Abstract
Cancer cells respond to matrix mechanical stiffness in a complex manner using a coordinated, hierarchical mechano-chemical system composed of adhesion receptors and associated signal transduction membrane proteins, the cytoskeletal architecture, and molecular motors1, 2. Mechanosensitivity of different cancer cells in vitro are investigated primarily with immortalized cell lines or murine derived primary cells, not with primary human cancer cells. Hence, little is known about the mechanosensitivity of primary human colon cancer cells in vitro. Here, an optimized protocol is developed that describes the isolation of primary human colon cells from healthy and cancerous surgical human tissue samples. Isolated colon cells are then successfully cultured on soft (2 kPa stiffness) and stiff (10 kPa stiffness) polyacrylamide hydrogels and rigid polystyrene (~3.6 GPa stiffness) substrates functionalized by an extracellular matrix (fibronectin in this case). Fluorescent microbeads are embedded in soft gels near the cell culture surface, and traction assay is performed to assess cellular contractile stresses using free open access software. In addition, immunofluorescence microscopy on different stiffness substrates provides useful information about primary cell morphology, cytoskeleton organization and vinculin containing focal adhesions as a function of substrate rigidity.
Keywords: Bioengineering, Issue 100, Primary human colon tumor cells, Soft Elastic Substrates, Traction force Microscopy, Mechanobiology, Immunofluorescence Microscopy, Cell mechanics
Introduction
In recent years it has become increasingly evident that mechanical micro-environment, in addition to bio-chemical factors, plays an important role in regulating cell functionalities. Cells can sense and respond to the substrate stiffness on which they are adhered to (as in 2D culture) or surrounded by (as in 3D culture) 3-7. By doing so, cells can modulate their differentiation3, morphology4, migration/motility5, bio-physical properties6, growth7, and other processes.
Cancer cells also respond to the 2D and 3D matrix stiffness using a coordinated, hierarchical mechano-chemical combination of adhesion receptors and associated signal transduction membrane proteins, the cytoskeletal architecture, and molecular motors1, 2. For example, mammary epithelial cells (MECs) form normal acinar parenchyma when cultured on 150 Pa substrates that is similar to the stiffness of healthy mammary tissue. Interestingly, they exhibit the hallmarks of a developing tumor, both structural and transcriptional, when cultured on stiffer substrates (> 5,000 Pa) that mimic the stiffness of a tumor stroma8. In addition, another experiment shows that breast tumorigenesis is accompanied by collagen crosslinking and ECM stiffening9. Recent experiments show that human colon carcinoma (HCT-8) cells display metastasis like phenotype (MLP) when they are cultured on 2D substrates having physiologically relevant stiffness (20–47 kPa), but not on very stiff (3.6 GPa) substrates10-12.These cells first form tumor-like cell clusters and then dissociate from one another, starting from the periphery. As this epithelial to rounded morphological (E to R transition) change occurs, they proliferate, reduce cell-cell and cell-ECM adhesion, and become migratory. HCT-8 cells cultured on very hard polystyrene substrates do not exhibit these malignant traits. Thus it has been hypothesized that HCT-8 cells become metastatic due to their exposure to appropriate micro-environment. It is worth noting that these experiments are carried out with immortalized cancer cell lines or murine derived primary cells, not with primary human cancer cells.
A recent study proposes that augmented cellular traction stress may be used as a potential biophysical signature for metastatic cells13.The study involves measuring traction force for different human cancer cell lines on polyacrylamide gels. It is found that metastatic cancer cells can exert significantly higher traction stress compared to non-metastatic cells in all cases13. However, these results directly contradict the earlier published findings on murine derived breast cancer cell lines14. Also, a recent study highlights remarkable differences between immortalized and primary human cells in their cytoskeletal remodeling protein profiling and cell survival protein expression15. Hence, it is important to revisit many of the biophysical assays including traction for primary human cancer cells. This will address the question whether the primary cells recapitulate immortalized cancer cell lines traction trend.
