Version Changes
Revised. Amendments from Version 1
The reviewers' comments, most of them minor points, were considered in the revised text. Major changes were made by adding a chapter about nuclear transport and assembly of ribosomes, a process with parallels to the proteasome system. Proteasome and ribosome dynamics in yeast are well explored, though the mechanism of nuclear export of proteasomes is not yet understood. In mammalian cells proteasome localizations are less consistent. Thus, we focused on the yeast system. We also addressed the sequestration of proteasomes in motile cytosolic storage granules, enigmatic structures which emerge during quiescence or upon cell cycle arrest. All figures were renewed to clarify that two models exist for nuclear import of proteasomes. In proliferating yeast proteasomal precursor and subcomplexes are mainly imported into the nucleus by the canonical NLS receptor importin / karyopherin alpha / beta (Figure 1). Upon exit from quiescence proteasome storage granules clear and mature proteasomes are imported either as Blm10-associated core particles or as holo-enzymes (Figure 2). Both models may coexist. Technical approaches using yeast genetics to decipher the nuclear import of yeast proteasomes were not discussed in detail. For that purpose, we would like refer to a related review by Burcoglu, Zhao and Enenkel about Nuclear Import of Yeast Proteasomes in Cells 2015.
Abstract
The ubiquitin-proteasome system is the major degradation pathway for short-lived proteins in eukaryotic cells. Targets of the ubiquitin-proteasome-system are proteins regulating a broad range of cellular processes including cell cycle progression, gene expression, the quality control of proteostasis and the response to geno- and proteotoxic stress. Prior to degradation, the proteasomal substrate is marked with a poly-ubiquitin chain. The key protease of the ubiquitin system is the proteasome. In dividing cells, proteasomes exist as holo-enzymes composed of regulatory and core particles. The regulatory complex confers ubiquitin-recognition and ATP dependence on proteasomal protein degradation. The catalytic sites are located in the proteasome core particle. Proteasome holo-enzymes are predominantly nuclear suggesting a major requirement for proteasomal proteolysis in the nucleus. In cell cycle arrested mammalian or quiescent yeast cells, proteasomes deplete from the nucleus and accumulate in granules at the nuclear envelope (NE) / endoplasmic reticulum ( ER) membranes. In prolonged quiescence, proteasome granules drop off the nuclear envelopeNE / ER membranes and migrate as droplet-like entitiesstable organelles throughout the cytoplasm, as thoroughly investigated in yeast. When quiescence yeast cells are allowed to resume growth, proteasome granules clear and proteasomes are rapidly imported into the nucleus.
Here, we summarize our knowledge about the enigmatic structure of proteasome storage granules and the trafficking of proteasomes and their substrates between the cyto- and nucleoplasm.
Most of our current knowledge is based on studies in yeast. Their translation to mammalian cells promises to provide keen insight into protein degradation in non-dividing cells, which comprise the majority of our body’s cells.
Keywords: Proteasome, Storage Granules; Dynamics; Nuclear Transport; Ubiquitin System; Blm10; Importin; Karyopherin; Quiescence
Introduction
Proteolysis determines the half-life of proteins and thus controls protein homeostasis. If protein homeostasis is disrupted, the incidence of protein misfolding and neurodegenerative diseases such as Huntington’s, Parkinson’s and Alzheimer’s increases ( Ciechanover & Brundin, 2003).
In eukaryotic cells two highly conserved degradation pathways exist: under starvation long-lived proteins are preferentially degraded within the lysosome, an organelle with membranes which protect the surrounding cytoplasm against lysosomal hydrolases ( Fuertes et al., 2003; Lee & Goldberg, 1996; Rendueles & Wolf, 1988); short-lived proteins are rather degraded by proteasomes, multimeric protease complexes which move between the nucleo- and cytoplasm ( Hershko & Ciechanover, 1998; Rock et al., 1994). Proteasomal substrates are often nuclear proteins such as proteins regulating cell cycle progression (cyclin-dependant kinases and their inhibitors), gene expression (transcriptions factors), DNA damage and stress response; although, misfolded proteins occurring during protein synthesis in the cytoplasm are also rapidly degraded by the proteasome ( Kirschner, 1999; Vabulas & Hartl, 2005; von Mikecz, 2006). As a result proteasomal proteolysis serves to eliminate obsolete proteins which compete with functional proteins for binding partners and are prone to associate with irreversible and toxic protein aggregates ( Goldberg, 2003).
Here, we want to address the dynamics of proteasomes, which select their substrates by specific determinants such as poly-ubiquitylation, a covalently linked chain of ubiquitin molecules ( Finley, 2009). This ubiquitin-dependent proteolysis undertakes up to 90% of protein degradation in growing yeast and cultured mammalian cells and consumes considerable amounts of ATP, since the activation and conjugation of ubiquitin to the protein substrate as well as the unfolding and translocation of the protein substrate into the proteasome is ATP-dependent ( Coux et al., 1996). Natively-disordered proteins also qualify as proteasome substrates and are cleaved without post-translational ubiquitin modification ( Erales & Coffino, 2014; Fishbain et al., 2015; Liu et al., 2003).
The advent of live cell imaging and GFP-labelling technologies in the 1990s ( Tsien, 1998) have greatly facilitated the study of proteasome dynamics in yeast and mammalian cells. Through these non-invasive techniques, the localization of the proteasome in growing yeast and highly proliferating cancer cells has been elucidated to be primarily nuclear ( Enenkel et al., 1998; Laporte et al., 2008; McDonald & Byers, 1997; Russell et al., 1999). In line with this finding, increasing evidence in the literature suggests that certain misfolded proteins are imported from the cytoplasm into the nucleus solely for proteasomal degradation ( Park et al., 2013; Prasad et al., 2010). Conversely, transient nuclear proteins are exported into the cytoplasm for proteolysis, indicating a dynamic movement of proteasomal substrates between the nucleus and cytoplasm ( Chen & Madura, 2014a). Under nutrient deprivation and during transition from proliferation to quiescence, yeast proteasomes gather in proteasome storage granules (PSGs) at the nuclear envelope (NE)/endoplasmic reticulum (ER) membrane ( Enenkel, 2014; Knecht & Rivett, 2000; Wojcik & DeMartino, 2003). With prolonged quiescence PSGs seem to pinch off the NE/ER, but are not associated with specific organelles or any detectable membrane and are defined as motile spherical structures in the cytoplasm ( Laporte et al., 2008). When cells resume growth, PSGs dissipate and proteasomes are rapidly imported into the nucleus to contribute their function in cell proliferation ( Laporte et al., 2008). The mechanism of PSG formation and clearance is still unknown but seems to be conserved, since PSG-like structures are observed in primary cell lines of non-dividing neuronal cells and in immortalized cell lines of cancer cells, if they are chemically arrested in cell cycle progression ( Bingol & Schuman, 2006; Kaganovich et al., 2008).
Our knowledge about proteasome dynamics in mammalian cells is poor. Thus, the focus of this review will be to critically integrate the literature about the dynamics of the proteasome, particularly based on studies in yeast. In our overview of the ubiquitin-proteasome system and common principles of nuclear transport, we cite and refer to original work and review articles written by investigators who did seminal work on these topics. In the paragraphs addressing detailed knowledge about proteasome dynamics we cite the original work.
