Abstract
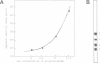
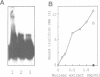
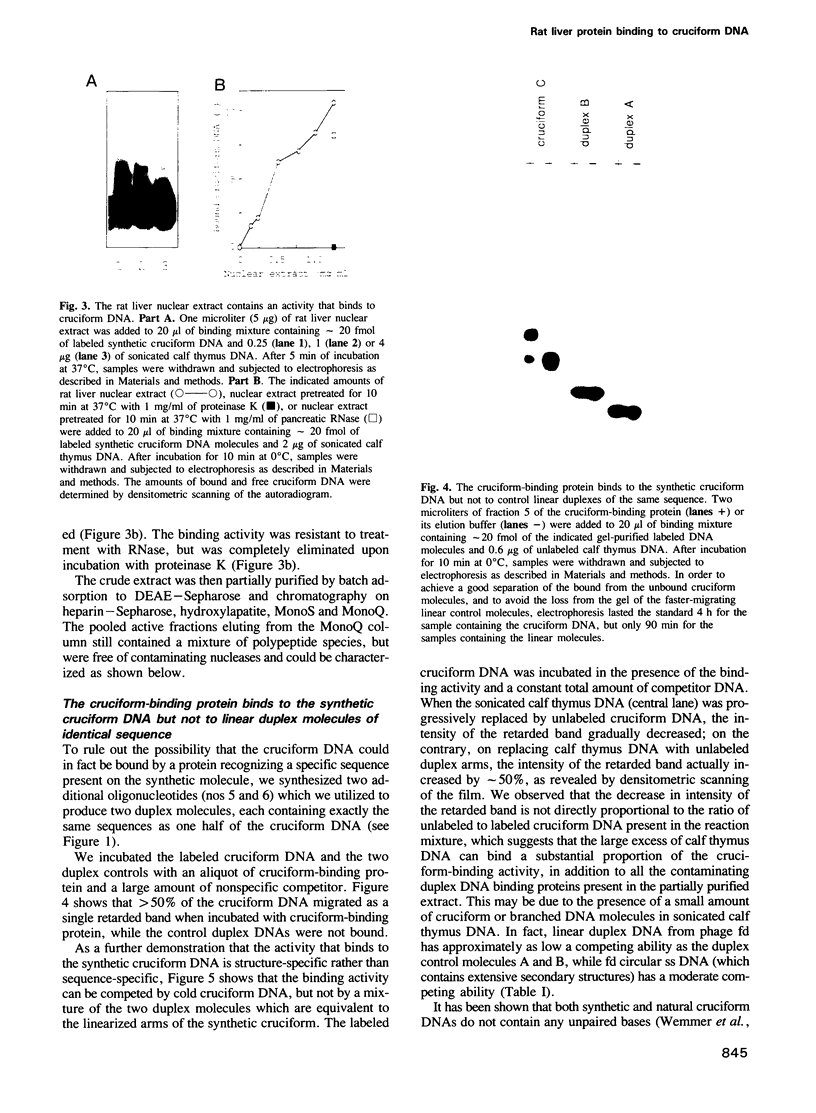
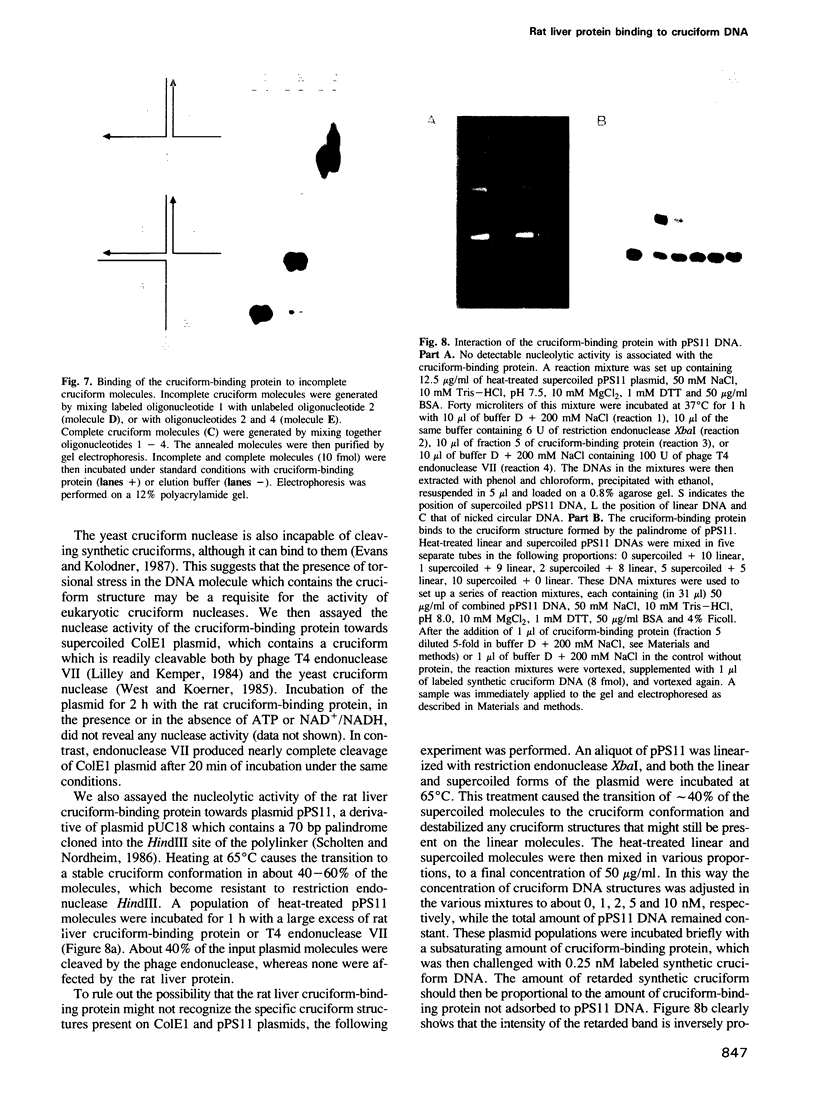
We constructed a synthetic cruciform DNA which closely resembles Holliday junctions, a DNA structure formed during recombination or following the transition from interstrand to intrastrand base pairing in palindromic DNA sequences. We identified and partially purified a protein from rat liver that specifically binds to this cruciform DNA molecule and does not bind to single-stranded or double-stranded DNAs of the same sequence. This protein also binds to the cruciform structure formed by a 70 bp palindromic sequence cloned in plasmid pUC18. No detectable nucleolytic activity is associated with the rat liver cruciform-binding protein, in contrast to all cruciform-recognizing proteins known so far.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell L., Byers B. Occurrence of crossed strand-exchange forms in yeast DNA during meiosis. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3445–3449. doi: 10.1073/pnas.76.7.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. H., Kolodner R. Construction of a synthetic Holliday junction analog and characterization of its interaction with a Saccharomyces cerevisiae endonuclease that cleaves Holliday junctions. J Biol Chem. 1987 Jul 5;262(19):9160–9165. [PubMed] [Google Scholar]
- Frappier L., Price G. B., Martin R. G., Zannis-Hadjopoulos M. Monoclonal antibodies to cruciform DNA structures. J Mol Biol. 1987 Feb 20;193(4):751–758. doi: 10.1016/0022-2836(87)90356-1. [DOI] [PubMed] [Google Scholar]
