Abstract
Heme serves as the prosthetic group for a wide variety of proteins known as hemoproteins, such as hemoglobin, myoglobin and cytochromes. It is involved in various molecular and cellular processes such as gene transcription, translation, cell differentiation and cell proliferation. The biosynthesis levels of heme vary across different tissues and cell types and is altered in diseased conditions such as anemia, neuropathy and cancer. This technique uses [4-14C] 5-aminolevulinic acid ([14C] 5-ALA), one of the early precursors in the heme biosynthesis pathway to measure the levels of heme synthesis in mammalian cells. This assay involves incubation of cells with [14C] 5-ALA followed by extraction of heme and measurement of the radioactivity incorporated into heme. This procedure is accurate and quick. This method measures the relative levels of heme biosynthesis rather than the total heme content. To demonstrate the use of this technique the levels of heme biosynthesis were measured in several mammalian cell lines.
Keywords: Molecular Biology, Issue 101, Heme, heme synthesis level, mammalian cells, [4-14C] 5-aminolevulinic acid ([14C] 5-ALA), cancer
Introduction
Heme, a complex of ferrous iron and protoporphyrin IX is a central molecule for transporting and utilizing oxygen in virtually all living organisms1-3. The unique structure of heme enables it to function as a carrier of diatomic gases and electrons, as well as to perform various other functions1-5. For example, heme binds to oxygen in hemoglobin and myoglobin for the transfer and storage of oxygen6,7. It also works as an electron carrier in the cytochromes during respiration and acts as an electron donor for redox reactions catalyzed by cytochrome P450 enzymes8,9. One of the most significant features of heme is that, it can play regulatory roles in cellular and molecular processes such as gene transcription, protein synthesis and micro-RNA biogenesis4. For example, it affects the transcription of many genes by controlling the activity of mammalian transcriptional repressor Bach1 and the mammalian nuclear receptor Rev-erbα10-15. Heme regulates the activation of heme activator protein (Hap) 1 which plays an important role in the activation of genes involved in respiration and controlling oxidative damage, in response to heme or oxygen16. Heme also regulates gene transcription in neuronal cells via nerve growth factor (NGF) signaling3,17-20. It also regulates protein synthesis in mammalian erythroid cells by modulating the activity of heme-regulated eIF2α kinase (HRI)21-24. Furthermore, heme affects the activity of key signaling proteins such as tyrosine kinase Jak2 and Src, which are essential for proper cell functioning and cell growth4,20,25. It was found that in HeLa cells heme inhibition causes cell cycle arrest and activation of markers associated with senescence and apoptosis26. Both Heme deficiency or increased levels of heme are associated with severe health effects in humans27. Recent molecular and epidemiological studies have shown a positive association of high heme intake and increased risk of diseases, such as type-2 diabetes, coronary heart disease and several cancers including lung cancer, colorectal cancer and pancreatic cancer27,28. Using a matched pair of normal and cancer lung cells authors' lab have found that cancer cells have increased levels of oxygen consumption, heme synthesis and proteins involved in heme uptake and oxygen utilization28. Interestingly, inhibition of heme synthesis decreased oxygen consumption, proliferation, migration and colony formation of cancer cells28. Thus, the fluctuation in the levels of endogenous heme plays an important role in the regulation of molecular and cellular processes3,4,28,29.
In mammals, the biosynthesis of heme occurs in eight steps, involving enzymes located in the mitochondria and the cytosol4 (Figure 1). Heme biosynthesis begins in the matrix of mitochondria with the condensation of glycine and succinyl-coA to form 5-aminolevulinic acid (5-ALA), catalyzed by ALA synthase (ALAS)4,31. This is the rate limiting step in heme biosynthesis in nonerythroid cells. 5-ALA is then exported out to the cytosol where the next four steps occur to form coproporphyrinogen III (CPgenIII), which is then imported back to the mitochondria, where it is converted into protoporphyrin IX (PPIX). Finally, one molecule of iron is incorporated to the protoporphyrin IX (PPIX) to produce heme, a reaction catalyzed by ferrochelatase (FECH)2,4.
