Abstract
The zebrafish has emerged as a valuable genetic model system for the study of developmental biology and disease. Zebrafish share a high degree of genomic conservation, as well as similarities in cellular, molecular, and physiological processes, with other vertebrates including humans. During early ontogeny, zebrafish embryos are optically transparent, allowing researchers to visualize the dynamics of organogenesis using a simple stereomicroscope. Microbead implantation is a method that enables tissue manipulation through the alteration of factors in local environments. This allows researchers to assay the effects of any number of signaling molecules of interest, such as secreted peptides, at specific spatial and temporal points within the developing embryo. Here, we detail a protocol for how to manipulate and implant beads during early zebrafish development.
Keywords: Developmental Biology, Issue 101, zebrafish, embryo, bead implantation, tissue manipulation, development, organogenesis
Introduction
Developmental biology researchers utilize a vast array of cellular, molecular, and genetic methods to uncover the mechanisms that control how an organism is formed. Among these approaches, tissue manipulation is a key tool in deciphering complex questions about cell fate, cellular movement, and the organization of tissues. One way to alter local tissue environments is through the surgical application of microbeads that are used to deliver a focal source of proteins or other signaling molecules 1. This type of experimental manipulation has been widely implemented in classical vertebrate embryology models, such as the frog and chick 2.
The zebrafish has become an important vertebrate model organism for the study of organogenesis and also provides many unique advantages for disease modeling 3-5 as they share high genetic conservation with humans 6. In particular, the optical transparency and external development of the zebrafish embryo offers an unmatched viewpoint for the observation of tissue ontogeny 3-5. The implementation of large-scale forward genetic screens has generated a powerful repository of zebrafish mutant strains for further study 7,8, and the identification of alternative screening techniques that can be efficiently conducted at reduced scale in single laboratories 9,10. Further experimental work with zebrafish has been facilitated through advances in transgenic methodologies and reverse genetic approaches 11,12, as well as chemical genetics 13-15.
Tissue manipulation techniques, such as the implementation of microbeads, have not been as widely employed in the zebrafish, but nevertheless provide a useful tool to further understand cell signaling during development. Microbead implantation has been used to interrogate the processes of organ formation in the zebrafish retina 16,17, heart 18, brain 19-22, neural crest 23, and fin 24,25. In these and other studies, beads have been applied during development to understand the diffusion of signaling molecules 26, how gradients affect cell migration 27 and axial patterning 28. More recently, microbeads have been utilized to evaluate regeneration mechanisms in zebrafish adults 29. In developmental studies, for example, zebrafish microbead work has provided insights into the mechanisms of limb formation through studies of the pectoral fin 25. The zebrafish pectoral fin bud is homologous to the forelimb bud in the mouse 30 and chick 31. The vertebrate limb bud has two essential signaling nodes: the zone of polarizing activity (ZPA) that establishes the anterior-to-posterior axis through the expression of Sonic hedgehog (Shh) and downstream Hox gene targets, and the apical ectodermal ridge (AER) present at the tip of the limb bud, which acts to establish proximal to distal identity of the limb through expression of Fibroblast growth factors (Fgfs). By implanting Fgf soaked microbeads into zebrafish Shh genetic mutants, investigators identified Fgf as essential to progression of the cell cycle and growth of the vertebrate limb 25. In addition to theFgf and Shh signaling cascades that establish positional identity, pioneering studies using the chick limb bud identified retinoic acid (RA) as a molecule that could mimic the action of the polarizing region to establish anterior to posterior identity 32. These experiments involved placing small strips of RA-soaked Whatman paper into the chick limb to assess digit patterning 32. Further, researchers have performed other elegant studies employing the use of microbeads, cell transplantation, and exogenous RA treatments in zebrafish to determine that RA acts to provide long-range positional cues within the zebrafish hindbrain and mesoderm 28. However, at present many questions remain about the roles of signaling factors like Fgf and RA during numerous aspects of vertebrate development. The signaling effects of RA, acting as a morphogen, impact many organs 33, such as the developing heart 34 and the renal progenitors, where RA specifies proximal kidney cells type fates 35-39. Further understanding of such topics could benefit significantly from experimental studies using tissue manipulation and microbead implantation techniques.
While fewer studies have been performed with microbead implantation in the zebrafish, when compared to models like the chick, those that have been carried out have been highly informative. One reason for the paucity of microbead implantation based-research in the zebrafish embryo is likely the notion that there are difficult technical challenges, based on the size of the embryo, which constitute an impediment to successfully performing such manipulations. However, microbead implantation in zebrafish embryos can be learned with practice and assisted through visual observation of the technique, and thus can be pursued as a means to interrogate the mechanisms of development. Here, we demonstrate the precise application of a microbead into the zebrafish embryo, which can be utilized for conducting a wide variety of assays on tissue formation and cellular morphogenesis.