The protocol described here is optimized for isolation of primary human colon cells (both healthy and cancerous), and for culturing them on soft substrates (polyacrylamide hydrogels) as well as on Petri dishes. The protocol is based on digestion and consequent enzymatic dissociation of surgical tissue sample into single cell suspension16. To our knowledge, this is the first demonstration of culturing isolated primary colon tumor and normal cells directly on soft hydrogel substrates with embedded fluorescent microbeads for traction cytometry. Transparent gel substrates also allow immunostaining. This assay revealed differences in F-actin organization and focal adhesions in primary human colon cells as substrate stiffness changes. This cell culture platform opens up the possibility of exploring various biophysical properties of primary human cells such as cell stiffness and traction as parameters for cancer prognostics.
Protocol
The protocol described below follows the guidelines of UIUC human research ethics committee.
1. Collection and Digestion of Surgical Tissue Sample
Collect the tumor tissue sample right after colon resection (Figure 1A and 1B). Collect tissue from an adjacent healthy site as well.
Transfer the tissue immediately to a 15 ml vial containing 12 ml HBSS solution. Keep the vial on ice inside an insulated foam box.
Transport the tissue containing vial to a tissue culture hood for further processing within 45 min. Keep the vial on an ice block inside the hood.
Pour the supplied tissue into a 6 well plate containing 6-7 ml of HBSS solution using a pipette. Note: Amount of tissue per well is not critical in this rinsing step.
Keep the 6 well plate on an ice block. Rinse the tissue in HBSS solution twice.
Cut tissue cleanly into separated sections with sterile scissor. Mince tissue into smaller sections no greater than 1-2 mm3 in size with a sterile scalpel blade.
Weigh a labelled 0.1% trypsin vial (containing 1 ml of 0.1% trypsin solution) before transferring the tissue. Transfer ~20 mg tissue into each of the 0.1% trypsin vials with a pipette.
Try to minimize the transfer of HBSS into the trypsin vial to avoid further dilution of trypsin.
Weigh the trypsin vials after tissue transfer; document the difference as the tissue mass.
Pour ~20 mg tissue and 1 ml trypsin into each well of a 24 well plate.
Put the 24 well plate on shaker in fridge at 4 °C for 16-20 hr. During this wait period or at an earlier time, prepare polyacrylamide gel substrates and functionalize them with ECM proteins as described in section 3.
2. Enzymatic Dissociation of Tissue Sample into Single Cell Suspension and Culturing Isolated Cells on ECM Functionalized Elastic Substrates and Rigid Polystyrene Dishes
After 16-20 hr of incubation in trypsin well at 4 °C on shaker, transfer tissue from 4 °C trypsin to 4 °C complete growth media vial (containing 2-3 ml of complete growth media) using a pipette. Ensure minimal transfer of trypsin into growth media. Note: Complete growth media formulation is as follows: RPMI 1640 base medium supplemented with horse serum to a final concentration of 10% and penicillin-streptomycin to 1% of total solution.
Place the tissue containing media vial in a warm water bath at 37 °C for 12-15 min.
Transfer the tissue to a 15 ml vial containing HBSS solution using a pipette, shake gently.
Repeat 2.3.
Remove tissue from HBSS solution and transfer to 0.1% collagenase solution (in HBSS) vial containing 1 ml solution. Incubate for 45 min at 37 °C and 5% CO2 environment in a humidified cell culture incubator. During the wait period, warm growth media in a 15 ml vial at 37 °C in water bath.
Pull out all tissue fragments into pipette minimizing collagenase carryover. Eject just the tissue fragments into pre-warmed culture media vials.
Place a 40 µm well-insert filter into each well of a 6 well plate. Moisten the filter with 1 ml cell culture media.
Triturate tissue in media with a 10 ml glass pipette several times until tissue is completely dissociated.
Maintain the pipette tip as close as possible to the base of the vial.
Deposit the solution onto the filter to remove debris and undissociated parts.
Centrifuge filtered cell suspension in media at 150 x g for 5 min at RT.
Remove the supernatant afterwards and resuspend cell pellet with 2 ml fresh culture media.
Count the cells using a hemocytometer (if required). Seed isolated primary cells on extracellular matrix (ECM) functionalized gels and polystyrene dishes at desired concentration.
3. Preparation and Functionalization of Different Stiffness Polyacrylamide (PA) Gels and Polystyrene Substrates
Note: For visual demonstration of Section 3, the viewers/readers are referred to a recent Journal of Visualized Experiments (JoVE) article17.