The level of heme biosynthesis depends primarily on the level of ALAS enzyme which is tightly controlled by intracellular iron and heme4. The biosynthesis of heme can be affected by genetic defects, availability of certain minerals and vitamins (e.g., riboflavin, zinc), exposure to toxins (e.g., aluminum, lead), anoxia, fever, and levels of certain steroids (e.g., estrogen)32-35. The level of heme synthesis is altered in various diseased conditions. Decreased heme biosynthesis can cause anemia as well as neurological diseases3,36. Alternatively, increased heme biosynthesis plays an important role in the progression of certain cancers28,37. Heme has been shown to be critical for the growth, differentiation and survival of mammalian adipose, erythroid and neuronal cells4,38-41. For example, heme deficiency leads to neurite damage in primary mouse cortical neurons via the inhibition of glutamate NMDA (N-methyl-D-aspartate) receptor17. Additionally, inhibition of heme synthesis causes programmed cell death in the human epithelial cervix carcinoma HeLa cells26,41. Therefore, measuring the heme biosynthesis levels in various cells under different conditions is important for studying etiology and progression of many diseases.
Here we describe a fast and sensitive method to measure the level of intracellular heme synthesis by using [4-14C] 5-aminolevulic acid. This is an alternative method to other methods using 55Fe or 59Fe. We prefer using 14C because its radiation is very weak. In contrast, strong protection is required for working with Fe isotopes. Furthermore, this method is intended to measure and compare heme synthesis in different cells in parallel in a quick manner. In order to measure absolute heme levels, one may use the previously established method involving the use of HPLC42,43.
Protocol
CAUTION: While working with radioactivity, take appropriate precautions to avoid contamination of the experimenter and the surroundings. Dispose off all waste following local radiation safety guidelines.
1. Preparation of Cells
- Seed cells in 3.5 cm plates such that the confluency reaches 80%-90% on the day of assay.
- Note that seeding confluency for cells depends upon the cell type and their growth rate. When treating cells with a reagent for a certain number of days, seed cells so that the confluency is 80%-90% on the day of heme measurement. If the number of cells obtained from 3.5 cm plates is insufficient, use 6 cm plates with same amount of radioactive ALA as in the next step. Note: Use of individual plates is recommended because it is easier to handle individual plates.
A day before heme measurement, add 0.1-0.3 µCi [14C] 5-ALA and cold ALA to reach a final concentration of 20 µM of total ALA to each culture dish in a work area fit for working with radioactive materials and put the plates back into the incubator. Ideally, incubate with radioactive ALA for 16 hr. Note: Radiolabeled 5-ALA required may vary between cell lines. A radioactive count per minute (cpm) of at least 100 is required to minimize the error associated with counting. It is advisable to determine a minimum amount of [14C] 5-ALA required for the cell lines under study. For most experiments 0.1-0.3 µCi is sufficient to get a good measurement. Addition of cold ALA is optional.
2. Heme Extraction
- Place the cell culture dishes on ice and take them to the area fit for radioactivity work.
- Perform the whole extraction and evaporation process in an explosion-proof hood. Because the stored ether can explode, use the ether in small bottles and evaporate any remaining liquids, do not store ether in an open bottle for more than one month. Do not use acetone near open flames and heaters as it is highly flammable. Store acetone containing reagents in glass bottles as it dissolves tissue culture plastics.
Aspirate off the medium using a pipette. Wash cells one time with 1 ml of 1x cold PBS. Aspirate off PBS.
Add 1 ml of 1x cold PBS to the plates and collect the cells at the bottom of the plate using a rubber policeman. Transfer the collected cells from each plate to a labeled pre-chilled 2 ml tube. Collect all cells in 2 ml tubes. Note: Use a fresh rubber policeman for each plate for scraping cells from each plate. For each plate you would require four 2 ml tubes, a 1.5 ml tube and a scintillation vial.
Spin at 15,000 x g for 1 min at 4 °C.
Take out PBS using a pipette, give a short spin and remove the residual PBS. Resuspend cells in 100 μl of 1x PBS. Vortex to resuspend cells and keep on ice.
Take 10 μl from step 5 and transfer to a 1.5 ml microcentrifuge tube and store on ice to measure protein concentration. Note: Cell lysis buffer should be added to the cells and kept on ice to measure protein concentration later.
Spin the remaining 90 µl from step 5 at 5,000 x g for 30 sec at 4 °C; remove PBS completely using a 200 µl pipette tip.
- Add 300 µl of heme extraction buffer (see Materials List) to each tube. Then vortex for 15 sec to mix and put on ice for 1 min, repeat this 3 times.
- Add heme extraction buffer (HEB) and vortex the tube quickly to resuspend the pellet, do only one or two tubes at a time because the pellet should be resuspended quickly without being left in the HEB for too long. Once HEB is added to all the tubes and pellet is resuspended then follow the 15 sec vortex and 1 min on ice cycle as in step 8. Note that pellet may not go completely into the HEB if the pellet is left in the HEB buffer for a longer time and not resuspended quickly.