Protocol
The procedures for working with zebrafish embryos described in this protocol were approved by the Institutional Animal Care and Use Committee at the University of Notre Dame.
1. Ringer’s Solution Preparation
Make a solution of 116 mM NaCl, 2.9 mM KCl, 3.6 mM CaCl2, and 5 mM Hepes with ultrapure H2O through continual stirring to avoid any salt precipitation.
After adding salts and while the solution is still stirring, add 100 units/penicillin per mL and 100 µg streptomycin per ml.
After adding all antibiotics and salts, perform filter sterilization of the solution and store in the sterile container. Note: N-Phenylthiourea (1-Phenyl-2-thiourea) or PTU can be added to Ringer’s Solution to block the formation of pigment in bead implanted embryos without negative consequences to embryo health. To prepare Ringer’s/pigmentation blocking solution, use a concentration of 0.003 % PTU. Due to the low concentration of PTU to be added it is advised that a larger volume of Ringer’s Solution be made, at least 1 L, to avoid adding miniscule amounts of PTU powder. Add PTU during step 1.2, while the solution is stirring.
2. Tricaine Solution Preparation
Add 2 g of Ethyl 3-aminobenzoate methanesulfonate salt (Tricaine) per L of solution plus 0.1 M Tris, from a 1 M stock balanced to a pH of 9.5 and made with ultrapure H2O. Note: Tricaine will be used to euthanize the zebrafish embryos at the desired time point to assay bead implantation phenotypes.
3. E3 Solution Preparation
Make a solution of 0.25 M NaCl, 10 mM KCl, 12.5 mM CaCl2,and 16.6 mM MgSO4 to 1 L of ultrapure water to create 1 L of E3.
Add 200 µl of Methylene Blue to the E3 solution to inhibit fungal growth.
4. Pulling Needles for Microbead Transfer
Use fire polished borosilicate glass with filament at an O.D.: 1.0 mm, I.D.: 0.5 mm, at a length of 10 cm.
Place borosilicated glass into a micropipette puller to prepare fine needles. Note: The following four dimensions can be used as a starting point to pull the needles: Heat = 540, Pull = 245, Velocity = 200, and Time = 125.
After pulling an adequate number of needles, store in covered plastic petri dishes, positioned on strips of modeling clay or adhesive tape to prevent breaking the needle tip, until use.
To trim the needle tip to the desired bore size, use a pair of fine forceps to score the end of the needle. Note: Needle bore size will vary depending on the microbead size being utilized as well as user handling preferences. A good starting point is to craft bore sizes in a range between 100 and 200 µm if microbeads ranging from 50-100 µm will be utilized.
Prior to use, insert the trimmed needle into the capillary tube holder to fashion the completed microbead transfer instrument.
5. Whisker/Lash Tool Preparation
Obtain a whisker, lash, or other small yet firm piece of natural or synthetic hair filament. Cut near the base of the filament at a 45° angle. Adhere the whisker to a P-1000 micropipette tip using permanent clear adhesive glue.
6. Microbead Implantation Tray Preparation
Mix 2 g of agarose with 100 ml of E3 solution to make a 2 % E3 agarose gel.
Heat the 2 % agarose solution for 1 min and 30 sec in a 250 ml Erlenmeyer flask, swirling the flask every 30 sec in order to avoid the gel from bubbling over.
Take the lid of a 60 mm x 15 mm petri dish and place it in a larger 150 mm x 15 mm dish with the interior facing upward.
Pour the hot agarose gel into the 150 mm x 15 mm petri dish, making sure to anchor down the lid of the smaller dish with the index finger. Note: Allow a thin layer of agarose below the lid of the smaller dish to remove condensation that will form when pouring the gel; this will make the beads easier to see under a dissection microscope.
As the agarose cools, but before it solidifies, place a well mold on the gel. This will allow for easier placement of the embryos.
Note: In order to avoid bubbles in the wells, place the mold slowly at an angle until it is floating on top of the gel.
Using a microspatula, carefully remove the well mold once the gel has solidified. Add E3 solution and store at 4 °C.
7. Embryo Collection
Prepare mating chambers, separated by dividers, by filling them up with system water, and then placing adult zebrafish male(s) and female(s) pairs in the chambers O/N.
Collect fertilized eggs using a fine wire mesh strainer (embryonic stages will vary slightly).
Deposit the fertilized eggs in a clean, 10 cm diameter petri dish by turning the strainer upside down and rinsing it with a fine stream of E3 until there are no eggs left in the strainer, and there approximately 25 ml of E3 solution in the dish.
After collection, divide the eggs into several dishes (approximately 50-60 eggs per dish) to avoid overcrowding, which might lead to developmental asynchrony within the clutch and make collecting desired times points difficult.
Incubate embryos in E3 solution at 28.5 °C to enable developmental assessment according to the standard zebrafish staging series 40. Note: The embryos should be transferred to E3/PTU at 24 hr post fertilization (hpf) to prevent pigment development if manipulations at older stages necessitate optical clarity.
