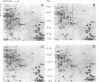
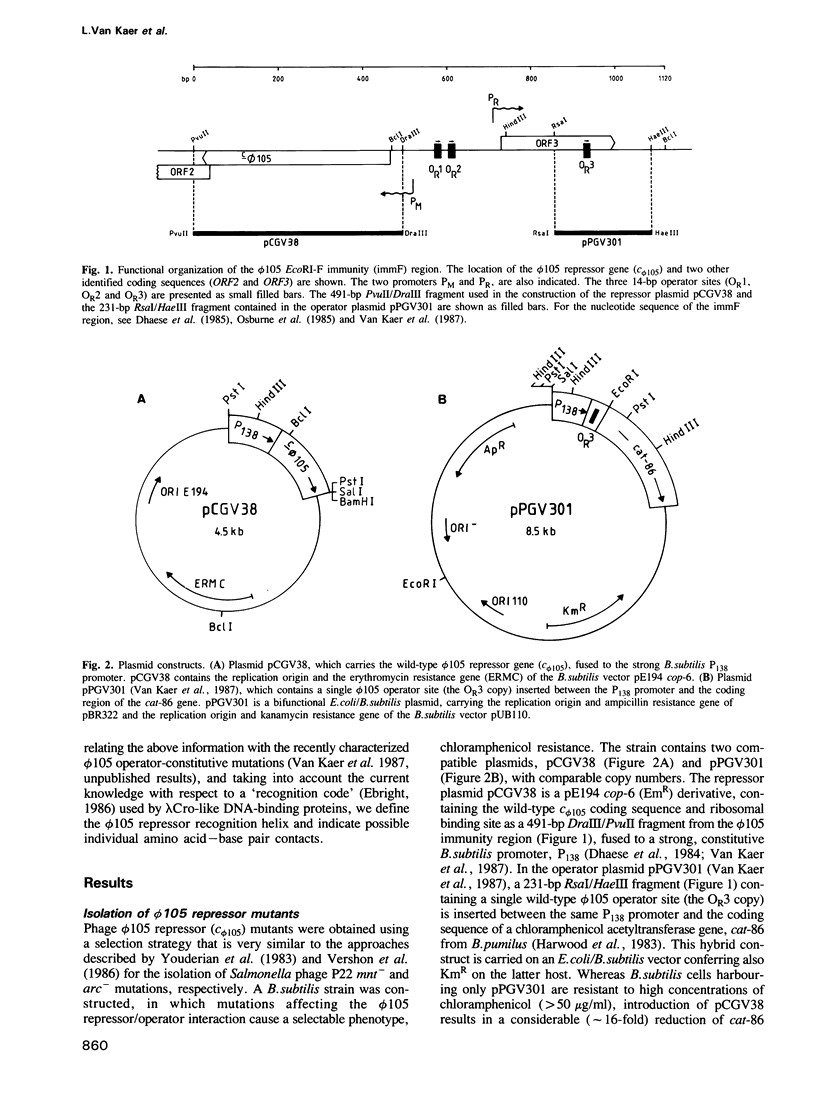
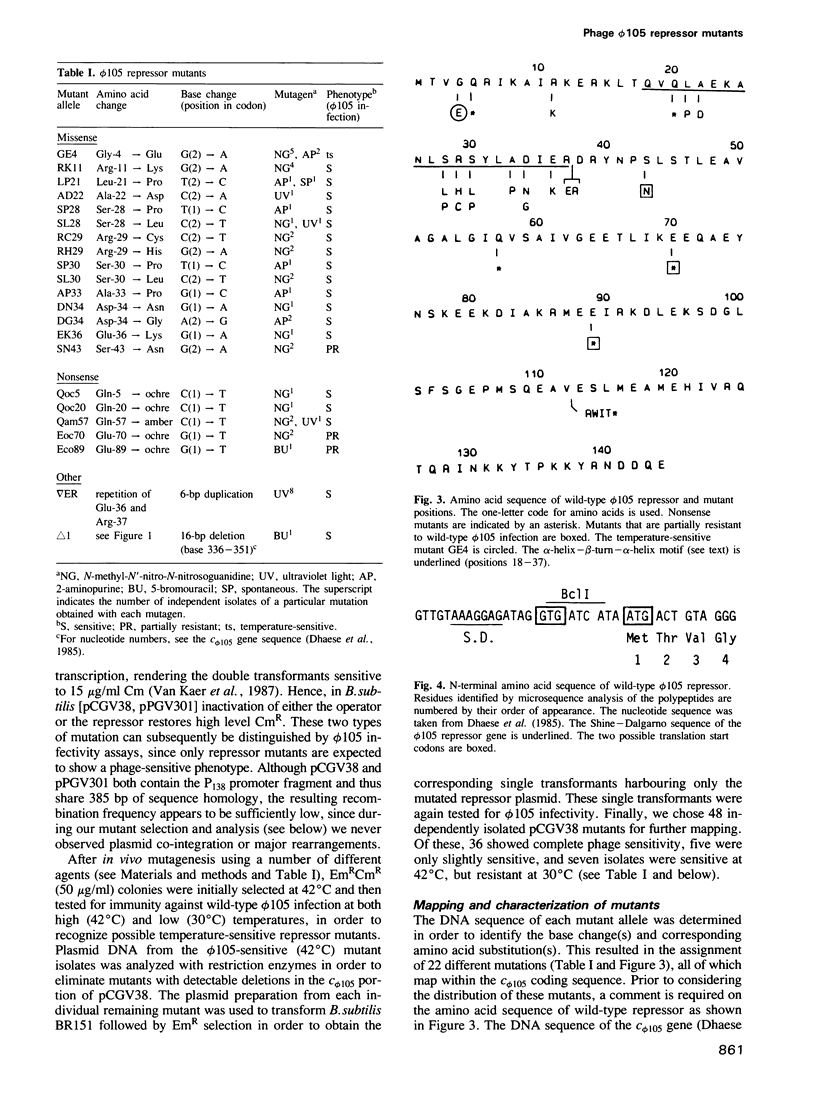
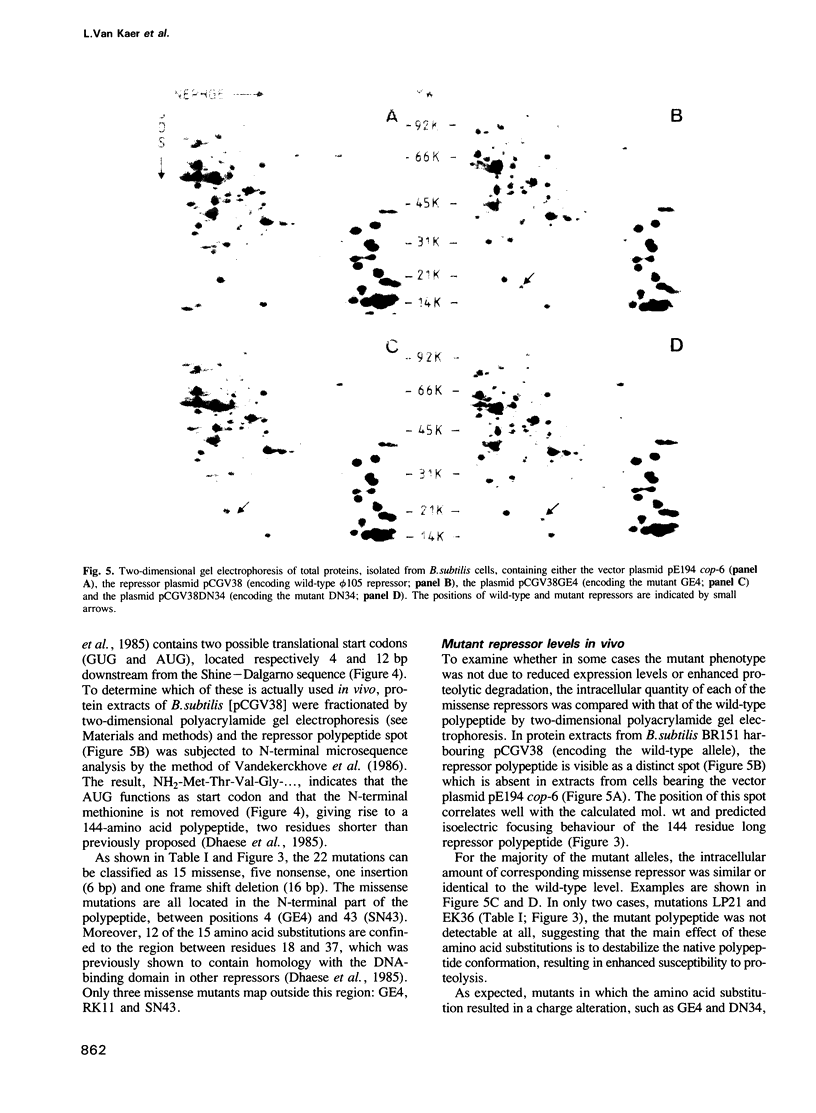
Abstract
The Bacillus subtilis phage phi 105 repressor specifically recognizes a 14-bp operator sequence which does not exhibit 2-fold rotational symmetry. To facilitate a genetic analysis of this sequence-dependent DNA binding a B. subtilis strain was constructed in which mutations affecting the phi 105 repressor-operator interaction cause a selectable phenotype, chloramphenicol resistance. After in vivo mutagenesis, we isolated and mapped 22 different mutations in the repressor coding sequence, 15 of which are missense substitutions. These are exclusively located in the N-terminal part (positions 1-43) of the 144 residue long polypeptide. Two nonsense mutants, at positions 70 and 89, respectively, still show partial repressor activity. These data suggest that the phi 105 repressor consists of at least two independently folding structural domains, of which the N-terminal is involved in operator binding. Twelve missense mutations are clustered in a region extending from Gln-18 to Arg-37, which we propose to be the DNA-binding alpha-helix--beta-turn--alpha-helix motif, common to all lambda Cro-like repressors. The second ('recognition') helix shows significant homology with the corresponding sequence in Tn3 resolvase, and there is also a striking similarity between the phi 105 operator and the consensus sequence for a Tn3 res half-site. Based on these observations, and on the previously isolated phi 105 0c mutants, we tentatively assign some specific contacts between base pairs from the first half of a phi 105 operator site and amino acids from the repressor's 'recognition helix'.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Bauw G., De Loose M., Inzé D., Van Montagu M., Vandekerckhove J. Alterations in the phenotype of plant cells studied by NH(2)-terminal amino acid-sequence analysis of proteins electroblotted from two-dimensional gel-separated total extracts. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4806–4810. doi: 10.1073/pnas.84.14.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Cully D. F., Garro A. J. Nucleotide sequence of the immunity region of Bacillus subtilis bacteriophage phi 105: identification of the repressor gene and its mRNA and protein products. Gene. 1985;38(1-3):153–164. doi: 10.1016/0378-1119(85)90214-8. [DOI] [PubMed] [Google Scholar]