Add 1.2 ml of diethyl ether to each tube and vortex for 30 sec. Spin at 15,000 x g for 5 min at 4 °C. Note: Diethyl ether runs out of pipette tips quickly. To control this, pipette diethyl ether up and down once or twice, doing so helps hold the diethyl ether in the pipette. Repeat this every time a fresh pipette tip is used.
Transfer top (ether) phase with 1 ml pipette to a new microcentrifuge tube. Add 0.5 ml of 2 N HCl, vortex to mix and spin as in step 9. Note: It is difficult to see the separation between ether and HCl phases. Therefore, add small amounts of Trypan blue in 2 N HCl solution to be used during the experiment, enough to give it a pale blue color. This will help distinguish the two phases. The lower phase will be blue and the upper ether phase will be colorless. Transfer the upper colorless ether phase to a new tube and discard the lower HCl (blue) phase.
Repeat step 10 two more times to wash the ether phase with 2 N HCl. Transfer only the top ether phase to a new tube during each repetition and discard the lower HCl phase.
After the third wash with HCl, transfer the top phase with 1 ml pipette to a scintillation vial. Dry the samples by evaporating the diethyl ether by keeping the samples in the chemical hood O/N.
3. Measurement of Radioactivity
Next day, add 5 ml of scintillation fluid to the dried samples from step 2.12.
Shake well to mix. Measure the radioactivity by using a scintillation counter.
4. Calculations
Lyse the cells from step 2.6 using equal amounts of CelLytic M buffer (see Materials List) and spin at 14,000 x g for 5 min at 4 °C.
Take an aliquot of the supernatant and measure the protein concentration using BCA protein assay kit. Total amount of protein in mg (TP) = (90 µl * conc. of protein in µg/µl) / 1,000 Heme synthesis level (radioactivity/TP) = pmol/mg of protein.
Representative Results
This method was used to compare the levels of heme synthesis in normal (HBEC30KT) vs. cancer (HCC4017) lung cells. Figure 2 shows a higher level of heme synthesis in cancer cells (HCC4017) than normal lung cells (HBEC30KT). The level of heme synthesis was also measured in normal and cancer cells in the presence of mitochondrial uncoupler carbonyl cyanide 3-chlorophenylhydrazone (CCCP). Cells were treated with 10 μM CCCP for 24 hr before the measurement of heme synthesis levels. As expected, the levels of heme synthesis (Figure 2) decreased in the presence of CCCP in both normal and cancer cells. It has been shown previously that heme synthesis can be inhibited by succinyl acetone (SA), a potent and specific inhibitor of 5-aminolevulinic acid dehydratase (ALAD) which is the second enzyme in heme biosynthetic pathway41. Using this method, the heme synthesis levels were measured in HeLa cells in the presence and absence of 0.5 mM SA. Figure 3 shows that the levels of heme synthesis decreased by more than 10-folds in the presence of SA. To further demonstrate the use of this technique, the levels of heme synthesis were measured and compared in 4 cancer and in HEK293T cell lines as shown in Figure 4.
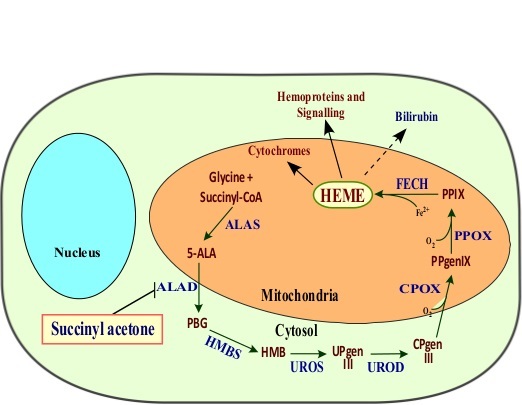 Figure 1.The heme biosynthesis pathway in humans. ALAS, 5-aminolevulinic acid synthase; 5-ALA, 5-aminolevulinic acid; ALAD, ALA dehydratase; PBG, porphobilinogen; HMBS, hydroxymethylbilane synthase; HMB, hydroxymethylbilane; UROS, uroporphyrinogen III synthase; UPgenIII, uroporphyrinogen III; UROD, uroporphyrinogen decarboxylase; CPgenIII, coproporphyrinogen III; CPOX, coproporphyrinogen III oxidase; PPgenIX, protoporphyrinogen IX; PPOX, protoporphyrinogen IX oxidase; PPIX, protoporphyrin IX; FECH, ferrochelatase. Please click here to view a larger version of this figure.
