Abstract
Knockdown resistance ( kdr) and CYP9K1 genotypes were detected by a MOLDI-TOF based SNP genotyping assay (Sequenom iPLEX) in samples of Anopheles gambiae collected at 13 sites throughout the Union of the Comoros and Dar es Salaam, Tanzania during February and March 2011. All A. gambiae specimens collected in the Comoros were homozygous for the susceptible kdr alleles (+/+) while 96% of A. gambiae from Dar es Salaam were homozygous for the East African kdr resistant genotype (E/E). In contrast, all specimens from Dar es Salaam and the Comoros were homozygous for the cyp3 allele (c3/c3) at the CYP9K1 locus; the locus has been implicated in metabolic resistance against pyrethroid insecticides in West Africa. All specimens had typical A. gambiae genotypes for SNPs within the divergence Islands on all three chromosomes. Although further spatial and temporal studies are needed, the distribution of kdr genotypes between the Comoros and Tanzania further supports isolation of the Comoros populations from A. gambiae populations on mainland Africa .
Keywords: DIS, Dar es Salaam, CYP9K1, kncokdown resistence, insecticide, resistence
Introduction
A majority of the human population residing in the Union of the Comoros (=94%) live in high malaria transmission zones 1. Anopheles gambiae and Anopheles funestus (Giles) are the major malaria vectors in the Comoros 2. Vector control efforts have concentrated on the adult stage using insecticide-treated bednets (ITNs) and indoor residual spraying (IRS) with DDT 1. ITN distribution was initiated in the Comoros in 2005 and by 2014 roughly 40% of the population has access to ITNs 1.
Limited insecticide resistance surveillance has been conducted on malaria vectors in Union of Comoros, with to date, published records stemming only from investigations in Mayotte (an island administered by France), where A. gambiae were susceptible to multiple insecticides except for a larvicide, temephos 3. Insecticide susceptibility studies have been conducted in neighboring East African countries such as in western Kenya (Chen et al. 2008. JME, Mathias et al. 2011, Malaria J, Ochomo et al. 2012 MVE), but little information is available on the coastal regions of Kenya. In Tanzania, information, based on small sample sizes, is available on the kdr allele frequency distribution in coastal districts of Muheza and Ilula (Dar es Salaam) 4 where about one third from Dar es Salaam were homozygous for the kdr-East (L1014S) mutation.
Here we present much needed data on kdr allele frequencies and include frequency data for a recently described pyrethroid metabolic resistance gene, CYP9K1. Allele frequencies for Anopheles gambiae collected at 13 sites in the Union of the Comoros, plus Dar es Salaam, Tanzania are presented ( Table 1).
Table 1. Sites, kdr, CYP9K1 information from Anopheles gambiae samples collected in the Comoros and Tanzania, February and March 2011.
Numbers (#) indicate site locations on the map in Figure 1.
| idx | Site | Lat | Lng |
Kdr
genotyped |
+/+ | E/+ | E/E | %E | CYP9K1
genotyped |
cyp3 | %cyp3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Assimpao | -12.24 | 44.32 | 7 | 7 | 0 | 7 | 7 | 100 | ||
| 2 | Boeninidi | -11.57 | 43.29 | 31 | 31 | 0 | 8 | 8 | 100 | ||
| 3 | Bouni | -11.49 | 43.40 | 32 | 32 | 0 | 8 | 8 | 100 | ||
| 4 | Fomboni | -12.28 | 43.73 | 28 | 28 | 0 | 5 | 5 | 100 | ||
| 5 | Hoani | -12.26 | 43.67 | 20 | 20 | 0 | 8 | 8 | 100 | ||
| 6 | Male | -11.89 | 43.51 | 17 | 17 | 0 | 14 | 14 | 100 | ||
| 7 | Miringoni | -12.30 | 43.64 | 16 | 16 | 0 | 7 | 7 | 100 | ||
| 8 | Moya | -12.31 | 44.44 | 68 | 68 | 0 | 8 | 8 | 100 | ||
| 9 | Mutsamudu | -11.61 | 43.39 | 30 | 30 | 0 | 8 | 8 | 100 | ||
| 10 | Ndremeani | -12.35 | 43.75 | 30 | 30 | 0 | 8 | 8 | 100 | ||
| 11 | Ossivo | -11.59 | 43.28 | 18 | 18 | 0 | 8 | 8 | 100 | ||
| 12 | Saliman | -11.68 | 43.27 | 4 | 4 | 0 | 4 | 4 | 100 | ||
| 13 | Wala | -12.34 | 43.67 | 29 | 29 | 0 | 8 | 8 | 100 | ||
| 14 | Wanani | -12.35 | 43.80 | 32 | 32 | 0 | 8 | 8 | 100 | ||
| 15 | Dar es Salaam | -6.83 | 39.27 | 25 | 1 | 24 | 98 | 8 | 8 | 100 | |
| Grand Total | 387 | 362 | 1 | 24 | 109 | 109 |
Figure 1. Collection sites in the Comoros and Tanzania.
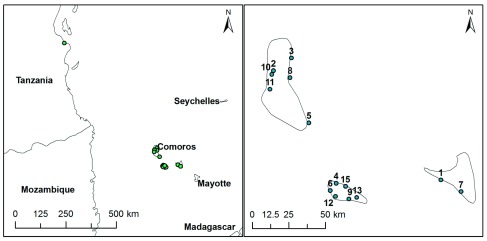
Site numbers corresponds to index number provided in Table 1.
Methods
A total of 362 indoor resting adults and larvae were collected from 13 locations from the three islands ( Figure 1) making up the Union of the Comoros between February and March, 2011. Larvae were individually rinsed twice in bottled mineral water and placed in 80% ethanol for downstream genomic DNA extraction. A collection of A. gambiae sensu lato from Furvela, Mozambique were collected using light traps inside houses. Mosquitoes from Dar es Salaam were obtained from Dr. Kija Ngh’abi at Ifakara Health Institute.