8. Bead Implantation
Incubate the microbeads in desired test molecule concentration or the corresponding vehicle solution for the required time in an appropriate volume. Note: The researcher can use sterile Petri dishes ranging from 3-6 cm in diameter, or multi-well plates, both of which enable visualization of the microbeads within the solution. Time may vary depending on the type and amount of molecule of interest, and the researcher should refer to manufacturer or published recommendations for duration of incubation and other aspects such as sensitivity to light and temperature.
Once the embryos have reached the desired developmental time point, remove the embryos from their chorions using fine forceps.
Incubate the embryos in the filtered Ringer solution with antibiotics for 10 min to acclimate them to the solution.
While the embryos are acclimating, transfer the microbeads into the microbead plate nestled within the implantation tray and rinse them in Ringer solution for 10 min. Note: Implant the beads within 50 min after reaching this point. This will decrease the likelihood that one will implant a bead with a smaller concentration of the molecule of interest.
Array the embryos into the wells of the microbead implantation tray, taking care to orient them so the area of interest where the microbead will be implanted is accessible.
Make a small incision on the embryo using the tungsten needle.
Transfer a microbead onto the embryo using the microcapillary transfer pipette: depressing the bulb of the pipette, gently pull the microbead into the capillary column, and then release the pressure to transfer the microbead at the desired destination.
Use the whisker/lash tool to position the microbead inside the embryo. Note: Be sure to position the microbead into the tissue away from the site of incision so the microbead will not be expelled from the embryo as it heals.
Representative Results
Several types of manipulation tools are useful for handling microbeads during research applications (Figure 1). In addition, a simple agarose mold with small wells can be utilized to hold the zebrafish embryo, which is vital to perform these experiments in a timely and efficient manner (Figure 1). The implantation of a microbead can be performed at the spatial position and temporal stage of interest, which allows researchers to narrow a specific window of action for the molecule of interest.
Here, single microbeads were implanted into zebrafish embryos at the tail bud stage or at later time points during somitogenesis stages (Figure 3). Two different sized microbeads, rinsed with Ringer’s solution alone, were used in this protocol demonstration (Figure 3A, 3B and 3C). This microbead implantation in the absence of any conjugated small molecules demonstrates that bead implantation can be achieved without gross morphological effects during the proper development of the zebrafish embryo (Figure 4).
The experimentalist may encounter difficulties with maneuvering the microbead into the puncture hole made by a tungsten needle atop the embryo. To achieve better handling of the microbead once placed on the embryo by the micro-needle, a whisker/lash tool can be used. This can be crafted with naturally shed feline or canine whiskers, although other natural or synthetic lash filaments can be utilized (Figure 1). A gentle push of the microbead with either the tungsten needle or the whisker tool should see the microbead sink into the Ringer’s solution of the mold. Once the microbead has sunk into the mold it should be moved large distances if necessary using the tungsten needle and once atop the embryo the needle or whisker tool can be used to position the microbead into the embryo. The success of the microbead implantation can be immediately gauged through visualization at a stereomicroscope, as the bead will remain stably positioned in the tissue. To evaluate bead position over time, each embryo sample can be imaged through live time-course photography or checked at periodic intervals (as depicted in Figure 4) to ensure that the bead location has not shifted over the duration of the experiment due to endogenous tissue morphogenesis. Understanding the position of the bead over time is critical for the most informed interpretation of results, such as evaluating the effect of a small molecule on a target tissue or developmental process.
The use of these above steps will provide an ideal environment in which to imbed microbeads into the zebrafish embryo at a desired time and position. In our experience, novice users of this microbead implantation protocol typically gained the skill necessary to perform consistent surgical implantation of microbeads into the zebrafish embryo after approximately a single week of practice.