- Dandanell G., Valentin-Hansen P., Larsen J. E., Hammer K. Long-range cooperativity between gene regulatory sequences in a prokaryote. 1987 Feb 26-Mar 4Nature. 325(6107):823–826. doi: 10.1038/325823a0. [DOI] [PubMed] [Google Scholar]
- Dhaese P., Hussey C., Van Montagu M. Thermo-inducible gene expression in Bacillus subtilis using transcriptional regulatory elements from temperate phage phi 105. Gene. 1984 Dec;32(1-2):181–194. doi: 10.1016/0378-1119(84)90046-5. [DOI] [PubMed] [Google Scholar]
- Dhaese P., Seurinck J., De Smet B., Van Montagu M. Nucleotide sequence and mutational analysis of an immunity repressor gene from Bacillus subtilis temperate phage phi 105. Nucleic Acids Res. 1985 Aug 12;13(15):5441–5455. doi: 10.1093/nar/13.15.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Systematic method for the detection of potential lambda Cro-like DNA-binding regions in proteins. J Mol Biol. 1987 Apr 5;194(3):557–564. doi: 10.1016/0022-2836(87)90681-4. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Molecular basis of DNA sequence recognition by the catabolite gene activator protein: detailed inferences from three mutations that alter DNA sequence specificity. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7274–7278. doi: 10.1073/pnas.81.23.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eismann E., von Wilcken-Bergmann B., Müller-Hill B. Specific destruction of the second lac operator decreases repression of the lac operon in Escherichia coli fivefold. J Mol Biol. 1987 Jun 20;195(4):949–952. doi: 10.1016/0022-2836(87)90499-2. [DOI] [PubMed] [Google Scholar]
- Fritz H. J., Bicknäse H., Gleumes B., Heibach C., Rosahl S., Ehring R. Characterization of two mutations in the Escherichia coli galE gene inactivating the second galactose operator and comparative studies of repressor binding. EMBO J. 1983;2(12):2129–2135. doi: 10.1002/j.1460-2075.1983.tb01713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Lauth M. R., Wells R. G., Wityk R. J., Salvo J. J., Reed R. R. Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of gamma delta and Tn3. Cell. 1982 Aug;30(1):19–27. doi: 10.1016/0092-8674(82)90007-1. [DOI] [PubMed] [Google Scholar]