- Adopting published protocols18, 19, activate 12 mm2 glass cover slips chemically to ensure covalent binding of the hydrogel.
- First, treat glass cover slips with 3- Aminopropyltrymethoxysilane (ATS) for 7 min at RT.
- Remove the ATS completely with DI water rinse and treat cover slips with 0.5% Glutaraldehyde (diluted in PBS from 70% Glutaraldehyde stock solution) solution for 30 min.
- Obtain 2 kPa gel solutions by mixing 5% w/v acrylamide solution and 0.05% N,N'-methylenebisacrylamide (bis) solution in 10 mM HEPES-buffered saline20. Use 8% w/v acrylamide and 0.13% N,N'-methylenebisacrylamide (bis) solution in 10 mM HEPES-buffered saline for 10 kPa gel20.
- In both cases, use 1:200 ammonium persulfate (10% w/v) and 1:2,000 N,N,N',N'-tetramethylethylenediamine (TEMED) as the initiator and catalyst for the polymerization process, respectively.
- Also, add 100 µl fluorescent beads to 2 kPa gel solution as fiduciary markers 21, 22.
Deposit a drop of 20 μl pre-polymer PA gel solution on the 12 mm2 activated glass cover slip. Place another 12 mm2 regular glass cover slip on the drop. Ensure that the drop spreads between the cover slips due to capillarity. Invert the sandwich for 2 kPa gels to ensure that most of the beads come near the cell culture surface.
Cure the PA gel for 45 min at RT.
Peel off the top cover slip using a single edge razor. Note: During peeling, detachment proceeds from one edge of the sandwich. The gel remains adherent to the activated glass slide.
Store the gels in PBS solution before use.
- Functionalize PA gels and glass with ECM molecules, human fibronectin at a concentration of 50 µg/ml following published methods 23.
- Briefly, incubate substrates with 2 ml pure hydrazine hydrate O/N.
- Remove hydrazine hydrate and rinse the substrates thoroughly with DI water.
- Wash the substrates with 5% acetic acid for 30 min.
- Remove acetic acid and rinse the substrates thoroughly with DI water.
- Keep the substrates immersed in DI water for 30 min.
- Incubate the substrates with oxidized fibronectin for 35 min at a concentration of 50 µg/ml.
- Rinse the substrates with PBS on shaker at low rpm for 10 min.
- Incubate all substrates at 37 °C in 2-3 ml culture media for 30 min before plating the cells. Note: Plate cells in a sparsely populated manner (1,000-3,000 cells/cm2). Each gel-covered glass slip needs to be contained in a 35 mm Petri dish. Allow cells to adhere completely before any microscopy (at least O/N).
4. Traction Force Microscopy and Immunofluorescence Microscopy Assays
- Traction Force Microscopy Assay
- Warm 0.25% trypsin-EDTA / 10% SDS solution at 37 °C in water bath for 10 minutes before traction experiments start.
- Remove one gel from the cell culture incubator at a time and place on the fluorescent inverted microscope stage (32X magnification).
- Find a single cell in field of view. Remove the Petri dish lid.
- Take a phase contrast image of cell (e.g., Figure 3).
- Switch the imaging mode to fluorescence and select appropriate filter. Don’t move the microscope stage or sample during this time.
- Take an image of the fluorescent beads displaced by cellular traction (Figure 4C).
- Add 1 ml trypsin/SDS solution to Petri dish to detach the cell from gel. Don’t move the microscope stage or sample during this time. Also, take a control image of the cell to ensure complete removal from gel.
- Take a reference (null force) image of the beads after the cell is removed.
- Repeat steps 4.1.2-4.1.8 for all other gels.
- Make an image stack using ImageJ from the two images acquired in step 4.1.6 and 4.1.8 for each case. To generate the image stack, use following sequence of commands in ImageJ after opening the images: Image → Stacks → Images to stack.
- To obtain the displacement field and traction, use ImageJ plugins following published methods 21, 22. Obtain codes and detailed tutorials for these plugins using the following link: https://sites.google.com/site/qingzongtseng/imagejplugins.