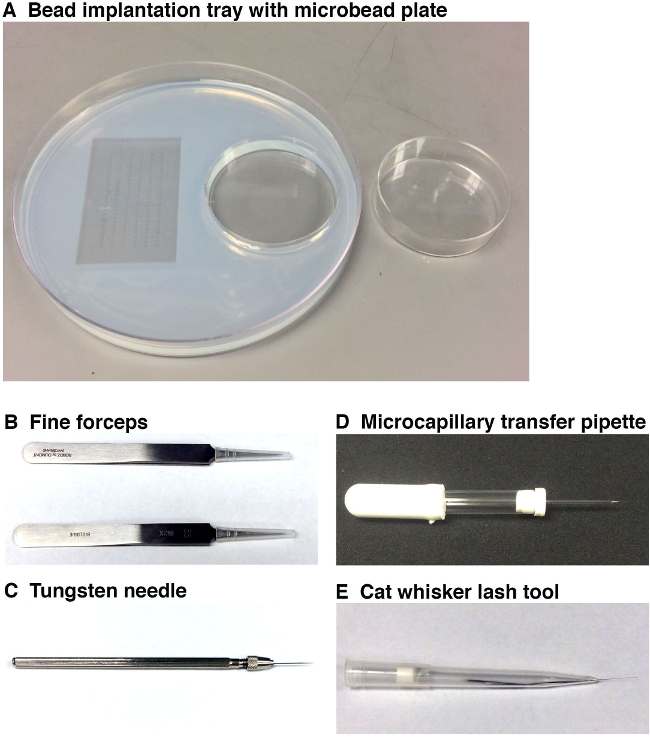 Figure 1. Tools used to perform microbead implantation. (A) The bead implantation tray (left, 15 cm diameter) with embryo molds (on left) and a microbead incubation tray (on right, 6 cm diameter). This working tray enables the researcher to place the embryos into the desired orientation for microbead implantation, and to have the prepared microbeads for implantation in close proximity at the microscope station. Microbeads should be incubated in small molecule(s) and/or vehicle and then rinsed in a separate microbead plate (right) before placing them into the bead implantation tray that is positioned under the microscope station. (B) Two pairs of fine forceps will be used to remove the chorion surrounding each zebrafish embryo and also to break the end of the microcapillary needle. (C) A tungsten needle will be employed to make a very fine puncture into the zebrafish embryo in which to position the microbead. (D) The microcapillary transfer pipette will be used to pick up and position a microbead on top of the zebrafish embryo near the site of the puncture. (E) The whisker/lash tool will allow the gentle positioning of the microbead into the incision site within the embryo that was previously made by the tungsten needle. Please click here to view a larger version of this figure.
Figure 1. Tools used to perform microbead implantation. (A) The bead implantation tray (left, 15 cm diameter) with embryo molds (on left) and a microbead incubation tray (on right, 6 cm diameter). This working tray enables the researcher to place the embryos into the desired orientation for microbead implantation, and to have the prepared microbeads for implantation in close proximity at the microscope station. Microbeads should be incubated in small molecule(s) and/or vehicle and then rinsed in a separate microbead plate (right) before placing them into the bead implantation tray that is positioned under the microscope station. (B) Two pairs of fine forceps will be used to remove the chorion surrounding each zebrafish embryo and also to break the end of the microcapillary needle. (C) A tungsten needle will be employed to make a very fine puncture into the zebrafish embryo in which to position the microbead. (D) The microcapillary transfer pipette will be used to pick up and position a microbead on top of the zebrafish embryo near the site of the puncture. (E) The whisker/lash tool will allow the gentle positioning of the microbead into the incision site within the embryo that was previously made by the tungsten needle. Please click here to view a larger version of this figure.
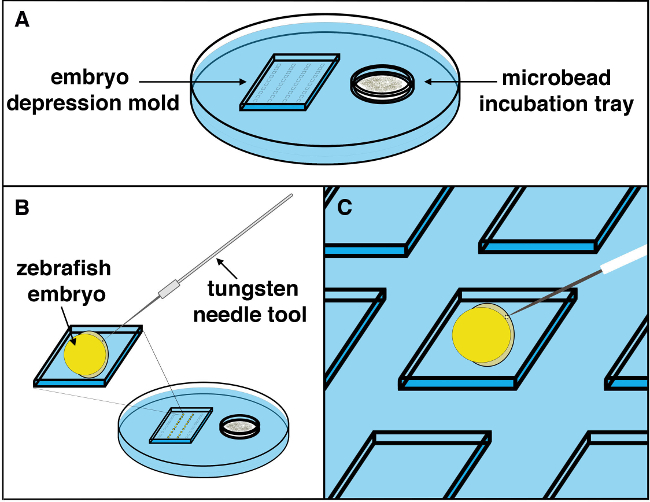 Figure 2. Schematic of the microbead implantation procedure. (A) A microbead implantation tray with microbeads soaking in the desired wash solution is set up under a stereomicroscope station, and the embryos positioned with tissue facing upwards. (B) Using a tungsten needle, a small incision into the embryo can be made with a slight yet forceful movement. (C) Using a microcapillary transfer pipette, the desired microbead can be picked up from the microbead incubation chamber in the tray, located on the right, and brought to the left compartment where the embryo working area is situated. The microbead should be deposited near the site of the incision, then gently positioned into the incision site made by the tungsten needle using the whisker/lash tool. Once a microbead is maneuvered into the incision site, it should be tucked into the tissue (away from the incision) to stably position the microbead and prevent subsequent movement out of the embryo as development continues. Please click here to view a larger version of this figure.
Figure 2. Schematic of the microbead implantation procedure. (A) A microbead implantation tray with microbeads soaking in the desired wash solution is set up under a stereomicroscope station, and the embryos positioned with tissue facing upwards. (B) Using a tungsten needle, a small incision into the embryo can be made with a slight yet forceful movement. (C) Using a microcapillary transfer pipette, the desired microbead can be picked up from the microbead incubation chamber in the tray, located on the right, and brought to the left compartment where the embryo working area is situated. The microbead should be deposited near the site of the incision, then gently positioned into the incision site made by the tungsten needle using the whisker/lash tool. Once a microbead is maneuvered into the incision site, it should be tucked into the tissue (away from the incision) to stably position the microbead and prevent subsequent movement out of the embryo as development continues. Please click here to view a larger version of this figure.