Discussion/analysis of the literature
The Ubiquitin System
Ubiquitylation is a post-translational modification commonly associated with proteasomal protein degradation. At least four ubiquitin molecules are required for a poly-ubiquitin chain to be recognized by the proteasome ( Thrower et al., 2000). Hershko and colleagues in the early 1980s showed that poly-ubiquitylation requires the ATP-dependent ubiquitin activation enzyme (E1), a family of ubiquitin conjugating enzymes (E2) and a family of ubiquitin protein ligases (E3) ( Hershko & Ciechanover, 1998). First, ATP hydrolysis is required to activate the AMP linkage to the C-terminal glycine of ubiquitin which enables the transfer of the ubiquitin moiety to the active site cysteine of the E1. Second, the E1-bound ubiquitin is linked to the active site cysteine residue of an E2 by transesterification. Finally, the E3 transfers the ubiquitin onto the substrate depending on the class of the E3 enzyme (RING, HECT and U-box ligases) ( Finley et al., 2012; Harper & Schulman, 2006). Elongation of the ubiquitin chain is achieved as succeeding ubiquitin molecules form isopeptide linkages with specific lysines of the preceding ubiquitin ( Hershko & Ciechanover, 1998). Prior to degradation, deubiquitinating activities within the proteasome cleave and recycle the ubiquitin molecules from the substrates ( Crosas et al., 2006; Hanna et al., 2006; Lam et al., 1997; Verma et al., 2002). Deubiquitinating enzymes in the cyto- and nucleoplasm provide an additional level on the plasticity on the repertoire of proteasomal substrates ( Sahtoe & Sixma, 2015). Intriguingly, GFP-labelled ubiquitin and the E1, named Uba1, is primarily nuclear in growing yeast and mammalian cells suggesting that ubiquitin-dependent proteolysis mainly occurs in the nucleus ( Huh et al., 2003; Salomons et al., 2010; Sugaya et al., 2014; Sugaya et al., 2015).
Proteasome assembly and composition
Composed of over 40 subunits, the proteasome is a protein complex of 2.5 MDa which consists of two main components: the 20S core particle (CP) and the 19S regulatory particle (RP) ( Coux et al., 1996).
Proteasome configurations centered on the CP can have either one or two RPs but also one or two alternative proteasome activating complexes giving rise to a variety of proteasome complex configurations. Proteasome holo-enzymes engaged in the degradation of poly-ubiquitylated proteins require the RP, thus occur either as RP-CP or RP-CP-RP, also termed the 26S and the 30S proteasome, respectively ( Eytan et al., 1989).
Structure of the 20S Core Particle
The proteasome belongs to the family of threonine proteases and its maturation follows the concept of zymogen activation upon which proteases are activated, once they arrive at their destination. With a molecular mass of 700 kDa, the CP is composed of seven distinct α and β subunits, each of which form heptameric rings stacked into a barrel composed of two outer α rings and two inner β rings ( Groll et al., 1997). The maturation of the CP involves the dimerization of two inactive precursor complexes, resembling two half-CPs. Half-CPs consist of an α ring and β ring with five of the seven β subunits synthesized with propeptides. With the dimerization of two half-CPs into the pre-holo CP, the autocatalytic processing of the propeptides is triggered and three β subunits contribute an active site threonine with different peptide cleavage specificities ( Li et al., 2007; Ramos et al., 1998). CP-dedicated chaperones, namely Pac/Pba/Poc 1-4 and Ump1, assist in CP assembly. Ump1 is a natively-disordered protein ( Kusmierczyk et al., 2008; Ramos & Dohmen, 2008), which is buried inside the pre-holo CP and later on becoming the first substrate of the nascent CP ( Sa-Moura et al., 2013; Uekusa et al., 2014). The α rings are the key players in CP gating. Normally CP α rings are closed, unless they are opened by the RP to allow access of protein substrates into the proteolytic cavity ( Groll et al., 2000).
Structure of the 19S Regulatory Particle
As “gate keeper” of the CP, the RP is the best understood proteasome activator ( Rechsteiner & Hill, 2005). The RP is divided into two parts, the base and the lid subcomplexes. The RP base is composed of six ATPases of the triple A family (ATPases Associated with diverse cellular Activities), named Rpt1-6, and five non-ATPases, Rpn1, Rpn2, Rpn10, Rpn13 and Ubp6. The base Rpn subunits are involved in the recognition of the poly-ubiquitin chain and the Rpt ATPase subunits guide the unfolding and translocation of the polypeptide substrate into the CP ( Finley et al., 1998). In contrast to the RP base subunits, the subunits comprising the RP lid are only of the non-ATPase class: Rpn3, Rpn5-9, Rpn11 and Rpn12 ( Glickman et al., 1998). The main known function of the RP lid is the processing of poly-ubiquitin chains. Rpn11 contributes isopeptidase activity to recycle ubiquitin moieties from the protein substrates. Ubp6 also has ubiquitin hydrolase activity and assists in trimming poly-ubiquitin chains ( Crosas et al., 2006; Hanna et al., 2006; Lam et al., 1997; Verma et al., 2002). In principle, the RP ensures that only targeted substrates are degraded by the proteasome, thereby conferring the ubiquitin- and ATP-dependence towards proteasomal protein degradation.
Two competing models exist for RP assembly ( Funakoshi et al., 2009; Le Tallec et al., 2009; Park et al., 2009; Roelofs et al., 2009). The first posits that RP assembly occurs in modules independent of the CP with the help of four RP-dedicated chaperones, named Hsm3, Nas2, Nas6 and Rpn14 ( Funakoshi et al., 2009). In contrast, the second model proposes that the CP serves as a scaffold for the heterohexameric ATPase ring of the RP base ( Park et al., 2009). The second model, however, appears less likely with regard to X-ray structure analysis showing that the RP-dedicated chaperones hinder the association between the RP base and CP α ring ( Barrault et al., 2012). The CP-independent assembly model is also supported by the finding that the assembly of RP base and lid can be reconstituted from recombinant proteins with the assistance of RP-dedicated chaperones but without the CP template ( Beckwith et al., 2013). However, the CP could serve as a platform for RP base assembly, if RP-dedicated chaperones are limiting.
Localization of the proteasome
At this point, it is important to acknowledge the importance of GFP labelling and the ease with which it has allowed localization studies to be conducted ( Enenkel, 2014; Groothuis & Reits, 2005). In our species of interest, Saccharomyces cerevisiae, which is an excellent model organism for eukaryotic cells, GFP labelling of proteasomes is achieved by homologous recombination techniques into the chromosomal locus to convert an endogenous proteasomal subunit to a GFP-tagged version ( Enenkel et al., 1999; Laporte et al., 2008; McDonald & Byers, 1997). Nearly all proteasomal genes are essential and could be modified by GFP fusions without interfering with their function; we prefer the CP subunits α4 and β5, the CP-dedicated chaperone Ump1, and the RP subunits Rpn1, Rpt1 and Rpn11 as GFP-labelled reporters, because their GFP fusion proteins are fully incorporated into the proteasomal subcomplexes. So far, ~30 subunits of the yeast proteasome were labelled with GFP. All of them reveal the same subcellular localization as thoroughly investigated by direct fluorescence microscopy in living yeast ( Laporte et al., 2008). The localization studies based on GFP labelling agree well with previous studies using indirect immunofluorescence microscopy of endogenous proteasomes in fixed yeast cells ( Enenkel et al., 1998; McDonald & Byers, 1997; Russell et al., 1999; Wilkinson et al., 1998), whereas direct and indirect localisations of proteasomes in higher eukaryotes are less consistent.