- Align the images in the stack using the ‘Template Matching’ plugin. Use following sequence of commands in ImageJ after opening the image stack generated in step 4.1.10: Plugins → Template Matching → Align Slices in Stack → OK. Save the image stack consequently. Use this new image stack in the following steps.
- Obtain the displacement field using the PIV (particle image velocimetry) plugin (Figure 4D). Use following sequence of commands after opening the image stack saved in step 4.1.11.1: Plugins → PIV → Iterative PIV (Basic) → OK → Accept this PIV and output → Ok. Save the PIV output in the same directory as in the original image stack. Use this as input in next step.
- Finally use the FTTC (Fourier transform traction cytometry) plugin to obtain the traction map (Figure 4E). Use following sequence of commands: Plugins → FTTC → Insert material properties, i.e., Young’s modulus and Poisson’s ratio of PA gel → OK → Select the PIV output file saved in step 4.1.11.2 → OK. Save FTTC results in the same directory as in the image stack and PIV output.
- Immunofluorescence Microscopy Assays
- Take the substrates to be immunostained to the laminar hood and remove the culture media.
- Rinse the substrates with PBS and fix the cells with 4% paraformaldehyde in PBS for 20 min at RT.
- Wash the substrates with PBS for 3 times (5 min each time). Consequently, incubate the substrates with 500 µl signal enhancer for 30 min and rinse with PBS.
- Incubate cells with monoclonal anti-vinculin antibody at a 1:250 dilution in PBS for 45 min at RT. Wash the substrates with PBS for 3 times (5 min each time).
- Incubate the samples with secondary antibody Alexa Fluor 488 goat anti-mouse IgG at a 1:200 dilution in PBS at RT for 30 min. Wash the substrates with PBS for 3 times (5 min each time).
- To visualize the F-actin structure, incubate cells with TRITC phalloidin conjugates at a concentration 50 µg/ml for 45 min at RT. Wash the substrates with PBS for 3 times (5 min each time).
- Image the samples using confocal scanning laser microscope (Figure 5A-5D).
Representative Results
The protocol described above is successfully employed for multiple tissue samples (n=12) from four different patients under guidelines of the institutional review board. Figure 1A illustrates a representative colorectal tumor right after surgery from which the tissue sections for cell cultures are obtained. A typical tissue section in HBSS solution after transfer to the laminar hood for further processing is shown in Figure 1B. A schematic of digestion and enzymatic dissociation of surgical tissue sample into single cell suspension is shown in Figure 2.
After successful tissue dissociation, isolated primary human cells are seeded on PA gels and polystyrene dishes. Phase contrast micrographs (Figure 3A and 3B) illustrate the representative morphology of primary human colon cancer and normal cells as a function of substrate rigidity. It is evident that primary colon cells (both healthy and cancerous) spread more on polystyrene dishes compared to the PA gels (Figure 3A and 3B).
Traction assays are performed on ECM (fibronectin) coated soft 2 kPa gels after confirmation of invasive adenocarcinoma from pathological H & E staining (Figure 4A). PA gels are uniformly coated with fibronectin as shown in Figure 4B. For ease of comparison, all traction experiments for tumor and healthy colon cells should be performed at the same time. Figure 4C shows a snapshot of the nanoscale fluorescent beads embedded inside the gel. From the displaced and reference bead images, the displacement fields are obtained using the ImageJ PIV plugin21, 22. A representative bead displacement field generated by invasive colon tumor cells on 2 kPa gel is displayed in Figure 4D. Figure 4E shows traction stress obtained using ImageJ FTTC plugin corresponding to displacement field in Figure 4D 21, 22.
Figure 5 shows the F-actin and vinculin containing focal adhesions of primary human colon cells on soft 2 kPa gels and rigid polystyrene substrates. No actin fiber was present in the less spread cells on soft gels (Figure 5A1-5A2). Punctate vinculin containing focal adhesions are present on soft gels (Figure 5B1-5B2). Conversely, primary cells show well spread morphology, well defined actin stress fibers and discrete elongated focal adhesions on rigid polystyrene substrates (Figures 5C-5D).
 Figure 1. (A) Tumor after colon resection. (B) Tissue sections from the tumor for cell isolation. Please click here to view a larger version of this figure.