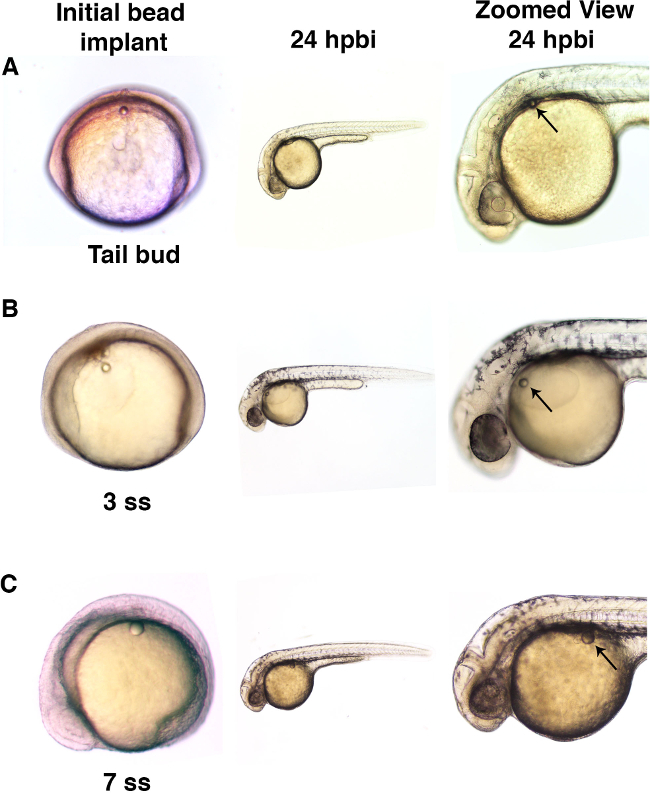 Figure 3. Microbeads can be implanted into embryos at different developmental stages. (A) Zebrafish embryos were implanted with a microbead (diameter of approximately 50 µm) into the trunk at the tail bud stage. When examined at the 24 hr post bead implant (hpbi), the embryos had developed normally and the microbead showed minimal movement during the passage of developmental time. (B) Zebrafish embryos implanted with microbeads (with a diameter of approximately 50 µm) into the yolk sac at the 3 somite stage (ss) displayed normal development with relatively minor bead movement over time. (C) Larger microbeads (diameter of approximately 70 µm) were implanted into the 7 ss embryo similarly showed normal subsequent development, as well as minimal if any movement from the site of implantation. Please click here to view a larger version of this figure.
Figure 3. Microbeads can be implanted into embryos at different developmental stages. (A) Zebrafish embryos were implanted with a microbead (diameter of approximately 50 µm) into the trunk at the tail bud stage. When examined at the 24 hr post bead implant (hpbi), the embryos had developed normally and the microbead showed minimal movement during the passage of developmental time. (B) Zebrafish embryos implanted with microbeads (with a diameter of approximately 50 µm) into the yolk sac at the 3 somite stage (ss) displayed normal development with relatively minor bead movement over time. (C) Larger microbeads (diameter of approximately 70 µm) were implanted into the 7 ss embryo similarly showed normal subsequent development, as well as minimal if any movement from the site of implantation. Please click here to view a larger version of this figure.
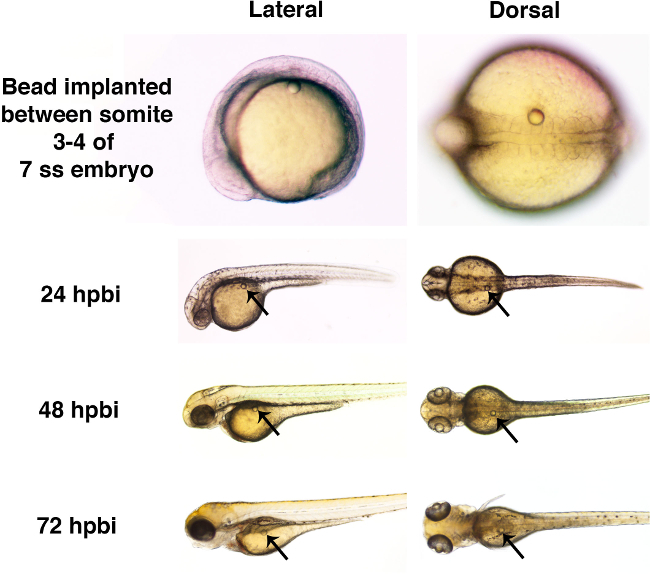 Figure 4. Gross development of the zebrafish embryo is unperturbed by the presence of a microbead through 72 hpbi. Zebrafish embryos were implanted with microbeads soaked in Ringer’s solution (with a diameter of approximately 50 µm) at the 7 somite stage (ss). Each microbead was surgically inserted into the trunk, between somites 3 and 4. The presence of the bead was not associated with overt developmental defects at 24 hr post bead implant (hpbi), 48 hpbi, or 72 hpbi. Please click here to view a larger version of this figure.