Samples were transported to the UC Davis Vector Genetics Laboratory for further genetic assay. DNA was extracted using a DNeasy extraction kit (Qiagen, Valencia, CA). Species were determined based on the combination of species diagnostic assays 5, 6 and a divergence island SNP (DIS) genotyping assay 7.
For DIS, kdr and CYP9K1 genotyping, we used the Sequenom iPLEX Gold Genotyping Reagent Set (Catalog number: Sequenom 10158) on a MassArray (Sequenom) mass spectrometer at the UC Davis Veterinary Genetics Laboratory. This assay was slightly modified from the original DIS assay 7 by adding the kdr and CYP9K1 markers, as described in Supplemental Document S1.
Results & discussion
A. gambiae from Dar es Salaam, Tanzania, had the kdr-East (L1014S) genotype at a frequency of 96%, which is higher than the frequency previously reported from Dar es Salaam by Kabula et al. 7 where respectively, 1/3 and 2/3 of their samples were homozygous and susceptible for kdr-East (L1014S). In contrast, all A. gambiae from the Comoros were homozygous for the susceptible kdr alleles. All A. gambiae from both Tanzania and the Comoros were homozygous for the cyp3 allele for the CYP9K1 gene. All specimens from Furvela, Mozambique were A. merus (30/35) or A. arabiensis (5/35) and were excluded from further analysis.
Significant pressure to select for resistance to pyrethroid insecticides in A. gambiae and other indoor biting and resting malaria vectors likely occurs throughout sub-Saharan Africa because of intense IRS and ITN usage. A recent study in Mali noted an adaptive introgression of kdr resistant alleles from A. gambiae stably incorporated into the A. coluzzii genome under high ITN coverage environments 8. A similar genomic signature of adaptive introgression was also observed in Ghana 9. A. gambiae populations in the Comoros have had the opportunity, via transport by boat or air, to acquire resistant A. gambiae genotypes from neighboring countries such as Tanzania where high levels of insecticide resistance have been reported 10. The failure of the Comoros population to acquire insecticide resistance alleles despite long term exposure to insecticide pressure 1 may potentially be due to several factors or combination of factor including: (1) ITN coverage (<25% compared to >60% Mali) is not high enough to drive selection for resistance, (2) these populations are very isolated from mainland populations, requiring them to develop resistance de novo rather than from gene flow from neighboring populations, and/or (3) A. gambiae on the Comoros may be exophilic.
Our study provides much needed information regarding the genetics of insecticide resistance in A. gambiae populations in the Comoros Islands. Although the malaria vectors in Comoros appear to be genetically predisposed to insecticide susceptibility, it is possible that these mosquitoes have developed phenotypic resistance via alternative mechanisms such as metabolic resistance other than CYP9K1 or behavior resistance (e.g. exophily). Further studies are needed to establish levels of phenotypic resistance against insecticides, as well as bionomics of the malaria vectors in this region to understand the impact of insecticide-based malaria control measures in the Comoros.
Acknowledgements
We thank Catelyn C. Nieman for assistance in DNA extraction and species diagnostic assay. We also thank Julia Malvick at the Veterinary Genetics Laboratory of UC Davis School of Veterinary Medicine for assistance in processing iPLEX SNP genotyping assay.
Funding Statement
The authors also acknowledge financial support from NIH grants: 5R21AI062929.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
v1; ref status: indexed
Supplementary materials
Supplemental Document S1.
Modification of the original DIS assay in 7.
.
References
- 1. WHO: World Malaria Report 2014. Switzerland: World Health Organization 2014.2014. Reference Source [Google Scholar]
- 2. Ayala D, Goff GL, Robert V, et al. : Population structure of the malaria vector Anopheles funestus (Diptera: Culicidae) in Madagascar and Comoros. Acta Trop. 2006;97(3):292–300. 10.1016/j.actatropica.2005.12.002 [DOI] [PubMed] [Google Scholar]
- 3. Pocquet N, Darriet F, Zumbo B, et al. : Insecticide resistance in disease vectors from Mayotte: an opportunity for integrated vector management. Parasit Vectors. 2014;7:299. 10.1186/1756-3305-7-299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kabula B, Kisinza W, Tungu P, et al. : Co-occurrence and distribution of East (L1014S) and West (L1014F) African knock-down resistance in Anopheles gambiae sensu lato population of Tanzania. Trop Med Int Health. 2014;19(3):331–341. 10.1111/tmi.12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scott JA, Brogdon WG, Collins FH: Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49(4):520–529. [DOI] [PubMed] [Google Scholar]
- 6. Favia G, Lanfrancotti A, Spanos L, et al. : Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Mol Biol. 2001;10(1):19–23. 10.1046/j.1365-2583.2001.00236.x [DOI] [PubMed] [Google Scholar]
- 7. Lee Y, Marsden CD, Nieman C, et al. : A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae. Mol Ecol Resour. 2014;14(2):297–305. 10.1111/1755-0998.12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Norris LC, Main BJ, Lee Y, et al. : Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc Natl Acad Sci U S A. 2015;112(3):815–820. 10.1073/pnas.1418892112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clarkson CS, Weetman D, Essandoh J, et al. : Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 2014;5:4248. 10.1038/ncomms5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kabula B, Tungu P, Malima R, et al. : Distribution and spread of pyrethroid and DDT resistance among the Anopheles gambiae complex in Tanzania. Med Vet Entomol. 2014;28(3):244–252. 10.1111/mve.12036 [DOI] [PMC free article] [PubMed] [Google Scholar]