Figure 1.The heme biosynthesis pathway in humans. ALAS, 5-aminolevulinic acid synthase; 5-ALA, 5-aminolevulinic acid; ALAD, ALA dehydratase; PBG, porphobilinogen; HMBS, hydroxymethylbilane synthase; HMB, hydroxymethylbilane; UROS, uroporphyrinogen III synthase; UPgenIII, uroporphyrinogen III; UROD, uroporphyrinogen decarboxylase; CPgenIII, coproporphyrinogen III; CPOX, coproporphyrinogen III oxidase; PPgenIX, protoporphyrinogen IX; PPOX, protoporphyrinogen IX oxidase; PPIX, protoporphyrin IX; FECH, ferrochelatase. Please click here to view a larger version of this figure.
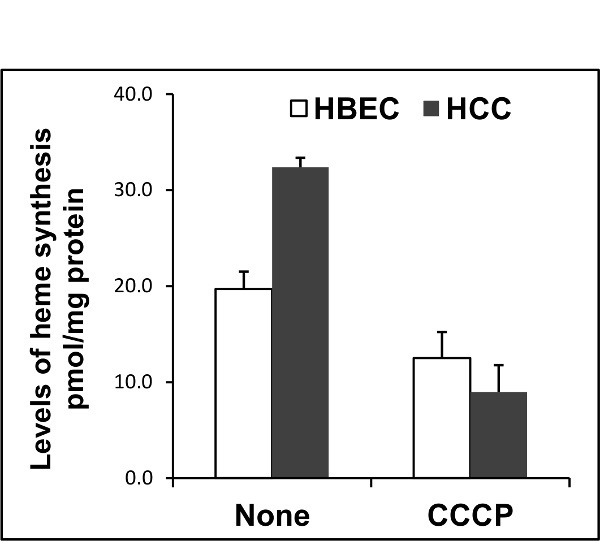 Figure 2. The levels of heme biosynthesis in normal (HBEC30KT) and cancer (HCC4017) cell lines, in absence and presence of 10 µM mitochondrial uncoupler CCCP. The background level was similar to the level of blank sample (scintillation fluid alone) and it was less than 2% of the control. Please click here to view a larger version of this figure.
Figure 2. The levels of heme biosynthesis in normal (HBEC30KT) and cancer (HCC4017) cell lines, in absence and presence of 10 µM mitochondrial uncoupler CCCP. The background level was similar to the level of blank sample (scintillation fluid alone) and it was less than 2% of the control. Please click here to view a larger version of this figure.
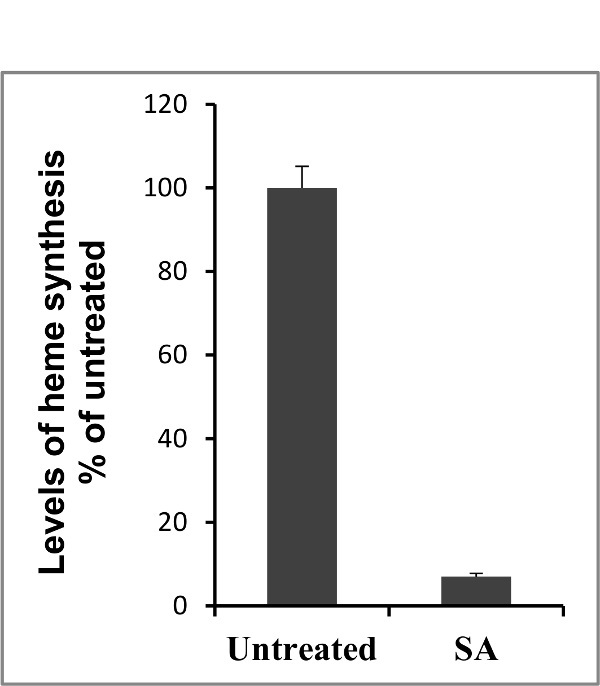 Figure 3.Levels of heme biosynthesis in HeLa cells in absence and presence of 0.5 mM of heme synthesis inhibitor succinyl acetone (SA). Cells were treated with SA for 3 days before heme measurement. The background level was similar to the level of blank sample (scintillation fluid alone) and it was less than 2% of the control. Please click here to view a larger version of this figure.
Figure 3.Levels of heme biosynthesis in HeLa cells in absence and presence of 0.5 mM of heme synthesis inhibitor succinyl acetone (SA). Cells were treated with SA for 3 days before heme measurement. The background level was similar to the level of blank sample (scintillation fluid alone) and it was less than 2% of the control. Please click here to view a larger version of this figure.