- Harwood C. R., Williams D. M., Lovett P. S. Nucleotide sequence of a Bacillus pumilus gene specifying chloramphenicol acetyltransferase. Gene. 1983 Oct;24(2-3):163–169. doi: 10.1016/0378-1119(83)90076-8. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Nelson H. C., Sauer R. T. Mutations in lambda repressor's amino-terminal domain: implications for protein stability and DNA binding. Proc Natl Acad Sci U S A. 1983 May;80(9):2676–2680. doi: 10.1073/pnas.80.9.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman W. J., Tanaka C. K. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 1986 Jul;81(3):802–806. doi: 10.1104/pp.81.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani M. H., Orosz L., Adhya S. A control element within a structural gene: the gal operon of Escherichia coli. Cell. 1983 Mar;32(3):783–788. doi: 10.1016/0092-8674(83)90064-8. [DOI] [PubMed] [Google Scholar]
- Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987 May;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehming N., Sartorius J., Niemöller M., Genenger G., v Wilcken-Bergmann B., Müller-Hill B. The interaction of the recognition helix of lac repressor with lac operator. EMBO J. 1987 Oct;6(10):3145–3153. doi: 10.1002/j.1460-2075.1987.tb02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A., Adhya S. Demonstration of two operator elements in gal: in vitro repressor binding studies. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6100–6104. doi: 10.1073/pnas.81.19.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B., Crapo L., Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968 Apr;59(4):1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Hecht M. H., Sauer R. T. Mutations defining the operator-binding sites of bacteriophage lambda repressor. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):441–449. doi: 10.1101/sqb.1983.047.01.052. [DOI] [PubMed] [Google Scholar]
- Newman B. J., Grindley N. D. Mutants of the gamma delta resolvase: a genetic analysis of the recombination function. Cell. 1984 Sep;38(2):463–469. doi: 10.1016/0092-8674(84)90501-4. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Oppenheim A. B., Noff D. Deletion mapping of trans dominant mutations in the lambda repressor gene. Virology. 1975 Apr;64(2):553–556. doi: 10.1016/0042-6822(75)90132-4. [DOI] [PubMed] [Google Scholar]