Figure 1. (A) Tumor after colon resection. (B) Tissue sections from the tumor for cell isolation. Please click here to view a larger version of this figure.
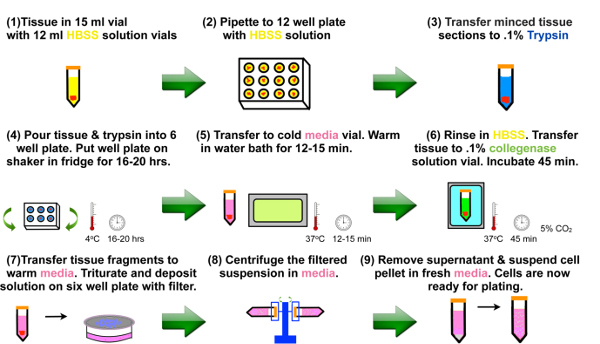 Figure 2.
Schematic of digestion and enzymatic dissociation of surgical tissue sample into single cell suspension.
Please click here to view a larger version of this figure.
Figure 2.
Schematic of digestion and enzymatic dissociation of surgical tissue sample into single cell suspension.
Please click here to view a larger version of this figure.
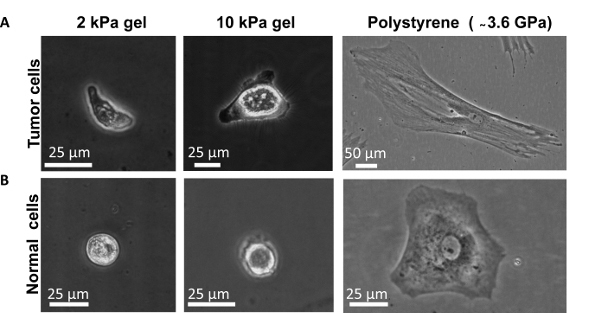 Figure 3.Successful primary human colon cell culture (both cancer and normal) on different stiffness gels and on polystyrene dish. Phase contrast images show typical morphology of (A) Tumor cells and (B) Normal cells on different stiffness substrates. Please click here to view a larger version of this figure.
Figure 3.Successful primary human colon cell culture (both cancer and normal) on different stiffness gels and on polystyrene dish. Phase contrast images show typical morphology of (A) Tumor cells and (B) Normal cells on different stiffness substrates. Please click here to view a larger version of this figure.
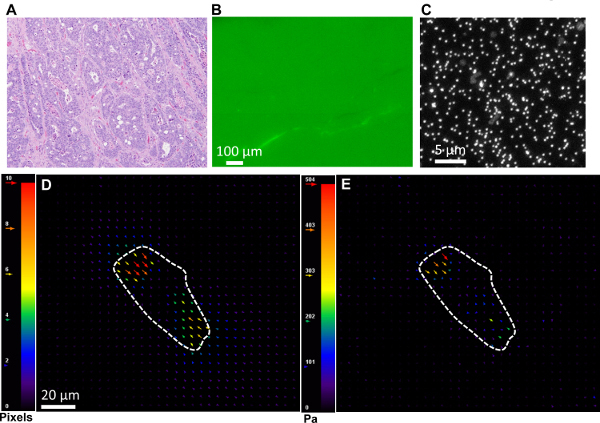 Figure 4. (A) H & E staining confirming invasive adenocarcinoma. (B) Fibronectin staining reveals that gels are uniformly coated with ECM molecules. (C) Nanoscale beads embedded inside gel as fiduciary markers. (D) Representative displacement field generated by tumor cell on soft PA gel using PIV plugin in ImageJ as described in 4.1.11.2. (E) Traction stress exerted by tumor cell on soft gel corresponding to displacement field in (D) obtained via FTTC plugin in ImageJ as described in 4.1.11.3. Please click here to view a larger version of this figure.
Figure 4. (A) H & E staining confirming invasive adenocarcinoma. (B) Fibronectin staining reveals that gels are uniformly coated with ECM molecules. (C) Nanoscale beads embedded inside gel as fiduciary markers. (D) Representative displacement field generated by tumor cell on soft PA gel using PIV plugin in ImageJ as described in 4.1.11.2. (E) Traction stress exerted by tumor cell on soft gel corresponding to displacement field in (D) obtained via FTTC plugin in ImageJ as described in 4.1.11.3. Please click here to view a larger version of this figure.
