Summary
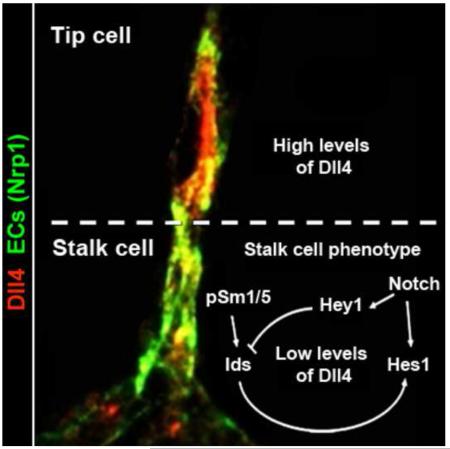
Gradients of VEGF induce single endothelial cells to become leading tip cells of emerging angiogenic sprouts. Tip cells then suppress tip cell features in adjacent stalk cells via Dll4/Notch mediated lateral inhibition. We report here that Smad1/Smad5 mediated BMP signaling synergizes with Notch signaling during selection of tip and stalk cells. Endothelium-specific inactivation of Smad1/Smad5 in mouse embryos results in impaired Dll4/Notch signaling and increased numbers of tip cell-like cells at the expense of stalk cells. Smad1/5 downregulation in cultured endothelial cells reduced the expression of several target genes of Notch and of other stalk cell-enriched transcripts (Hes1, Hey1, Jagged1, VEGFR1 and Id1-3). Moreover, Id proteins act as competence factors for stalk cells and form complexes with Hes1, which augment Hes1 levels in the endothelium. Our findings provide in vivo evidence for a regulatory loop between BMP/TGFβ-Smad1/5 and Notch signaling that orchestrates tip versus stalk cell selection and vessel plasticity.
Keywords: sprouting angiogenesis, tip cell, stalk cell, mouse embryo, Smad, BMP, Dll4/Notch, lateral inhibition, polarity, directed migration
Graphical Abstract
Introduction
Increasing demands for blood supply during embryogenesis, wound healing and certain diseases require the formation of new blood vessels by sprouting angiogenesis (De Smet et al., 2009; Eilken and Adams, 2010). Sprouting angiogenesis involves the selection of a leading tip cell and the trailing stalk cells in a vessel, elongation of the new sprout, anastomosis, perfusion and ultimately the stabilization of the newly formed vessel. Gradients of VEGF-A trigger the selection of single endothelial cells (ECs) to become the leading tip cells that guide emerging sprouts (Gerhardt et al., 2003; Ruhrberg et al., 2002). In response to VEGF-A/VEGFR2-mediated signaling, tip cells become enriched in Delta-like 4 (Dll4), a ligand for Notch, and instruct adjacent ECs to become stalk cells via Dll4/Notch1-mediated lateral inhibition (Hellstrom et al., 2007; Harrington et al., 2008; Lobov et al., 2007). In the stalk cells, Hes1 and Hey1, target genes for Notch signaling, down regulate the levels of VEGFR2 and Dll4, thereby decreasing transiently the responsiveness to the tip cell inducing stimuli. This mechanism balances the numbers of tip cells required for effective sprouting and network formation (Hellstrom et al., 2007; Leslie et al., 2007; Noguera-Troise et al., 2006; Ridgway et al., 2006; Siekmann and Lawson, 2007). The tip and stalk cell phenotypes are remarkably transient and exchangeable as ECs dynamically shuffle position along the angiogenic sprout, and compete for the tip cell position (Jakobsson et al., 2010). A continuous re-evaluation of VEGFR/Dll4/Notch signaling is required when migrating ECs meet new neighbors, yet it remains obscure what other signals regulate this tip and stalk cell shuffling. Hence, this requires the identification of candidate pathways that may trigger this cellular competition.