- Osburne M. S., Craig R. J., Rothstein D. M. Thermoinducible transcription system for Bacillus subtilis that utilizes control elements from temperate phage phi 105. J Bacteriol. 1985 Sep;163(3):1101–1108. doi: 10.1128/jb.163.3.1101-1108.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pakula A. A., Young V. B., Sauer R. T. Bacteriophage lambda cro mutations: effects on activity and intracellular degradation. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8829–8833. doi: 10.1073/pnas.83.23.8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P., Albrechtsen B., Løve Larsen J. E. DNA-protein recognition: demonstration of three genetically separated operator elements that are required for repression of the Escherichia coli deoCABD promoters by the DeoR repressor. EMBO J. 1986 Aug;5(8):2015–2021. doi: 10.1002/j.1460-2075.1986.tb04458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kaer L., Van Montagu M., Dhaese P. Transcriptional control in the EcoRI-F immunity region of Bacillus subtilis phage phi 105. Identification and unusual structure of the operator. J Mol Biol. 1987 Sep 5;197(1):55–67. doi: 10.1016/0022-2836(87)90609-7. [DOI] [PubMed] [Google Scholar]
- Velleman M., Dreiseikelmann B., Schuster H. Multiple repressor binding sites in the genome of bacteriophage P1. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5570–5574. doi: 10.1073/pnas.84.16.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon A. K., Bowie J. U., Karplus T. M., Sauer R. T. Isolation and analysis of arc repressor mutants: evidence for an unusual mechanism of DNA binding. Proteins. 1986 Dec;1(4):302–311. doi: 10.1002/prot.340010404. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Brown E. L., Ptashne M. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell. 1984 Sep;38(2):361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. A new-specificity mutant of 434 repressor that defines an amino acid-base pair contact. 1987 Apr 30-May 6Nature. 326(6116):888–891. doi: 10.1038/326888a0. [DOI] [PubMed] [Google Scholar]
- Youderian P., Vershon A., Bouvier S., Sauer R. T., Susskind M. M. Changing the DNA-binding specificity of a repressor. Cell. 1983 Dec;35(3 Pt 2):777–783. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]