Seminal studies on proteasome localization in vertebrate cells were performed by Werner Franke’s and Wolfgang Baumeister’s laboratories in the early 1990s. Proteasomes were mainly detected in the nuclei of Xenopus laevis oocytes and cultured mammalian cells ( Amsterdam et al., 1993; Hugle et al., 1983; Kleinschmidt et al., 1983). Later investigations reported a shift towards cytoplasmic proteasomes dependent on the type of the cell line and the density of the cell culture ( Wojcik & DeMartino, 2003). Proteasome localization also varies with the growth phase in yeast ( Laporte et al., 2008; Weberruss et al., 2013). In growing yeast at logarithmic phase (OD~1), proteasomes are primarily nuclear. During the transition from proliferation to quiescence and the entrance into stationary phase (OD>3), proteasomes deplete from the nucleus and accumulate at the NE/ER in membraneless droplet-like structures. These enigmatic structures of proteasome accumulations were initially observed by Isabelle Sagot and her co-workers, who coined the term proteasome storage granules (PSGs) ( Laporte et al., 2008). With prolonged quiescence, one to two PSGs with a diameter of ~ 0.2 to 0.5 µm seem to pinch off the NE into the cytoplasm. The PSGs are motile and stable in yeast cultures and are kept in quiescence for several weeks. If quiescent yeast cells are allowed to resume growth by replacing the glucose-depleted medium with glucose-rich medium, the PSG rapidly clears and the proteasomes are relocated into the nucleus within a few minutes.
Studies with mammalian cancer cell lines also exploited GFP-labelling techniques and fluorescence recovery after photobleaching experiments. The experiments suggested that nuclear transport of GFP-labelled CP across the NE was inefficient. Only the mitotic breakdown of the NE and its reassembly after mitosis allowed nuclear uptake of proteasomes ( Reits et al., 1997). However, this nuclear uptake mechanism cannot explain the predominant nuclear localization of proteasomes in yeast cells which divide without mitotic breakdown of the NE. Proteasomes are the second most abundant protein complexes in eukaryotic cells and require continuous synthesis within the cytoplasm and nuclear import during cell division ( Weberruss et al., 2013). The most common route for protein complexes to cross the NE in an organism with closed mitosis is through nuclear pore complexes (NPC). Before we address this pathway for yeast proteasomes, we will shortly summarize the concept of nuclear transport through the NPC, a pathway conserved from yeast to human.
Nuclear import in proliferating yeast cells
The NE is embellished with NPCs which regulate the entry of molecules into and out of the nucleus. Their principal function is to allow free diffusion of small molecules, such as water/ions/peptides, and to block non-specific translocation of macromolecules that exceed 40kDa or a diameter larger than 5nm ( Aitchison & Rout, 2012; Wente & Rout, 2010). Translocation of larger macromolecules requires specific interactions with the NPC. Protein cargoes therefore associate with soluble transport factors, called karyopherins/importins/exportins, that themselves interact with phenylalanine-glycine rich nucleoporins (FG-Nups) decorating the NPC ( Wozniak et al., 1998). Importins and exportins identify their protein cargoes by nuclear localization sequences (NLSs) and nuclear export signals (NESs), which ensure their nuclear import and export, respectively. In the literature, there are variations of nuclear import and export signals, only some of which comply with the classical import/export concept. The classical concept applies for nuclear import of proteasomes. Thus, we will focus on the key components required for the classical pathway ( Gorlich & Kutay, 1999).
The classical nuclear import cycle starts with the association of the importin/karyopherin αβ heretodimer, called Srp1/Kap95 in yeast, with the cargo NLS. Two types of classical NLSs exist: the monopartite NLS which contains five basic amino acid residues and the bipartite NLS in which two clusters of basic residues are spaced by 10–12 indifferent residues. Importin α has the NLS-binding grooves, and importin β mediates the interaction with FG-Nups. The directionality of nuclear transport is dictated by the Ran-GTP/GDP gradient across the NE. Ran is a small GTPase, named Gsp1 in yeast. Ran exists in its GTP-bound state in the nucleus and in its GDP-bound state in the cytoplasm due to the actions of the Ran guanine nucleotide exchange factor (RanGEF) and the RanGTPase activating protein (RanGAP) in the nucleo- and cytoplasm, respectively ( Gorlich & Kutay, 1999; Moore & Blobel, 1993). In the nucleus, the cargo-importin αβ complex encounters RanGTP, which results in the release of the cargo ( Rexach & Blobel, 1995). Cargo-free importin αβ is recycled into the cytoplasm for the next round of nuclear import.
Nuclear import of proteasomes during cell proliferation
Our studies in yeast strongly suggest that newly synthesized proteasomes are imported from the cytosol into the nucleus as inactive precursor complexes and that the maturation of nuclear CP proceeds to completion post-import ( Lehmann et al., 2002). Although electron microscopy studies have shown that the NPC could expand to accommodate the longitudinal passage of the 30S proteasome, the permeability barriers towards macromolecules such as CP precursor complexes and RP assembly modules must be overcome by specific importins/karyopherins ( Pante & Kann, 2002). Several classical NLSs exist within the N-termini of distinct α subunits which were proposed to be either accessible rendering the CP in an import-competent conformation, or to be masked rendering the CP in an import-incompatible conformation ( Tanaka et al., 1990). Indeed, recent EM structure analysis revealed flexible and less structured α ring surfaces in Ump1-associated CP precursor complexes ( Kock et al., 2015; Wani et al., 2015), consistent with our finding that importin α recognizes CP precursor complexes but not mature CP with closed α rings ( Lehmann et al., 2002). Our model upon which CP precursor complexes are imported into the nucleus was supported by the following observations ( Figure 1A). First, when tagged with GFP, Ump1 is predominantly nuclear in spite of the fact that CP precursor complexes are assembled from nascent subunits in the cytoplasm. Second, in importin α mutants namely srp1-49 but not in srp1-31, several groups found that the CP is mislocalized to the cytoplasm, providing another piece of evidence for the classical import pathway of proteasomes ( Chen & Madura, 2014b; Chen et al., 2011; Lehmann et al., 2002; Pack et al., 2014; Wendler et al., 2004). Unprocessed and incompletely processed β5 subunits, crucial determinants of CP precursor complexes and pre-holo-CP, respectively, accumulate in srp1-49 mutants, while precursors of β5 subunits are hardly detectable in wild type cells ( Lehmann et al., 2002). Third, when CP maturation is delayed by UMP1 deletion, CP reporter proteins accumulate in the nucleus. Half of the reporter proteins is incorporated into incompletely matured CP, most likely the pre-holo-CP ( Fehlker et al., 2003; Lehmann et al., 2008). If mature CP were imported into the nucleus, CP precursor complexes would have accumulated in the cytoplasm of ump1Δ cells.