Figure 4. Gross development of the zebrafish embryo is unperturbed by the presence of a microbead through 72 hpbi. Zebrafish embryos were implanted with microbeads soaked in Ringer’s solution (with a diameter of approximately 50 µm) at the 7 somite stage (ss). Each microbead was surgically inserted into the trunk, between somites 3 and 4. The presence of the bead was not associated with overt developmental defects at 24 hr post bead implant (hpbi), 48 hpbi, or 72 hpbi. Please click here to view a larger version of this figure.
Discussion
Over the past century, the understanding of body plan patterning and organogenesis has undergone monumental advances. Tissue manipulation techniques were critical in uncovering key information about these vital processes. Genetic modification is one of the most commonly used methods for ascertaining gene function, and methods of mosaic analysis, such as cell transplantation, provide useful approaches to understand autonomy of gene function. Microbead implantation provides another venue to interrogate how particular molecules alter developmental dynamics, as this method enables the researcher to alter a local tissue environment by introducing signaling molecules or inhibitors. A range of different microbeads are commercially available, that have varying sizes and other physical properties exist (e.g., charge) such that they can be employed to for the experimental conditions of interest. Thus, by implanting microbeads that are soaked in a protein or chemical of interest within an organism, researchers can investigate localized effects during development and find associations between the gene or molecule of interest and particular biological phenotype(s).
Studies such as the one performed by Wada and colleagues used microbead implantation in order to assess the effect of increased Hedgehog signaling in skeletal patterning in the anterior neurocranium (ANC) in zebrafish 23. Previous studies have shown that Shh is required for ANC formation 14. Using Shh-coated microbeads researchers identified that this signal promotes cartilage formation in the ANC. The microbead implantation process was used to demonstrate a clear link between Hedgehog signaling and cartilage formation in the ANC. Another key example of this tissue manipulation technique in zebrafish is observed in studies where scientists investigated the transcriptional control of the Erythroblastoma Twenty-Six (ETS) domain factors ETS-related molecule (Erm) and Polyoma enhancer activator 3 (Pea3) by Fgf signaling during early zebrafish brain development 19. Through microbead implantation experiments, they were able to show that Fgf8 and Fgf3 can ectopically activate expression of erm and pea3. These examples illustrate the utility of microbeads to provide insights into the developmental mechanisms that collaborate during tissue formation, which can be well-characterized by the use of methods to assess gene expression 41. Thus, microbead transplantation can be a viable method to explore other tissues, such as the intermediate mesoderm (IM) which gives rise to the kidney. Specifically, it would aid in renal developmental studies, to investigate how different molecules affect nephron segmentation 42 and tubulogenesis 43,44, processes that are only superficially understood at present. Furthermore, microbead implantation has begun to be used to study regeneration processes in zebrafish 29] and could be adapted for use with any number of organ regeneration models, such as following laser ablation of embryonic tissues like the nephron 45 or in conjunction with methods that have been formulated to perform research with the corresponding adult structures 46-49. Finally, microbead implantation has the potential to be used in models of human disease, such as cancer 50,51 or tissue degeneration 52,53.
In the present protocol, we demonstrate the method of microbead implantation in zebrafish embryos, which has also been similarly described by other researchers, but not shown through video protocols 1. With minimal practice we were able to implant microbeads at an approximate rate of 8-10 embryos/hr, which relates the feasibility of this procedure once the researcher has some experience. The results shown herein demonstrate that beads of various dimensions can be implanted at early stages, and that with care, this tissue manipulation technique can be implemented with minimal physical disruption to the embryo. One improvement that should be emphasized is the use of a whisker/lash tool to position the microbead into the embryo. This relatively inexpensive and simple piece of equipment is about the same diameter as the microbead, is easy to obtain and helps speed up the implantation process. The whisker/lash can be cut to a desired length to produce a firm yet delicate bead-handling tool, depending on researcher dexterity and preference. Finally, while we described here how to physically manipulate microbeads and zebrafish embryos to perform implantation, this protocol does not outline specific handling procedures for different drugs or peptides. In general, chemically treated microbeads should be implanted into the animal with expediency, to avoid undesired effects in other areas of the organism, and researchers should be well informed about the possible safety concerns associated with any such chemicals before initiating their studies.
In sum, we have demonstrated a relatively simple and efficient microbead implantation method with a wide range of applications using materials that are readily available in the lab. Ultimately, we hope that this manual will aid researchers with the delicate nature of zebrafish tissue manipulation.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by NIH grant DP2OD008470.