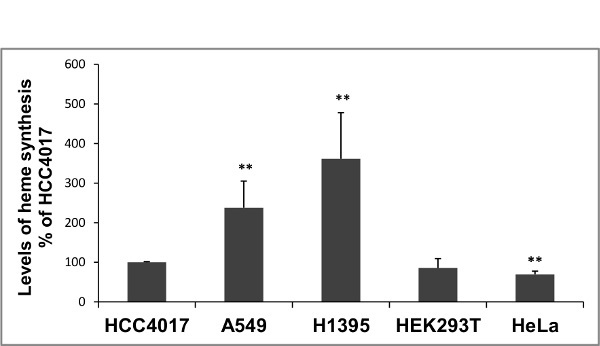 Figure 4. Comparison of the levels of heme biosynthesis in 4 cancer and HEK293T cell lines. Assay was performed as per the protocol. HCC4017 were grown in ACL4 media, A549 and H1395 in RPMI1640 supplemented with 5% FBS, HEK293T and HeLa in DMEM supplemented with 10% FBS. For statistical analysis, the heme synthesis levels in cancer cells and HEK293T cells were compared to the heme synthesis level in HCC4017 cells, by using Welch 2-sample t-test. **, p value < 0.005. The background level was similar to the level of blank sample (scintillation fluid alone) and it was less than 2% of the control. Please click here to view a larger version of this figure.
Figure 4. Comparison of the levels of heme biosynthesis in 4 cancer and HEK293T cell lines. Assay was performed as per the protocol. HCC4017 were grown in ACL4 media, A549 and H1395 in RPMI1640 supplemented with 5% FBS, HEK293T and HeLa in DMEM supplemented with 10% FBS. For statistical analysis, the heme synthesis levels in cancer cells and HEK293T cells were compared to the heme synthesis level in HCC4017 cells, by using Welch 2-sample t-test. **, p value < 0.005. The background level was similar to the level of blank sample (scintillation fluid alone) and it was less than 2% of the control. Please click here to view a larger version of this figure.
Discussion
Heme plays a key role in the generation of cellular energy via respiration26. Altered heme metabolism is known to be associated with various diseases including cancer28,41. Inhibition of heme synthesis is known to cause cell cycle arrest and apoptosis in Hela cells26,41. It has been shown that high heme synthesis level is associated with the progression of lung cancer cells28. Therefore, it would be of great importance to measure the levels of heme biosynthesis in cells under different conditions. The method discussed here does not quantify the total amount of heme therefore, it does not provide the absolute value of heme present in the cells. There are a variety of colorimetric methods and kits available to measure the total amounts of heme in the cells. For example, heme can be extracted from cells and mitochondria in a mixture of acetone and HCl and mixed with 50% (v/v) acetonitrile. Then the mixture can be applied to a 3.9 x 300 mm C18 Bondclone column followed by elution of heme in a gradient of acetonitrile containing 0.05% (v/v) trifluoroacetic acid and identifying heme compounds at 400 nm44,45.
Previously, to measure the levels of heme biosynthesis in mammalian tissues and cell extracts [14C]-glycine and [14C] 5- ALA were used46,47. Although labeled glycine enters the heme biosynthesis pathway at the first step but it is drained off by other metabolic pathways resulting in its limited incorporation into heme. Glycine is not a specific metabolite for heme biosynthesis pathway48. On the other hand, labeled 5-ALA is specific to heme biosynthesis pathway. This feature confers a quantitative advantage of using 5-ALA over glycine46,48,49. Therefore, use of radioactive 5-ALA is a more specific and accurate method to measure and compare the levels of heme biosynthesis.
The most critical steps to get consistent results are: 1) make sure the cell confluency does not go higher than 100% and not below 70% across various conditions; 2) the amount of radioactivity added per plate should be identical because this method is very sensitive; 3) heme extraction buffer should not be old, it is recommended to make fresh every time. The volume of radioactivity required per plate is very small and it is difficult to maintain the identical amounts across the samples. Therefore, it is recommended to make a working stock solution in the culture medium, if using the same medium for all the samples, else PBS (phosphate buffered saline) can be used. Working stock solution can be prepared in such a way that the volume dispensed per plate is under 10 μl. Also, to be more accurate, the medium from the replicate samples for one condition can be taken out in a 50 ml tube and the appropriate amounts of radioactivity can be added from the working stock solution, then dispense back equal amounts of medium to the plates. If the number of cells are limited, such as in primary cultures, 12-well or 24-well plates can be used to culture the cells and reduce the amount of radioactivity added per well proportionately.