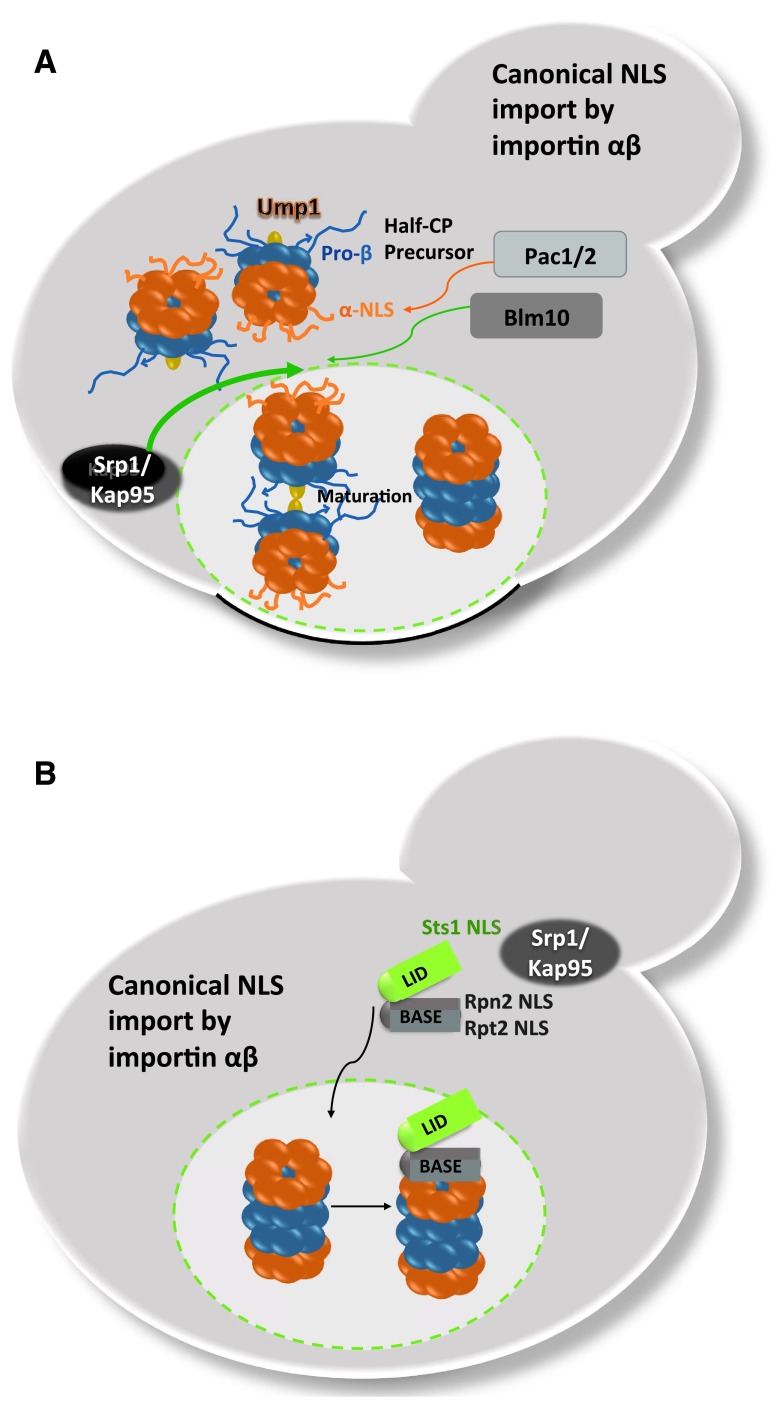
Figure 1. Model of nuclear proteasome assembly based on nuclear import of CP precursor complexes and RP subcomplexes in proliferating yeast cells.
( A) Ump1-containing CP precursor complexes are mainly imported into the nucleus by Srp1/Kap95, the classical importin/karyopherin αβ pathway. The α rings with the classical NLS are depicted in red. The β rings with propeptides are depicted in blue. The CP-dedicated chaperone and maturation factor Ump1 is depicted in yellow. The completion of CP maturation occurs in the nucleus with the degradation of Ump1. CP-dedicated chaperones Pac 1/2 are masking the NLS within the α ring, possibly preventing premature nuclear import. Blm10 serves as an alternative import receptor. ( B) Nuclear import of RP base and lid subcomplexes by the classical importin/karyopherin αβ pathway. Rpn2/Rpt2 and Sts1 confer classical NLS to the RP base and lid complex, respectively. Sts1 is short-lived and most likely degraded with nuclear RP-CP assembly.
However, the CP-dedicated chaperones Pac/Pba/Poc 1-4 binding to the α ring are cytosolic ( Huh et al., 2003). Particularly, Pac/Pba/Poc 1/2 seem to prevent premature nuclear import of CP precursor complexes by blocking the access to the NLSs within α subunits ( Kock et al., 2015; Stadtmueller et al., 2012), possibly allowing cytosolic CP maturation. Again, the deletion of Ump1 results in a predominant nuclear localization of Pac/Pba/Poc 1-4 supporting the model of nuclear import of CP precursor complexes (unpublished results, ( Le Tallec et al., 2007)). Here, it is interesting to mention recent localization studies monitoring GFP-labelled β7 subunits in mammalian HeLa cells. This reporter subunit of the CP was found to be exclusively cytoplasmic but became nuclear upon DNA damage ( Kulichkova et al., 2015). Possibly, the deletion of UMP1 in yeast is comparable with DNA damage in human cancer cells and requests an abundance of nuclear proteasomes.
In the case of the RP, functional NLSs were identified in RP base subunits Rpn2 and Rpt2 and are recognized by importin α ( Figure 1B). The deletion of the Rpn2 NLS caused a temperature sensitive phenotype and mislocalizations of the RP base into cytosolic foci, whereas the deletion of the Rpt2 NLS was compensated by the presence of the Rpn2 NLS. At permissive temperatures, neither the Rpn2 nor the Rpt2 NLS deletion had severe impact on nuclear proteasome localization suggesting a redundancy of proteasomal NLSs ( Wendler et al., 2004). Isono et al. (2007) later confirmed that Rpn2 provides a crucial NLS to aid nuclear import of the RP base and that the lid is separately imported. The nuclear import of the RP lid also requires importin α, though no classical NLS has been identified within RP lid subunits; rather Sts1, a short-lived protein that itself contains a classical NLS, associates with Rpn11 to facilitate nuclear import of the RP lid by importin αβ ( Chen et al., 2011). In accordance, deletion of the Sts1 NLS has downstream effects on the nuclear localization of RP lid in addition to RP base and CP, which suggests that proteasomes could also be transported as holo-enzymes ( Chen & Madura, 2014b). In order to ensure comparable stoichiometry of proteasomal subcomplexes in the nucleus and similar kinetics by which they are imported into the nucleus, it is reasonable that importin αβ is used as common nuclear import receptor.
Recent fluorescence correlation spectroscopy studies also support the conclusion that proteasomes can be imported into the nucleus as holo-enzymes ( Pack et al., 2014). However, the maturation state of the GFP-labelled proteasomes was unclear. Possibly, pre-holo-CP are the real nuclear transport intermediates which degrade Ump1 and Sts1 upon the arrival in the nucleus with the completion of proteasome maturation.