Many signaling components of the bone morphogenetic protein (BMP) and transforming growth factor type beta (TGFβ) pathways play pivotal but often poorly defined roles in angiogenesis in development and disease (David et al., 2009; Pardali et al., 2010). Mutations in the genes encoding the endothelial specific TGFβ co-receptor Endoglin/CD105 (ENG) and Activin receptor-like kinase 1 (ALK1), a type I receptor, cause hereditary haemorrhagic telangiectasia. Moreover, targeted inactivation of ALK1 can complement anti-VEGF therapies to inhibit normal and tumor angiogenesis in mouse and humans (Hu-Lowe et al., 2011). Smad1 and Smad5 are intracellular effector proteins of BMP and TGFβ/Endoglin/ALK1 signaling in ECs. The genetic inactivation of Smad1 or Smad5 in mice results in early embryonic lethality due to several embryonic and extraembryonic defects that include cardiovascular malformations (Chang et al., 1999; Lechleider et al., 2001; Tremblay et al., 2001; Yang et al., 1999). The endothelium specific inactivation of Smad5 results however in normal and viable animals (Umans et al., 2007), which suggests that Smad1 functionally compensates for Smad5 absence in angiogenic endothelium.
Cross-signaling between Notch and BMP/Smad pathways has been documented in various cell types (Bai et al., 2007; Dahlqvist et al., 2003). For instance, Smad-mediated BMP signaling acts as a competence factor for the robust expression of target genes of Notch, and the crosstalk of both signaling cascades is required for the inhibition of the projection neuron fate in the future photoreceptors in Drosophila (Quillien et al., 2011). In ECs, Smad1 and Smad5 form upon receptor-mediated activation a complex with the Notch intracellular domain (NICD) to potentiate downstream target gene expression for both pathways (Itoh et al., 2004). Hes and Hey/Herp are primary target genes of Notch signaling and encode basic helix-loop-helix (bHLH) proteins that function as transcriptional repressors of e.g. VEGFR2/3, Dll1, Dll4 and Jagg1 (Henderson et al., 2001; Kobayashi and Kageyama, 2010; Kobayashi et al., 2009). Downstream of BMP/Smad signaling, members of the Id family of HLH proteins negatively regulate cell differentiation and stimulate cell cycle progression (Norton and Atherton, 1998; Zebedee and Hara, 2001). In cultured cells Id1 stimulates EC migration and tube formation (Valdimarsdottir et al., 2002), and Hey1 antagonizes BMP/Id1-induced migration of ECs by promoting Id protein degradation (Itoh et al., 2004). Conversely, in neuronal progenitor cells Id proteins interact directly with Hes1 through their HLH domain and suppress the DNA-binding activity of Hes1, thereby releasing the negative feedback loop of Hes1 on its own promoter and stabilizing Hes1 expression (Bai et al., 2007). Interestingly, the formation of Id/Hes1 heteromers preserves the ability of Hes1 to affect other target genes that ultimately leads to inhibition of precocious neurogenesis. Thus, these roles for TGFβ family members and Notch signaling summarized above prompted us to study the importance of Smad1/5 in embryonic angiogenesis, specifically in the regulation of Dll4/Notch mediated suppression of the tip cell behavior.
Here we present evidence that crosstalk between Smad1/5 and Notch signaling orchestrates angiogenic sprouting in mid-gestation mouse embryos by securing the right balance between tip and stalk cells. Genetic co-inactivation of Smad1 and Smad5 in ECs results in defective vascular remodeling, excessive sprouting, impaired tip cell polarity and embryonic lethality. We demonstrate that Smad1/5 regulate directed EC migration, and synergistically activate the expression of target genes of Dll4/NICD in stalk cells. Furthermore, downstream of Smad1/5, Id proteins strengthen Notch signaling by forming heteromers with Hes1 proteins, leading to increased/stabilized Hes1 levels in the endothelium. Hence, Smad1/5 act as crucial regulators of stalk cell competence and blood vessel plasticity.