References
- Picker A, et al. Lieschke GJ, et al. Tissue micromanipulation in zebrafish embryos. Zebrafish, Methods in Molecular Biology. 2009. [DOI] [PubMed]
- Wilson J, Tucker AS. Fgf and Bmp signals repress the expression of Bapx1 in the mandibular mesenchyme and control the position of the developing jaw joint. Dev Biol. 2004;266:138–150. doi: 10.1016/j.ydbio.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith JR, Jobin C. Think small: zebrafish as a model system of human pathology. J Biomed Biotechnol. 2012;2012:817341. doi: 10.1155/2012/817341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Nüsslein-Volhard C. Large scale genetics in a small vertebrate, the zebrafish. Int J Dev Biol. 1996;40:221–227. [PubMed] [Google Scholar]
- Amsterdam A, Hopkins N. Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet. 2006;22:473–478. doi: 10.1016/j.tig.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Pickart MA, Klee EW. Zebrafish approaches enhance the translational research tackle box. Transl Res. 2014;163:65–78. doi: 10.1016/j.trsl.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Kroeger PT, Jr, Poureetezadi SJ, McKee R, Jou J, Miceli R, Wingert RA. Production of haploid zebrafish embryos by in vitro fertilization. J Vis Exp. 2014;89:e51708. doi: 10.3791/51708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Wolfe SA. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev Cell. 2011;21:48–64. doi: 10.1016/j.devcel.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Auer TO, Del Bene F. CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods. 2014;69:142–150. doi: 10.1016/j.ymeth.2014.03.027. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci USA. 2000;97:12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessman CA. The developing zebrafish (Danio rerio): A vertebrate model for high-throughput screening of chemical libraries. Birth Defects Res C Embryo Today. 2011;93:e52063. doi: 10.1002/bdrc.20212. [DOI] [PubMed] [Google Scholar]
- Donahue EK, Wingert RA. A manual small molecule screen approaching high–throughput using zebrafish embryos. J Vis Exp. 2014. p. 52063. [DOI] [PMC free article] [PubMed]
- Hyatt GA, Schmitt EA, Marsh-Armstrong N, McCaffery P, Dräger UC, Dowling JE. Retinoic acid establishes ventral retinal characteristics. Development. 1996;122:195–204. doi: 10.1242/dev.122.1.195. [DOI] [PubMed] [Google Scholar]
- Picker A, Brand M. Fgf signals from a novel signaling center determine axial patterning of the prospective neural retina. Development. 2005;132:4951–4962. doi: 10.1242/dev.02071. [DOI] [PubMed] [Google Scholar]
- Reifers F, Walsh EC, Leger S, Stainier DYR, Brand M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar.) Development. 2000;127:225–235. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev. 2001;107:105–117. doi: 10.1016/s0925-4773(01)00456-7. [DOI] [PubMed] [Google Scholar]
- Reim G, Brand M. spiel-ohne-grenzen/pou2. mediates regional competence to respond to Fgf8 during zebrafish early neural development. Development. 2002;129:917–933. doi: 10.1242/dev.129.4.917. [DOI] [PubMed] [Google Scholar]
- Jaszai J, Reifers F, Picker A, Langenberg T, Brand M. Isthmus-to-midbrain transformation in the absence of midbrain-hindbrain organizer activity. Development. 2003;130:6611–6623. doi: 10.1242/dev.00899. [DOI] [PubMed] [Google Scholar]
- Hirati Y, Okamoto H. Canopy1, a novel regulator of FGF signaling around the midbrain-hindbrain boundary in zebrafish. Curr Biol. 2006;16:421–427. doi: 10.1016/j.cub.2006.01.055. [DOI] [PubMed] [Google Scholar]
- Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132:3977–3988. doi: 10.1242/dev.01943. [DOI] [PubMed] [Google Scholar]
- Abe G, Ide H, Tamura K. Function of FGF signaling in the developmental process of the median fin fold in zebrafish. Dev Biol. 2007;304:355–366. doi: 10.1016/j.ydbio.2006.12.040. [DOI] [PubMed] [Google Scholar]
- Prykhozhij SV, Neumann CJ. Distinct roles of Shh and Fgf signaling in regulating cell proliferation during zebrafish pectoral fin development. BMC Dev Biol. 2008;8:91. doi: 10.1186/1471-213X-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholpp S, Brand M. Endocytosis controls spreading and effective signaling range of Fgf8 protein. Curr Biol. 2014;14:1834–1841. doi: 10.1016/j.cub.2004.09.084. [DOI] [PubMed] [Google Scholar]
- Hardt S, et al. The Bmp gradient of the zebrafish gastrula guides migrating lateral cells by regulating cell-cell adhesion. Curr Biol. 2007;17:475–487. doi: 10.1016/j.cub.2007.02.013. [DOI] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 .creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:e2522–e2523. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittijn SA, et al. Zebrafish development and regeneration: new tools for biomedical research. Int J Dev Biol. 2009;53:835–850. doi: 10.1387/ijdb.082615sb. [DOI] [PubMed] [Google Scholar]