Parallels between nuclear transport of proteasomes and ribosomes
Ribosome 40S and 60S subunits are the most abundant protein complexes in eukaryotic cells and are composed of more than 70 ribosomal subunits and four different ribosomal RNAs ( Marguerat et al., 2012). Their assembly begins in the nucleolus and requires about 300 evolutionarily conserved nonribosomal trans-acting factors, which transiently associate with pre-ribosomal subunits at distinct assembly stages. Transport factors are required to import ribosomal proteins into the nucleus for pre-ribosomal subunit assembly and to passage pre-ribosomal subunits in a functionally inactive state through the NPC into the cytoplasm, where they undergo final maturation before initiating translation (for references see ( Gerhardy et al., 2014)). Different GFP-tagged ribosomal protein and a pre-RNA reporter are established that reliably monitor the movement of pre-ribosomal particles from the nucleus into the cytoplasm in yeast ( Altvater et al., 2014; Milkereit et al., 2001; Tschochner & Hurt, 2003). Nuclear import of ribosomal proteins is mediated by importins belonging to the karyopherin β family ( Rout et al., 1997). Export competent pre-ribosomal particles are separately exported by the general nuclear export factor Xpo1/Crm1. In addition, multiple trans-acting factors are engaged to shield the highly negative charge of the ribosomal RNA for entry into the disordered FG-Nups of the NPC. Most of the trans-acting factors are released and reused for another round of ribosome assembly. Failures in recycling a factor back into the nucleolus leads to its depletion resulting in delayed pre-ribosomal RNA processing, assembly defects and impaired nuclear export (for references on the original work see ( Gerhardy et al., 2014)).
Certainly, proteasome and ribosome assembly differ as mature proteasomes do not contain RNA. However, parallels exist with regard to the tight coupling between assembly and transport of inactive precursor complexes.
The Enigma of Proteasome Storage Granules
When cells experience nutrient exhaustion or enter quiescence, a drastic change in proteasome localization is observed. In prolonged quiescence, proteasomes deplete from the nucleus and reside in motile and reversible PSGs in the cytoplasm ( Laporte et al., 2008). Upon addition of glucose, cells receive the signal to resume proliferation, and PSGs dissolve rapidly, and proteasomes are relocated in the nucleus. How PSGs are organized is not understood. Premature PSG formation in proliferating cells was found to depend on vacuolar ATPases and linked premature PSG formation with disregulation of the intracellular pH. In view of that, PSGs could serve as storage depots for mature proteasomes in quiescence, to protect the proteasome from cellular stress and elimination by autophagocytosis ( Peters et al., 2013). The storage of proteasomes during quiescence would also alleviate energy-consuming synthesis of new proteasomes with cell proliferation ( Laporte et al., 2008).
The formation of PSG-like structures is also observed by chemical inhibition of proteasomes in mammalian cells or temperature sensitive proteasome mutants in yeast, conditions which result in cell cycle arrest. In spite of the differences between chemically-induced cell cycle arrest and quiescence, inhibited proteasomes are sequestered into juxta nuclear quality control compartments (JUNQs), situated at the cytoplasmic side of the NE and behaving similar to PSGs. When the cell cycle-arrested mutants were allowed to resume growth at permissive temperatures or upon withdrawal of proteasome inhibition, JUNQs were seen to dissolve like the PSG. In the context of these studies poly-ubiquitylated proteins were found to be accumulated in the JUNQ. Thus, it was proposed that the JUNQ represents a major site for ubiquitin-dependent proteolysis ( Kaganovich et al., 2008), though it has to be taken into account that JUNQ formation was induced by proteasome inhibition. The poly-ubiquitylated reporter proteins used in the studies on JUNQ functions by Kaganovich et al. (2008) were also detected within the PSG suggesting that JUNQ and PSG describe the same structure ( Weberruss et al., 2013). All studies on the JUNQ and PSGs agree that these enigmatic structures serve protective functions. Their presence protects cells against proteo- and genotoxic stress and confers cell fitness during aging. Post-translation modifications such as N-acetylation also play a role in PSG organization, but their targets are unknown ( Saunier et al., 2013; van Deventer et al., 2015; Weberruss et al., 2013).
Nuclear import of proteasomes upon exit from quiescence
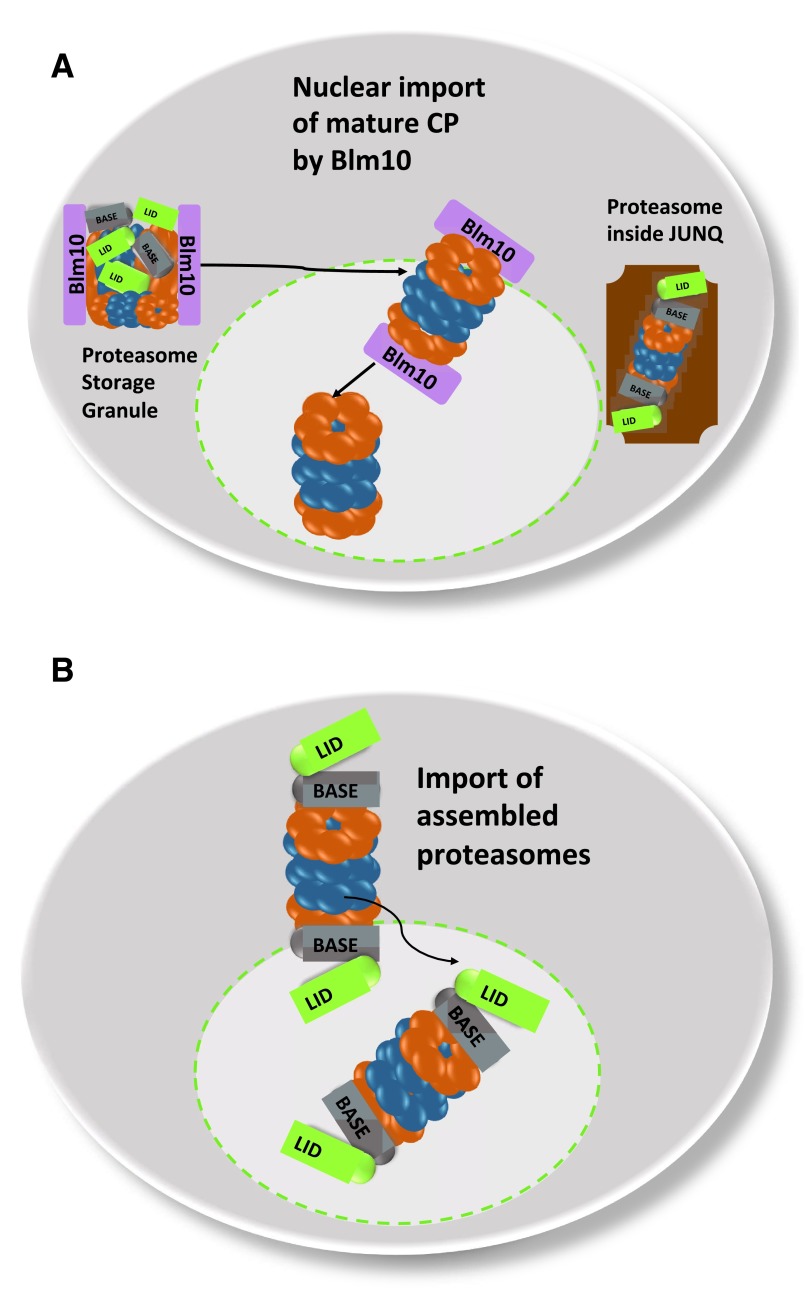
Though the CP and RP co-localize in the PSG, they seem to be loosely associated. Conflicting reports exist about the stability of RP-CP assemblies in lysates of quiescent cells ( Bajorek et al., 2003; Hanna et al., 2012; Weberruss et al., 2013). The finding that RP-CP assemblies are less stable coincides with the decline in ATP during quiescence as well as the reduced proclivity of the proteasome to degrade poly-ubiquitylated substrates. Instead of an association of the CP with the RP, most CP is seen interacting with Blm10, a conserved 240 kDa HEAT repeat protein ( Weberruss et al., 2013). Upon exit from quiescence, the PSGs rapidly clear and mature proteasomes are imported into the nucleus within a few minutes. The imported proteasomes must be matured and assembled, as time does not permit the new synthesis of precursor complexes ( Laporte et al., 2008). Here, Blm10 plays an important role and represents the first characterized nuclear transporter which particularly facilitates nuclear import of mature CP ( Figure 2A). Quiescent blm10Δ mutants exhibit a significant delay in resuming cell growth due to the deficit in mature CP in the nucleus. Furthermore, Blm10 binds FG-Nups and GTP-bound Ran and dissociates from the CP upon interaction with RanGTP, suggesting that Blm10 shares functional similarities with Kap95, the classical importin β ( Weberruss et al., 2013). Along this line, Blm10 belongs to the HEAT repeat family with α-solenoid fold, a structural feature shared by β karyopherins/importins ( Huber & Groll, 2012). During cell proliferation, Blm10 is also expressed but to a much lesser extent ( Weberruss et al., 2013). Only a minor fraction of the CP, pre-holo-CP and CP precursor complexes is associated with Blm10 in growing yeast. The Blm10-bound fraction significantly increases under geno-and proteotoxic stress suggesting a high demand for nuclear proteasomes under these growth conditions ( Doherty et al., 2012; Fehlker et al., 2003; Lehmann et al., 2008). Since Blm10 associates with constitutively open or disordered CP α rings, Blm10 also plays a role in regulating α-ring gating during CP maturation ( Lehmann et al., 2008). The wider α ring conformation of CP-precursor complexes seems to be preferentially bound to Blm10 and importin αβ by representing import intermediates. Thus, the Blm10-dependent import pathway complements the canonical nuclear import pathway, which also allows nuclear import of assembled proteasomes ( Chen et al., 2011; Pack et al., 2014), especially upon the exit from quiescence ( Figure 2B).