This protocol is straightforward, simple, and provides accurate measures of the levels of heme synthesis. Sometimes, it may be difficult to resuspend the cell pellet in the heme extraction buffer (step 2.8) completely. To avoid this be quick in step 2.8, add heme extraction buffer to 1-2 samples at a time, vortex and put them on ice. Make sure the heme extraction buffer is not too old. If the standard deviation among the replicate samples is too high, pay attention to the following steps: 1) PBS was removed completely in step 2.7; 2) avoid spilling the sample when in diethyl ether phase, follow the tip in step 2.9; 3) make sure the radioactivity added per plate and the confluency of the replicate samples were the same.
Using radioactive 5-ALA method, the levels of heme synthesis were compared as shown previously28 in normal nonmalignant (HBEC) vs. cancer (HCC) cells, developed from the same patient. The technique described here requires the use of [14C] 5-ALA, a precursor in heme biosynthesis pathway (Figure 1), and measures the radioactivity incorporated into heme. In this technique, porphyrins, including heme, were extracted from the cells in a mixture of acetone, concentrated HCl and water50,51. Heme was further separated from porphyrins by extracting in diethyl ether and washing the ether phase with 2 N HCl50,51. Then, the radioactivity incorporated into heme was determined using a scintillation counter. Results can be normalized by the total amount of protein or cell number. Heme synthesis in HeLa cells was inhibited by using succinyl acetone (SA) and a decrease in the level of heme synthesis was observed as shown previously26 (Figure 3).
This method is very useful for the comparison of the levels of heme synthesis under different conditions or between cell lines (Figure 4). It requires a small number of cells and provides specific measurement and comparison of the levels of heme synthesis in different cells. It is not intended for the accurate measurement of absolute heme levels in the cells. It is best used to compare the levels of heme synthesis in cells with different properties, such as different cancer cells. Because cobalt, instead of iron, can be incorporated into heme in the last step of heme synthesis and be extracted 51, this method may not yield accurate comparison of heme synthesis, if the growth media contain different levels of cobalt or other metal ions. Nonetheless, the heme synthetic pathway from 5-aminolevulinic acid to porphyrins and heme is very specific, measuring the flux levels of the whole pathway is likely to be more reflective of metabolic activities relevant to heme, regardless of the metal ions. In contrast, the method using 59Fe detects only the part of the pathway incorporating Fe to form heme52. Although this is more specific, in the presence of significant levels of metal ions, the detected heme synthesis levels may not reflect the metabolic potential of certain cells such as cancer cells. Furthermore, Fe can be sequestered into other cellular locations beside mitochondria to make heme. By and large, we believe that the use of [4-14C] 5-ALA is a more practical method for measuring heme levels when comparing different cells with varying properties.
Disclosures
We have nothing to disclose.
Acknowledgments
The HCC4017 and HBEC30KT cell lines were kindly provided by Dr. John Minna’s lab. This work was supported by the Cecil H. and Ida Green funds to Dr. Li Zhang.
References
- Furuyama K, Kaneko K, Vargas PD. Heme as a magnificent molecule with multiple missions: heme determines its own fate and governs cellular homeostasis. Tohoku J Exp Med. 2007;213:1–16. doi: 10.1620/tjem.213.1. [DOI] [PubMed] [Google Scholar]
- Hamza I, Dailey HA. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta. 2012;1823:1617–1632. doi: 10.1016/j.bbamcr.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hon T, Ye W, Zhang L. Heme deficiency interferes with the Ras-mitogen-activated protein kinase signaling pathway and expression of a subset of neuronal genes. Cell Growth Differ. 2002;13:431–439. [PubMed] [Google Scholar]
- Zhang L. HEME BIOLOGY: The Secret Life of Heme in Regulating Diverse Biological Processes. Singapore: World Scientific Publishing Company. 2011.