 Figure 5. Immunofluorescent staining of F-actin and vinculin on soft gel and on hard polystyrene substrates. No actin fiber was present in the less spread (A1) tumor cell or (A2) normal cell on soft 2 kPa gels. Punctate vinculin containing focal adhesions are present on soft gels (B1-B2). Conversely, primary cells show well spread morphology, well defined actin stress fibers (C1-C2) and discrete elongated focal adhesions (D1-D2) on rigid polystyrene substrates. Please click here to view a larger version of this figure.
Figure 5. Immunofluorescent staining of F-actin and vinculin on soft gel and on hard polystyrene substrates. No actin fiber was present in the less spread (A1) tumor cell or (A2) normal cell on soft 2 kPa gels. Punctate vinculin containing focal adhesions are present on soft gels (B1-B2). Conversely, primary cells show well spread morphology, well defined actin stress fibers (C1-C2) and discrete elongated focal adhesions (D1-D2) on rigid polystyrene substrates. Please click here to view a larger version of this figure.
Discussion
Cellular traction stress has recently emerged as a potential biophysical indicator of metastatic state13. However, no experimental traction data with primary tumor cells exists in literature to date. Also, directly culturing isolated primary colon cells on different stiffness polyacrylamide gels is not reported yet. Hence, we establish an optimized primary colon cell culture conditions on gels and polystyrene (Figure 2). Fluorescent microbeads encapsulation near cell culture surface during soft gel preparation enables the measurement of displacement field and traction generated by primary human cells (Figure 4C, 4D and 4E). In addition, immunofluorescence assays can provide important information regarding cytoskeleton organization and focal adhesions as substrate stiffness changes (Figure 5A-5D). Note that different cancer and normal immortalized cell lines also show augmented spreading, actin stress bundle formation and mature elongated focal adhesions with increased substrate stiffness4, 24-25.
The protocol can be easily adopted/modified for isolation of primary cells from surgical tissues of different human organs or other animals. The incubation time of tissue in the dissociation agents — trypsin and collagenase — needs to be optimized empirically for each application. In addition, the filter size for removing debris and undissociated tissue parts is also an important parameter to choose based on expected maximum cell size in suspended state (40 µm in this protocol).
One limitation of the method is that a few soft gels may lack consistent localization of the fluorescent beads near cell culture surface (Section 3.3) which may adversely affect the following traction cytometry measurements (Section 4.1). This can be easily circumvented by examining the gel surface for bead density and distribution using fluorescent microscopy before cell culture. Consequently, gels with relatively non-homogenous bead distribution can be discarded. Another approach is to use a recently proposed method that allows precise localization of the beads near the cell culture surface17.
Caution should be taken to start processing the surgical samples as soon as they are received, preferably within 45 minutes. The most critical step in the protocol is the optimum exposure time of the tissue in the trypsin and collagenase solution to achieve isolation yield, yet retaining sufficient cell viability. Before culturing the cells on soft hydrogels, the bead density and distribution needs to be confirmed using fluorescent microscopy. Also, a null force image needs to be acquired after confirming that the cell is completely detached from the gel surface.
In summary, a protocol is described that results dissociated single cells suspension from surgical cancerous/normal tissue sample. This isolation of single primary tumor cells from surgical specimens followed by their culture on soft and hard substrates allows various downstream biophysical measurements, e.g., cell stiffness using AFM, intracellular rheology using microinjected particles, cell migration/motility, and vesicle dynamics analysis. These measurements may be used for cancer prognosis. The protocol has been successfully used for the extraction and culture of murine cardiomyocytes as well26. The method presented here may be used to isolate primary human cells from surgical tissues of various organs and to culture them directly on different stiffness substrate for mechanobiology studies.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This project was funded by the National Science Foundation ECCS grant 10-02165, and the Interdisciplinary Innovation Initiative Program, University of Illinois grant 12035. M.Y. A. was funded at UIUC from NIH National Cancer Institute Alliance for Nanotechnology in Cancer ‘Midwest Cancer Nanotechnology Training Center’ Grant R25 CA154015A. Immnunostaining and confocal microscopy imaging were carried out at the Institute for Genomic Biology (IGB), UIUC. M.Y.A. acknowledges the discussions with B. J. Williams of UIUC regarding the isolation experiments. M.Y.A. acknowledges C. Nemeh and Abdul Bhuiya of UIUC for assistance in schematic and materials list preparation.