Figure 2. Model of nuclear import of mature proteasomes upon the exit from quiescence.
( A) In quiescence mature proteasomes are stored in PSG, reversible and motile granules in the cytoplasm. The PSG is formed at the NE/ER with the transition from proliferation to quiescence. The PSG clears with the resumption of growth and mature CP is imported into the nucleus by Blm10. In cell cycle arrested cells as induced by proteasome inhibition, proteasomes reside within JUNQ in the nuclear periphery. JUNQ rapidly clear with the release of proteasome inhibition. ( B) Assembled holo-proteasomes with RP-CP-RP configuration pass the nuclear pore.
For the RP, the import pathway upon exit from quiescence is yet to be solidified. A possible candidate for a RP-dedicated nuclear import receptor is Rpn2 which exhibits a similar α-solenoid fold as Blm10 and importin β, all of which belong to the family of HEAT-repeat proteins ( Huber & Groll, 2012; Kajava, 2002).
Conclusions
In this review, we discussed the recent literature on the dynamics of the ubiquitin-proteasome system with a major focus on the proteasome. During cell proliferation a high traffic volume of proteasomes and proteasomal substrates arises between the cyto- and nucleoplasm. In cell-cycle arrested and quiescent cells, proteasomes exit the nucleus and accumulate with poly-ubiquitylated proteins in motile and reversible PSGs in the nuclear periphery. While the basic concepts of nuclear import of proteasomes during cell proliferation and upon exit from quiescence are well explored, little is known about the nuclear export of proteasomes during the transition from proliferation to quiescence. We may wonder why proteasomes exit the nucleus during quiescence. Which kind of substrates will be available in the cytoplasm, once proteasomes are sequestered into the PSG? Possibly, PSG-resident proteasomes are starving for newly synthesized proteins which arise with the resumption of cell proliferation.
The dynamics of proteasomes and their substrates are fascinating and will inspire our discussions and experiments in the future.
Acknowledgement
We thank Zhu Chao (Jerry) Gu and Julianne Burcoglu for critical reading the manuscript.
Funding Statement
This work was supported by grants from NSERC (4422666-2011) awarded to M.C. and C.E. and from CIHR (325477) awarded to C.E.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 3 approved]
References
- Aitchison JD, Rout MP: The yeast nuclear pore complex and transport through it. Genetics. 2012;190(3):855–883. 10.1534/genetics.111.127803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altvater M, Schütz S, Chang Y, et al. : Dissecting ribosome assembly and transport in budding yeast. Methods Cell Biol. 2014;122:437–461. 10.1016/B978-0-12-417160-2.00020-5 [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Pitzer F, Baumeister W: Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc Natl Acad Sci U S A. 1993;90(1):99–103. 10.1073/pnas.90.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Finley D, Glickman MH: Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol. 2003;13(13):1140–1144. 10.1016/S0960-9822(03)00417-2 [DOI] [PubMed] [Google Scholar]
- Barrault MB, Richet N, Godard C, et al. : Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proc Natl Acad Sci U S A. 2012;109(17):E1001–1010. 10.1073/pnas.1116538109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith R, Estrin E, Worden EJ, et al. : Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol. 2013;20(10):1164–1172. 10.1038/nsmb.2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Schuman EM: Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441(7097):1144–1148. 10.1038/nature04769 [DOI] [PubMed] [Google Scholar]
- Chen L, Madura K: Degradation of specific nuclear proteins occurs in the cytoplasm in Saccharomyces cerevisiae. Genetics. 2014a;197(1):193–197. 10.1534/genetics.114.163824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Madura K: Yeast importin-α (Srp1) performs distinct roles in the import of nuclear proteins and in targeting proteasomes to the nucleus. J Biol Chem. 2014b;289(46):32339–32352. 10.1074/jbc.M114.582023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Romero L, Chuang SM, et al. : Sts1 plays a key role in targeting proteasomes to the nucleus. J Biol Chem. 2011;286(4):3104–3118. 10.1074/jbc.M110.135863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P: The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40(2):427–446. 10.1016/S0896-6273(03)00606-8 [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL: Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. 10.1146/annurev.bi.65.070196.004101 [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, et al. : Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127(7):1401–1413. 10.1016/j.cell.2006.09.051 [DOI] [PubMed] [Google Scholar]
- Doherty KM, Pride LD, Lukose J, et al. : Loss of a 20S proteasome activator in Saccharomyces cerevisiae downregulates genes important for genomic integrity, increases DNA damage, and selectively sensitizes cells to agents with diverse mechanisms of action. G3 (Bethesda). 2012;2(8):943–959. 10.1534/g3.112.003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C: Proteasome dynamics. Biochim Biophys Acta. 2014;1843(1):39–46. 10.1016/j.bbamcr.2013.03.023 [DOI] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel PM: Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 1998;17(21):6144–6154. 10.1093/emboj/17.21.6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel PM: GFP-labelling of 26S proteasomes in living yeast: insight into proteasomal functions at the nuclear envelope/rough ER. Mol Biol Rep. 1999;26(1–2):131–135. 10.1023/A:1006973803960 [DOI] [PubMed] [Google Scholar]
- Erales J, Coffino P: Ubiquitin-independent proteasomal degradation. Biochim Biophys Acta. 2014;1843(1):216–221. 10.1016/j.bbamcr.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eytan E, Ganoth D, Armon T, et al. : ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc Natl Acad Sci U S A. 1989;86(20):7751–7755. 10.1073/pnas.86.20.7751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlker M, Wendler P, Lehmann A, et al. : Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003;4(10):959–963. 10.1038/sj.embor.embor938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D: Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Tanaka K, Mann C, et al. : Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem Sci. 1998;23(7):244–245. 10.1016/S0968-0004(98)01222-5 [DOI] [PubMed] [Google Scholar]