- Mense SM, Zhang L. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- Ingram DJ, Kendrew JC. Orientation of the haem group in myoglobin and its relation to the polypeptide chain direction. Nature. 1956;178:905–906. doi: 10.1038/178905a0. [DOI] [PubMed] [Google Scholar]
- Perutz MF. X-ray analysis of hemoglobin. Science. 1963;140:863–869. doi: 10.1126/science.140.3569.863. [DOI] [PubMed] [Google Scholar]
- Chance B. The nature of electron transfer and energy coupling reactions. FEBS Lett. 1972;23:3–20. doi: 10.1016/0014-5793(72)80272-2. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, MacDonald TL. Mechanisms of cytochrome P-450 catalysis. Faseb J. 1990;4:2453–2459. doi: 10.1096/fasebj.4.8.2185971. [DOI] [PubMed] [Google Scholar]
- Igarashi K, et al. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for beta-globin locus control region complex. J Biol Chem. 1998;273:11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- Ogawa K, et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. Embo J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyake T, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder SH, Jaffrey SR, Zakhary R. Nitric oxide and carbon monoxide: parallel roles as neural messengers. Brain Res Brain Res Rev. 1998;26:167–175. doi: 10.1016/s0165-0173(97)00032-5. [DOI] [PubMed] [Google Scholar]
- Sun J, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. Embo J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. Embo J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon T, Lee HC, Hu Z, Iyer VR, Zhang L. The heme activator protein Hap1 represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae. Genetics. 2005;169:1343–1352. doi: 10.1534/genetics.104.037143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T, et al. Neurite degeneration induced by heme deficiency mediated via inhibition of NMDA receptor-dependent extracellular signal-regulated kinase 1/2 activation. J Neurosci. 2007;27:8475–8485. doi: 10.1523/JNEUROSCI.0792-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T, et al. Early failure of N-methyl-D-aspartate receptors and deficient spine formation induced by reduction of regulatory heme in neurons. Mol Pharmacol. 2011;79:844–854. doi: 10.1124/mol.110.069831. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Hon T, Zhang L. Heme deficiency suppresses the expression of key neuronal genes and causes neuronal cell death. Brain Res Mol Brain Res. 2005;137:23–30. doi: 10.1016/j.molbrainres.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Smith AG, Raven EL, Chernova T. The regulatory role of heme in neurons. Metallomics. 2011;3:955–962. doi: 10.1039/c1mt00085c. [DOI] [PubMed] [Google Scholar]
- Raghuram S, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor Rev-erbalpha. Genes Dev. 2009;23:2201–2209. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Hon T, Zhang L. Heme initiates changes in the expression of a wide array of genes during the early erythroid differentiation stage. Biochemical and biophysical research communications. 1999;258:87–93. doi: 10.1006/bbrc.1999.0586. [DOI] [PubMed] [Google Scholar]
- Yao X, Balamurugan P, Arvey A, Leslie C, Zhang L. Heme controls the regulation of protein tyrosine kinases Jak2 and Src. Biochemical and biophysical research communications. 2010;402:30–35. doi: 10.1016/j.bbrc.2010.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Zhang L. Heme controls the expression of cell cycle regulators and cell growth in HeLa cells. Biochem and biophys res comm. 2004;315:546–554. doi: 10.1016/j.bbrc.2004.01.092. [DOI] [PubMed] [Google Scholar]
- Hooda J, Shah A, Zhang L. Heme, an essential nutrient from dietary proteins, critically impacts diverse physiological and pathological processes. Nutrients. 2014;6:1080–1102. doi: 10.3390/nu6031080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooda J, et al. Enhanced heme function and mitochondrial respiration promote the progression of lung cancer cells. PloS one. 2013;8:e63402. doi: 10.1371/journal.pone.0063402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamna H, Walter PB, Ames BN. The role of heme and iron-sulfur clusters in mitochondrial biogenesis, maintenance, and decay with age. Arch Biochem Biophys. 2002;397:345–353. doi: 10.1006/abbi.2001.2671. [DOI] [PubMed] [Google Scholar]
- Atamna H, Killilea DW, Killilea AN, Ames BN. Heme deficiency may be a factor in the mitochondrial and neuronal decay of aging. Proc Natl Acad Sci U S A. 2002;99:14807–14812. doi: 10.1073/pnas.192585799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P. Cell biology of heme. Am J Med Sci. 1999;318:241–256. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- Brawer JR, Naftolin F, Martin J, Sonnenschein C. Effects of a single injection of estradiol valerate on the hypothalamic arcuate nucleus and on reproductive function in the female rat. Endocrinol. 1978;103:501–512. doi: 10.1210/endo-103-2-501. [DOI] [PubMed] [Google Scholar]