Results
Smad1 and Smad5 mediated signaling is required for the developing vasculature
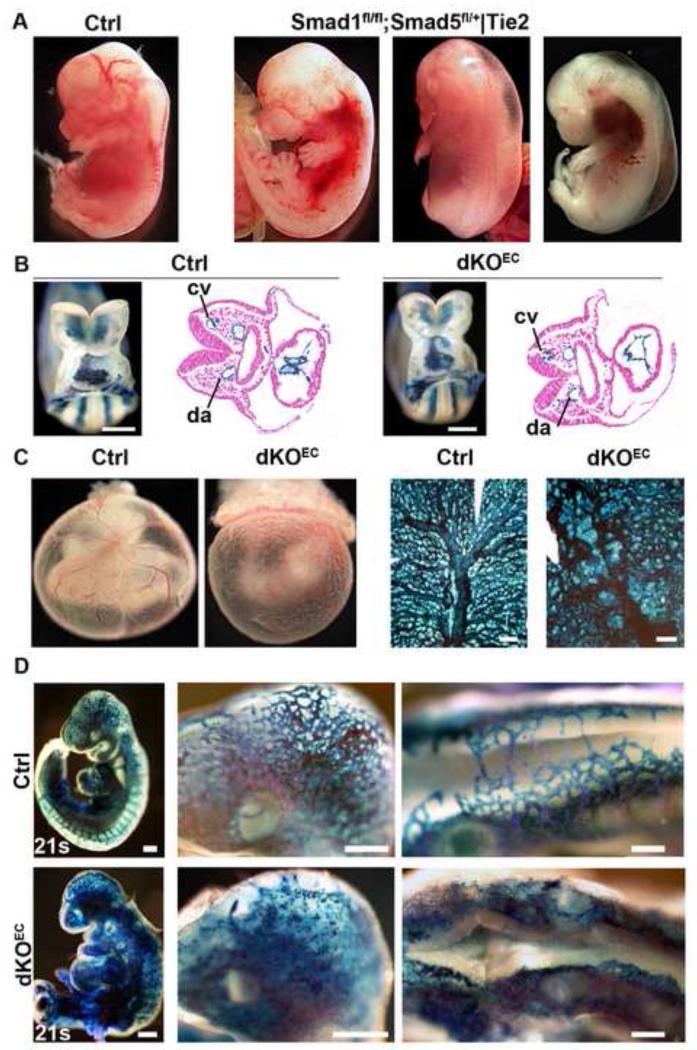
Endothelium-specific (Tie2-Cre) Smad1;Smad5 knockout (KO) mice were generated to investigate the role of these cognate Bmp-Smads during angiogenesis. Doing so, we also observed a crucial gene dosage effect for Smad1 and Smad5 mediated signaling in the endothelium. Compound Smad1 and Smad5 heterozygosity (Tie2-Cre+/0;Smad1fl/wt;Smad5fl/wt) was compatible with normal development and postnatal life (not shown). A further decrease in the number of Smad1 (Tie2-Cre+/0;Smad1fllfl;Smad5fl/wt) or Smad5 (Tie2-Cre+/0;Smad1fl/wt;Smad5fl/fl) alleles resulted in normal onset of angiogenesis at embryonic day (E)9.5 (not shown), but such embryos still died later during gestation (E14.5). This is due to severe angiogenesis and lymphangiogenesis defects characterized by severe bleedings, oedema or a combination of both, and cardiac defects (Fig. 1a, data not shown). Blood and lymphatic vessels were hyperplastic and abnormal (not shown). Embryos lacking all four Smad1 and Smad5 alleles in ECs (Tie2-Cre+/0;Smad1fl/fl;Smad5fl/fl) developed earlier and more severe angiogenesis defects leading to embryonic lethality already at E10.5 (Figure 1b-d). In this study we further analyzed the endothelium-specific Smad1;Smad5 double knockout embryos (dKOEC). Such embryos underwent vasculogenesis at E8.5 with normal formation of dorsal aorta and cardinal vein as visualized upon breeding into a R26R background (Figure 1b). Remodeling of the embryonic and extraembryonic primitive vascular plexi occurred in control E9.5 embryos, but this was severely impaired in dKOEC stage-matched littermates (Figure 1c-d). Severely affected mutant embryos had vestigial heart development and increased apoptosis mainly in non-ECs from the 24-25 somite (S) stage onwards (Figure S1a-b). Therefore, embryos with less than 24 S were analyzed, unless stated otherwise. Altogether, these results illustrate that Smad1/5 mediated signaling is dispensable for vasculogenesis, but is essential for embryonic angiogenesis. At least one functional Smad1 or Smad5 allele is required to safeguard the onset of sprouting angiogenesis, but later events in vessel development or stability may require higher demands (more intact alleles) for Smad1/5-mediated signaling.
Figure 1. Normal vasculogenesis but impaired angiogenesis in dKOEC embryos.
a, E13.5 control embryo (left panel) and embryos containing one functional allele of Smad1 or Smad5 in ECs. b-d, Embryos lacking the four alleles of Smad1 and Smad5 in endothelium. b, Control and mutant E8.5 whole-mount and sectioned R26R reporter embryo stained with X-gal (blue). c-d, Whole mount and flat mounted X-gal stained yolk sacs of control and mutant E9.5 R26R reporter embryos. In d, magnified views are shown of the head and roof of the hindbrain (middle and right panels). Abbreviations: cv, cardinal vein; da, dorsal aorta; s, somites. Scale bars: 250 μm (b,c,d left panels) and 300 μm (d middle and left panels). See also Figure S1.