- Frisque R. J. Nucleotide sequence of the region encompassing the JC virus origin of DNA replication. J Virol. 1983 Apr;46(1):170–176. doi: 10.1128/jvi.46.1.170-176.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Scholten P. M., Nordheim A. Diethyl pyrocarbonate: a chemical probe for DNA cruciforms. Nucleic Acids Res. 1986 May 27;14(10):3981–3993. doi: 10.1093/nar/14.10.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Spadaro A., Schaffer J. E., Murray A. W., Maxam A. M., Schaffer P. A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985 May;5(5):930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer D. E., Wand A. J., Seeman N. C., Kallenbach N. R. NMR analysis of DNA junctions: imino proton NMR studies of individual arms and intact junction. Biochemistry. 1985 Oct 8;24(21):5745–5749. doi: 10.1021/bi00342a009. [DOI] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Parsons C. A., Picksley S. M. Purification and properties of a nuclease from Saccharomyces cerevisiae that cleaves DNA at cruciform junctions. J Biol Chem. 1987 Sep 15;262(26):12752–12758. [PubMed] [Google Scholar]
- Wolgemuth D. J., Hsu M. T. DNA replication-mediated recombination of molecules of adenovirus 2 DNA. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5076–5080. doi: 10.1073/pnas.78.8.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Kaufmann G., Martin R. G. Mammalian DNA enriched for replication origins is enriched for snap-back sequences. J Mol Biol. 1984 Nov 15;179(4):577–586. doi: 10.1016/0022-2836(84)90156-6. [DOI] [PubMed] [Google Scholar]
- Zyskind J. W., Cleary J. M., Brusilow W. S., Harding N. E., Smith D. W. Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1164–1168. doi: 10.1073/pnas.80.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]