- Finley D, Ulrich HD, Sommer T, et al. : The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192(2):319–360. 10.1534/genetics.112.140467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbain S, Inobe T, Israeli E, et al. : Sequence composition of disordered regions fine-tunes protein half-life. Nat Struct Mol Biol. 2015;22(3):214–221. 10.1038/nsmb.2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes G, Villarroya A, Knecht E: Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int J Biochem Cell Biol. 2003;35(5):651–664. 10.1016/S1357-2725(02)00382-5 [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Tomko RJ Jr, Kobayashi H, et al. : Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137(5):887–899. 10.1016/j.cell.2009.04.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardy S, Menet AM, Peña C, et al. : Assembly and nuclear export of pre-ribosomal particles in budding yeast. Chromosoma. 2014;123(4):327–344. 10.1007/s00412-014-0463-z [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, et al. : The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18(6):3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL: Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–899. 10.1038/nature02263 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U: Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Köhler A, et al. : A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7(11):1062–1067. 10.1038/80992 [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, et al. : Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386(6624):463–471. 10.1038/386463a0 [DOI] [PubMed] [Google Scholar]
- Groothuis TA, Reits EA: Monitoring the distribution and dynamics of proteasomes in living cells. Methods Enzymol. 2005;399:549–563. 10.1016/S0076-6879(05)99037-X [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, et al. : Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127(1):99–111. 10.1016/j.cell.2006.07.038 [DOI] [PubMed] [Google Scholar]
- Hanna J, Waterman D, Boselli M, et al. : Spg5 protein regulates the proteasome in quiescence. J Biol Chem. 2012;287(41):34400–34409. 10.1074/jbc.M112.390294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Schulman BA: Structural complexity in ubiquitin recognition. Cell. 2006;124(6):1133–1136. 10.1016/j.cell.2006.03.009 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A: The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Huber EM, Groll M: The 19S cap puzzle: a new jigsaw piece. Structure. 2012;20(3):387–388. 10.1016/j.str.2012.02.006 [DOI] [PubMed] [Google Scholar]
- Hügle B, Kleinschmidt JA, Franke WW: The 22 S cylinder particles of Xenopus laevis. II. Immunological characterization and localization of their proteins in tissues and cultured cells. Eur J Cell Biol. 1983;32(1):157–163. [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, et al. : Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Isono E, Nishihara K, Saeki Y, et al. : The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol Biol Cell. 2007;18(2):569–580. 10.1091/mbc.E06-07-0635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J: Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454(7208):1088–1095. 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV: What curves alpha-solenoids? Evidence for an alpha-helical toroid structure of Rpn1 and Rpn2 proteins of the 26 S proteasome. J Biol Chem. 2002;277(51):49791–49798. 10.1074/jbc.M204982200 [DOI] [PubMed] [Google Scholar]
- Kirschner M: Intracellular proteolysis. Trends Cell Biol. 1999;9(12):M42–45. 10.1016/S0962-8924(99)01666-9 [DOI] [PubMed] [Google Scholar]
- Kleinschmidt JA, Hügle B, Grund C, et al. : The 22 S cylinder particles of Xenopus laevis. I. Biochemical and electron microscopic characterization. Eur J Cell Biol. 1983;32(1):143–156. [PubMed] [Google Scholar]
- Knecht E, Rivett J: Intracellular Localization of Proteasomes. In Proteasomes: The World of Regulatory Proteolysis.W Hilt, and DH Wolf, eds. (Georgetown, Texas, USA: Landes Bioscience, Eurekah.com).2000;176–185. Reference Source [Google Scholar]
- Kock M, Nunes MM, Hemann M, et al. : Proteasome assembly from 15S precursors involves major conformational changes and recycling of the Pba1-Pba2 chaperone. Nat Commun. 2015;6:6123. 10.1038/ncomms7123 [DOI] [PubMed] [Google Scholar]
- Kulichkova VA, Artamonova TO, Zaykova JJ, et al. : Simultaneous EGFP and tag labeling of the β7 subunit for live imaging and affinity purification of functional human proteasomes. Mol Biotechnol. 2015;57(1):36–44. 10.1007/s12033-014-9799-0 [DOI] [PubMed] [Google Scholar]
- Kusmierczyk AR, Kunjappu MJ, Funakoshi M, et al. : A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15(3):237–244. 10.1038/nsmb.1389 [DOI] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, et al. : Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385(6618):737–740. 10.1038/385737a0 [DOI] [PubMed] [Google Scholar]
- Laporte D, Salin B, Daignan-Fornier B, et al. : Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol. 2008;181(5):737–745. 10.1083/jcb.200711154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Courbeyrette R, et al. : 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27(4):660–674. 10.1016/j.molcel.2007.06.025 [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Guérois R, et al. : Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol Cell. 2009;33(3):389–399. 10.1016/j.molcel.2009.01.010 [DOI] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL: Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271(44):27280–27284. 10.1074/jbc.271.44.27280 [DOI] [PubMed] [Google Scholar]
- Lehmann A, Janek K, Braun B, et al. : 20 S proteasomes are imported as precursor complexes into the nucleus of yeast. J Mol Biol. 2002;317(3):401–413. 10.1006/jmbi.2002.5443 [DOI] [PubMed] [Google Scholar]
- Lehmann A, Jechow K, Enenkel C: Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Rep. 2008;9(12):1237–1243. 10.1038/embor.2008.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kusmierczyk AR, Wong P, et al. : beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007;26(9):2339–2349. 10.1038/sj.emboj.7601681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Corboy MJ, DeMartino GN, et al. : Endoproteolytic activity of the proteasome. Science. 2003;299(5605):408–411. 10.1126/science.1079293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, et al. : Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151(3):671–683. 10.1016/j.cell.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald HB, Byers B: A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol. 1997;137(3):539–553. 10.1083/jcb.137.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Gadal O, Podtelejnikov A, et al. : Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell. 2001;105(4):499–509. 10.1016/S0092-8674(01)00358-0 [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G: The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365(6447):661–663. 10.1038/365661a0 [DOI] [PubMed] [Google Scholar]
- Pack CG, Yukii H, Toh-e A, et al. : Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat Commun. 2014;5:3396. 10.1038/ncomms4396 [DOI] [PubMed] [Google Scholar]