- Daniell WE, et al. Environmental chemical exposures and disturbances of heme synthesis. Environ Health Perspect. 1997;105(1):37–53. doi: 10.1289/ehp.97105s137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara T, et al. Hepatic heme metabolism in rats with fever induced by interleukin 1beta. Res Commun Mol Pathol Pharmacol. 1999;104:115–126. [PubMed] [Google Scholar]
- Vijayasarathy C, Damle S, Prabu SK, Otto CM, Avadhani NG. Adaptive changes in the expression of nuclear and mitochondrial encoded subunits of cytochrome c oxidase and the catalytic activity during hypoxia. Eur J Biochem. 2003;270:871–879. doi: 10.1046/j.1432-1033.2003.03447.x. [DOI] [PubMed] [Google Scholar]
- Anderson KESS, Bishop DF, Desnick RJ. The Metabolic and Molecular Bases of Inherited Disease. New York: The McGraw-Hill Companies, Inc; 2009. Disorders of heme biosynthesis: X-linked sideroblastic anemia and the porphyrias; pp. 1–53. [Google Scholar]
- Salvo ML, Contestabile R, Paiardini A, Maras B. Glycine consumption and mitochondrial serine hydroxymethyltransferase in cancer cells: the heme connection. Med Hypotheses. 2013;80:633–636. doi: 10.1016/j.mehy.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Ishii DN, Maniatis GM. Haemin promotes rapid neurite outgrowth in cultured mouse neuroblastoma cells. Nature. 1978;274:372–374. doi: 10.1038/274372a0. [DOI] [PubMed] [Google Scholar]
- Padmanaban G, Venkateswar V, Rangarajan PN. Haem as a multifunctional regulator. Trends Biochem Sci. 1989;14:492–496. doi: 10.1016/0968-0004(89)90182-5. [DOI] [PubMed] [Google Scholar]
- Rutherford TR, Clegg JB, Weatherall DJ. K562 human leukaemic cells synthesise embryonic haemoglobin in response to haemin. Nature. 1979;280:164–165. doi: 10.1038/280164a0. [DOI] [PubMed] [Google Scholar]
- Ye W, Zhang L. Heme deficiency causes apoptosis but does not increase ROS generation in HeLa cells. Biochemical and biophysical research communications. 2004;319:1065–1071. doi: 10.1016/j.bbrc.2004.05.089. [DOI] [PubMed] [Google Scholar]
- Bonkovsky HL, et al. High-performance liquid chromatographic separation and quantitation of tetrapyrroles from biological materials. Anal Biochem. 1986;155:56–64. doi: 10.1016/0003-2697(86)90224-1. [DOI] [PubMed] [Google Scholar]
- Sinclair PR, Gorman N, Jacobs JM. Measurement of heme concentration. Curr Protoc Toxicol. 2001;8:Unit 8.3. doi: 10.1002/0471140856.tx0803s00. [DOI] [PubMed] [Google Scholar]
- Barros MH, Carlson CG, Glerum DM, Tzagoloff A. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 2001;492:133–138. doi: 10.1016/s0014-5793(01)02249-9. [DOI] [PubMed] [Google Scholar]
- Shinjyo N, Kita K. Up-regulation of heme biosynthesis during differentiation of Neuro2a cells. J Biochem. 2006;139:373–381. doi: 10.1093/jb/mvj040. [DOI] [PubMed] [Google Scholar]
- Israels LG, Yoda B, Schacter BA. Heme binding and its possible significance in heme movement and availability in the cell. Ann N Y Acad Sci. 1975;244:651–661. doi: 10.1111/j.1749-6632.1975.tb41559.x. [DOI] [PubMed] [Google Scholar]
- Yannoni CZ, Robinson SH. Early-labelled haem in erythroid and hepatic cells. Nature. 1975;258:330–331. doi: 10.1038/258330a0. [DOI] [PubMed] [Google Scholar]
- Robinson SH. Formation of bilirubin from erythroid and nonerythroid sources. Semin Hematol. 1972;9:43–53. [PubMed] [Google Scholar]
- Granick S, Granick D. Nucleolar necklaces in chick embryo myoblasts formed by lack of arginine. J Cell Biol. 1971;51:636–642. doi: 10.1083/jcb.51.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell DB, Barrett J, Clezy PS. The prosthetic group of cytochrome oxidase. 1. Purification as porphyrin alpha and conversion into haemin alpha. Biochem J. 1961;78:793–797. doi: 10.1042/bj0780793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair P, Gibbs AH, Sinclair JF, de Matteis F. Formation of cobalt protoporphyrin in the liver of rats. A mechanism for the inhibition of liver haem biosynthesis by inorganic cobalt. Biochem J. 1979;178:529–538. doi: 10.1042/bj1780529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Haile DJ, Wessling-Resnick M. Copper-induced ferroportin-1 expression in J774 macrophages is associated with increased iron efflux. Proc Natl Acad Sci U S A. 2004;101:2700–2705. doi: 10.1073/pnas.0306622101. [DOI] [PMC free article] [PubMed] [Google Scholar]


