative RNA splicing. Nature. 1984 Dec 20;312(5996):729–734. doi: 10.1038/312729a0. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Pan Z. Q., Prives C., Manley J. L. Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell. 1987 Jul 17;50(2):227–236. doi: 10.1016/0092-8674(87)90218-2. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. In vitro splicing of simian virus 40 early pre mRNA. Nucleic Acids Res. 1986 Feb 11;14(3):1219–1235. doi: 10.1093/nar/14.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris K., Norris F., Christiansen L., Fiil N. Efficient site-directed mutagenesis by simultaneous use of two primers. Nucleic Acids Res. 1983 Aug 11;11(15):5103–5112. doi: 10.1093/nar/11.15.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Aebi M., Hornig H., Weissmann C., Sharp P. A. Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit beta-globin genes. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8349–8353. doi: 10.1073/pnas.82.24.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. An RNA processing activity that debranches RNA lariats. Science. 1985 Jul 12;229(4709):135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Pikielny C. W., Rosbash M., Green M. R. Alternative branch points are selected during splicing of a yeast pre-mRNA in mammalian and yeast extracts. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2022–2026. doi: 10.1073/pnas.83.7.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Patel R. S., Fonda D., Hynes R. O. Multiple sites of alternative splicing of the rat fibronectin gene transcript. EMBO J. 1987 Sep;6(9):2573–2580. doi: 10.1002/j.1460-2075.1987.tb02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Proudfoot N. J., Shander M., Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982 Jul;29(3):903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- Ulfendahl P. J., Pettersson U., Akusjärvi G. Splicing of the adenovirus-2 E1A 13S mRNA requires a minimal intron length and specific intron signals. Nucleic Acids Res. 1985 Sep 11;13(17):6299–6315. doi: 10.1093/nar/13.17.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Leung H., Weiner A. M. The natural 5' splice site of simian virus 40 large T antigen can be improved by increasing the base complementarity to U1 RNA. Mol Cell Biol. 1987 Aug;7(8):3018–3020. doi: 10.1128/mcb.7.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]
- van Santen V. L., Spritz R. A. mRNA precursor splicing in vivo: sequence requirements determined by deletion analysis of an intervening sequence. Proc Natl Acad Sci U S A. 1985 May;82(9):2885–2889. doi: 10.1073/pnas.82.9.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]