- Panté N, Kann M: Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13(2):425–434. 10.1091/mbc.01-06-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Roelofs J, Kim W, et al. : Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459(7248):866–870. 10.1038/nature08065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Kukushkin Y, Gupta R, et al. : PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154(1):134–145. 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Peters LZ, Hazan R, Breker M, et al. : Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J Cell Biol. 2013;201(5):663–671. 10.1083/jcb.201211146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Kawaguchi S, Ng DT: A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21(13):2117–2127. 10.1091/mbc.E10-02-0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos PC, Dohmen RJ: PACemakers of proteasome core particle assembly. Structure. 2008;16(9):1296–1304. 10.1016/j.str.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Ramos PC, Hockendorff J, Johnson ES, et al. : Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92(4):489–499. 10.1016/S0092-8674(00)80942-3 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Hill CP: Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15(1):27–33. 10.1016/j.tcb.2004.11.003 [DOI] [PubMed] [Google Scholar]
- Reits EA, Benham AM, Plougastel B, et al. : Dynamics of proteasome distribution in living cells. EMBO J. 1997;16(20):6087–6094. 10.1093/emboj/16.20.6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendueles PS, Wolf DH: Proteinase function in yeast: biochemical and genetic approaches to a central mechanism of post-translational control in the eukaryote cell. FEMS Microbiol Rev. 1988;4(1):17–45. 10.1111/j.1574-6968.1988.tb02706.x-i1 [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G: Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83(5):683–692. 10.1016/0092-8674(95)90181-7 [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, et al. : Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–771. 10.1016/S0092-8674(94)90462-6 [DOI] [PubMed] [Google Scholar]
- Roelofs J, Park S, Haas W, et al. : Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459(7248):861–865. 10.1038/nature08063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD: A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89(5):715–725. 10.1016/S0092-8674(00)80254-8 [DOI] [PubMed] [Google Scholar]
- Russell SJ, Steger KA, Johnston SA: Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J Biol Chem. 1999;274(31):21943–21952. 10.1074/jbc.274.31.21943 [DOI] [PubMed] [Google Scholar]
- Sá-Moura B, Simões AM, Fraga J, et al. : Biochemical and biophysical characterization of recombinant yeast proteasome maturation factor ump1. Comput Struct Biotechnol J. 2013;7:e201304006. 10.5936/csbj.201304006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahtoe DD, Sixma TK: Layers of DUB regulation. Trends Biochem Sci. 2015;40(8):456–467. 10.1016/j.tibs.2015.05.002 [DOI] [PubMed] [Google Scholar]
- Salomons FA, Acs K, Dantuma NP: Illuminating the ubiquitin/proteasome system. Exp Cell Res. 2010;316(8):1289–1295. 10.1016/j.yexcr.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Saunier R, Esposito M, Dassa EP, et al. : Integrity of the Saccharomyces cerevisiae Rpn11 protein is critical for formation of proteasome storage granules (PSG) and survival in stationary phase. PLoS One. 2013;8(8):e70357. 10.1371/journal.pone.0070357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmueller BM, Kish-Trier E, Ferrell K, et al. : Structure of a proteasome Pba1-Pba2 complex: implications for proteasome assembly, activation, and biological function. J Biol Chem. 2012;287(44):37371–37382. 10.1074/jbc.M112.367003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya K, Ishihara Y, Inoue S: Nuclear localization of ubiquitin-activating enzyme Uba1 is characterized in its mammalian temperature-sensitive mutant. Genes Cells. 2015. 10.1111/gtc.12257 [DOI] [PubMed] [Google Scholar]
- Sugaya K, Ishihara Y, Inoue S, et al. : Characterization of ubiquitin-activating enzyme Uba1 in the nucleus by its mammalian temperature-sensitive mutant. PLoS One. 2014;9(5):e96666. 10.1371/journal.pone.0096666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Yoshimura T, Tamura T, et al. : Possible mechanism of nuclear translocation of proteasomes. FEBS Lett. 1990;271(1–2):41–46. 10.1016/0014-5793(90)80367-R [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, et al. : Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19(1):94–102. 10.1093/emboj/19.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E: Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13(5):255–263. 10.1016/S0962-8924(03)00054-0 [DOI] [PubMed] [Google Scholar]
- Tsien RY: The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. 10.1146/annurev.biochem.67.1.509 [DOI] [PubMed] [Google Scholar]
- Uekusa Y, Okawa K, Yagi-Utsumi M, et al. : Backbone 1H, 13C and 15N assignments of yeast Ump1, an intrinsically disordered protein that functions as a proteasome assembly chaperone. Biomol NMR Assign. 2014;8(2):383–386. 10.1007/s12104-013-9523-1 [DOI] [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU: Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310(5756):1960–1963. 10.1126/science.1121925 [DOI] [PubMed] [Google Scholar]
- van Deventer S, Menendez-Benito V, van Leeuwen F, et al. : N-terminal acetylation and replicative age affect proteasome localization and cell fitness during aging. J Cell Sci. 2015;128(1):109–117. 10.1242/jcs.157354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, et al. : Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298(5593):611–615. 10.1126/science.1075898 [DOI] [PubMed] [Google Scholar]
- von Mikecz A: The nuclear ubiquitin-proteasome system. J Cell Sci. 2006;119(Pt 10):1977–1984. 10.1242/jcs.03008 [DOI] [PubMed] [Google Scholar]
- Wani PS, Rowland MA, Ondracek A, et al. : Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association. Nat Commun. 2015;6:6384. 10.1038/ncomms7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberruss MH, Savulescu AF, Jando J, et al. : Blm10 facilitates nuclear import of proteasome core particles. EMBO J. 2013;32(20):2697–2707. 10.1038/emboj.2013.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler P, Lehmann A, Janek K, et al. : The bipartite nuclear localization sequence of Rpn2 is required for nuclear import of proteasomal base complexes via karyopherin alphabeta and proteasome functions. J Biol Chem. 2004;279(36):37751–37762. 10.1074/jbc.M403551200 [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP: The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2(10):a000562. 10.1101/cshperspect.a000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CR, Wallace M, Morphew M, et al. : Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17(22):6465–6476. 10.1093/emboj/17.22.6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C, DeMartino GN: Intracellular localization of proteasomes. Int J Biochem Cell Biol. 2003;35(5):579–589. 10.1016/S1357-2725(02)00380-1 [DOI] [PubMed] [Google Scholar]
- Wozniak RW, Rout MP, Aitchison JD: Karyopherins and kissing cousins. Trends Cell Biol. 1998;8(5):184–188. 10.1016/S0962-8924(98)01248-3 [DOI] [PubMed] [Google Scholar]