- Niswander L, Jeffrey S, Martin GR, Tickle C. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371:609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- Laufer E, Nelson CE, Johnson RL, Morgan BA, Tabin C. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell. 1994;79:993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Tickle C, Alberts B, Wolpert L, Lee J. Local application of retinoic acid to the limb bud mimics the action of the polarizing region. Nature. 1982;296:564–566. doi: 10.1038/296564a0. [DOI] [PubMed] [Google Scholar]
- Samarut E, Fraher D, Laudet V, Gibert Y. ZebRA: an overview of retinoic acid signaling during zebrafish development. Biochimica et Biophysica Acta. 2015;1849:73–83. doi: 10.1016/j.bbagrm.2014.05.030. [DOI] [PubMed] [Google Scholar]
- Lengerke C, et al. Interactions between Cdx genes and retinoic acid modulate early cardiogenesis. Dev Biol. 2011;354:134–142. doi: 10.1016/j.ydbio.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, et al. The cdx and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:1922–1938. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, Davidson AJ. Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev Dyn. 2011;240:2011–2027. doi: 10.1002/dvdy.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach GF, Wingert RA. Kidney organogenesis in the zebrafish: insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip Rev Dev Biol. 2013;2:559–585. doi: 10.1002/wdev.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cheng CN, Verdun VA, Wingert RA. Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom., and Notch signaling. Dev Biol. 2014;386:111–122. doi: 10.1016/j.ydbio.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CN, Wingert RA. Nephron proximal tubule patterning and corpuscles of Stannius formation are regulated by the sim1a. transcription factor and retinoic acid in the zebrafish. Dev Biol. 2015;399:100–116. doi: 10.1016/j.ydbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Cheng CN, Li Y, Marra AN, Verdun V, Wingert RA. Flat mount preparation for observation and analysis of zebrafish embryo specimens stained by whole mount in situ hybridization. J Vis Exp. 2014. p. e51604. [DOI] [PMC free article] [PubMed]
- Cheng CN, Wingert RA. Chapter 9: Renal system development in the zebrafish: a basic nephrogenesis model. In: Carver E, Lessman C, editors. Zebrafish: Topics in Reproduction & Development. Nova Scientific Publishers; 2014. [Google Scholar]
- Gerlach GF, Wingert RA. Zebrafish pronephros tubulogenesis and epithelial identity maintenance are reliant on the polarity proteins Prkc iota .and zeta. Dev Biol. 1016;396:183–200. doi: 10.1016/j.ydbio.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee R, Gerlach GF, Jou J, Cheng CN, Wingert RA. Temporal and spatial expression of tight junction genes during zebrafish pronephros development. Gene Expr Patterns. 2014;16:104–113. doi: 10.1016/j.gep.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Holzemer NF, Wingert RA. Laser ablation of the zebrafish pronephros to study renal epithelial regeneration. J Vis Exp. 2011. p. e2845. [DOI] [PMC free article] [PubMed]
- McCampbell KK, Wingert RA. New tides: using zebrafish to study renal regeneration. Transl Res. 2014;163:109–122. doi: 10.1016/j.trsl.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach GF, Schrader LN, Wingert RA. Dissection of the adult zebrafish kidney. J Vis Exp. 2011;54:e2839. doi: 10.3791/2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCampbell KK, Springer KN, Wingert RA. Analysis of nephron composition and function in the adult zebrafish kidney. J Vis Exp. 2014. p. e51644. [DOI] [PMC free article] [PubMed]
- McCampbell KK, Springer K, Wingert RA. Atlas of cellular dynamics during zebrafish adult kidney regeneration. Stem Cells Int. 2015;In press doi: 10.1155/2015/547636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SL, et al. Hypoxia-induced pathological angiogenesis mediates tumor cell dissemination, invasion, and metastasis in a zebrafish tumor model. Proc Natl Acad Sci USA. 2009;106:19485–19490. doi: 10.1073/pnas.0909228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhi P, et al. Hypoxia-induced metastasis model in embryonic zebrafish. Nat Prot. 2010;5:1911–1918. doi: 10.1038/nprot.2010.150. [DOI] [PubMed] [Google Scholar]
- Cao R, Jensen LDE, Söll I, Hauptmann G, Cao Y. Hypoxia-induced retinal angiogenesis in zebrafish as a model to study retinopathy. PLoS One. 1371;3:e2748. doi: 10.1371/journal.pone.0002748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, et al. Hypoxia-induced retinopathy model in adult zebrafish. Nat Prot. 2010;5:1903–1910. doi: 10.1038/nprot.2010.149. [DOI] [PubMed] [Google Scholar]