References
- Ingber DE. Can cancer be reversed by engineering the tumor microenvironment. Semin. Cancer Biol. 2008;18:356–364. doi: 10.1016/j.semcancer.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Weaver VM. Mechanics, malignancy, metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113–127. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Yeung T, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Wang YL. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94(25):3661–3665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys. J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Dembo M, Wang YL. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Cell. Physiol. 2000;279:1345–1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, et al. Mechanical force affects expression of an in vitro metastasis-like phenotype in HCT-8 cells. Biophys. J. 2010;99:2460–2469. doi: 10.1016/j.bpj.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MY, Saif MTA. Substrate Stiffness Mediated Metastasis Like Phenotype of Colon Cancer Cells is Independent of Cell to Gel Adhesion. Cell. Mol. Bioeng. 2014.
- Ali MY, Chuang CY, Saif MTA. Reprogramming cellular phenotype by soft collagen gels. Soft Matter. 2014. [DOI] [PMC free article] [PubMed]
- Kraning-Rush CM, Califano JP, Reinhart-King CA. Cellular traction stresses increase with increasing metastatic potential. PLoS ONE. 2012;7(2):e32572. doi: 10.1371/journal.pone.0032572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra I, Undyala V, Kandow C, Thirumurthi U, Dembo M, Beningo KA. An in vitro correlation of mechanical forces and metastatic capacity. Phys. Biol. 2011;8(1) doi: 10.1088/1478-3975/8/1/015015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alge CS, Hauck SM, Priglinger SG, Kampik A, Ueffing M. Differential protein profiling of primary versus immortalized human RPE cells identifies expression patterns associated with cytoskeletal remodeling and cell survival. J. Proteome Res. 2006;5(4):862–878. doi: 10.1021/pr050420t. [DOI] [PubMed] [Google Scholar]
- Oikonomou E, Kothonidis K, Zografos G, Nasioulas G, Andera L, Pintzas A. Newly established tumourigenic primary human colon cancer cell lines are sensitive to TRAIL-induced apoptosis in vitro and in vivo. Br. J. Cancer. 2007;97(1):73–84. doi: 10.1038/sj.bjc.6603835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll SG, Ali MY, Saif MTA. A novel method for localizing reporter fluorescent beads near the cell culture surface for traction force microscopy. J. Vis. Exp. 2014. [DOI] [PMC free article] [PubMed]
- Wang YL, Pelham R. Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Methods in Enzymology. 1998;298:489–496. doi: 10.1016/s0076-6879(98)98041-7. [DOI] [PubMed] [Google Scholar]
- Tang X, Ali MY, Saif MTA. A novel technique for micro-patterning proteins and cells on polyacrylamide gels. Soft Matter. 2012;8:3197–3206. doi: 10.1039/C2SM25533B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Current Protocols in Cell Biology. 2010. pp. 10–16. [DOI] [PubMed]
- Tseng Q, et al. Spatial organization of the extracellular matrix regulates cell–cell junction positioning. Proc. Natl. Acad. Sci. USA. 2012;109(5):1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A, et al. Augmentation of integrin-mediated mechanotransduction by hyaluronic acid. Biomaterials. 2014;35(1):71–82. doi: 10.1016/j.biomaterials.2013.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damljanovic V, Lagerholm BC, Jacobson K. Bulk and micropatterned conjugation of extracellular matrix proteins to characterized polyacrylamide substrates for cell mechanotransduction assays. Biotechniques. 2005;39(6):847–851. doi: 10.2144/000112026. [DOI] [PubMed] [Google Scholar]
- Tilghman RW, et al. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS ONE. 2010;5(9):e12905. doi: 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BJ, Anand SV, Rajagopalan J, Saif MTA. A self-propelled biohybrid swimmer at low Reynolds number. Nat. Commun. 2014;5 doi: 10.1038/ncomms4081. [DOI] [PubMed] [Google Scholar]