Smad1/5 signaling regulates sprouting angiogenesis
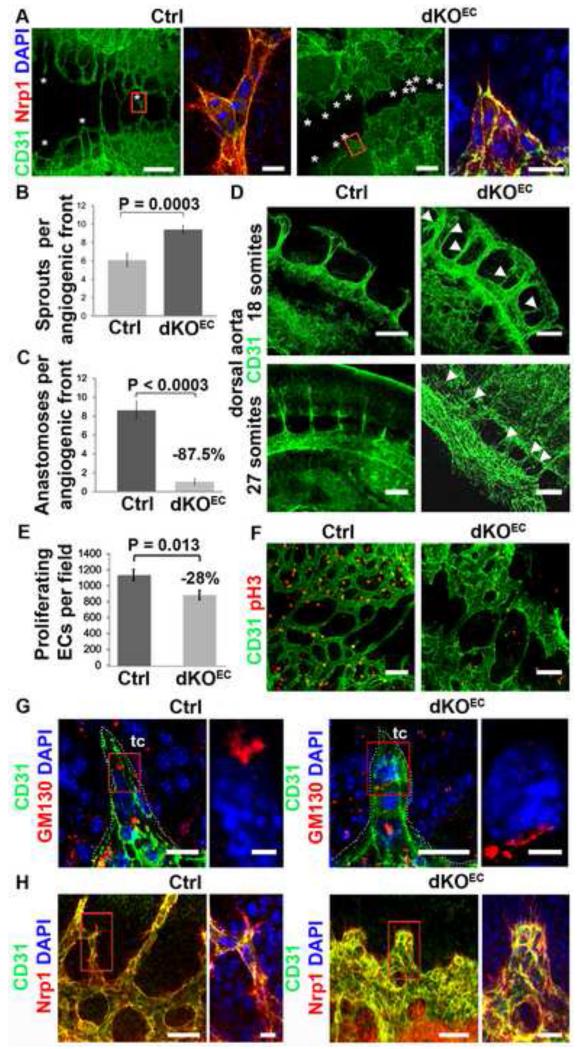
The early lethality of the dKOEC embryos prevented the use of traditional organ-based sprouting angiogenesis models such as the midbrain of E10.5 embryos or the retina of neonatal mice to investigate tip and stalk cell formation. Therefore, we took advantage of the stereotypical formation of sprouts in the thin roof of the hindbrain of E9.5 embryos (Figure 1d and Figure S1c) to study early sprouting angiogenesis. In control embryos, sprouts emerged bilaterally from the perineural vascular plexi at the level of the otic vesicles and anastomosed medially in the roof of the hindbrain in a rostro-caudal fashion. Mutant embryos formed large sinusoid-like vessels instead of the normal ramified network of capillaries. Interestingly, more sprouts formed on both sides of mutant embryos, yet few or none anastomosed medially (Figure 2a-c). These sprouts were, however, pyramid-shaped and broader than in stage-matched controls (Figure 2a, boxed areas). Sprouting from the dorsal aorta was also increased in mutants, resulting in numerous ectopic intrasomitic vessels (Figure 2d) in addition to the intersomitic vessels that were guided between morphologically normal somites (not shown). siRNA mediated downregulation of Smad1 and Smad5 (Smad1/5KD) in Human Umbilical Vein Endothelial Cells (HUVECs) resulted in a tubulogenesis assay in Matrigel substrate in an increased number of branching points when compared with the non-targeting (NT) siRNA-transfected cells (Figure S2a-c). We next determined whether impaired pericyte recruitment or EC proliferation contributed to the sprouting defect in the mutant vessels. Pericytes were recruited to the dorsal aorta and major vessels of the yolk sac of both control and mutant littermates (Figure S2d). Vessels of the perineural vascular plexus were still devoid of pericyte coverage in both control and mutant embryos at this developmental stage (not shown), indicating that impaired pericyte recruitment does not underlie the angiogenic defect observed in this region in mutants. However, there was a significant reduction in the number of phospho-H3/CD31-positive proliferating ECs, which may affect sprout elongation in dKOEC embryos (Figure 2e,f). Altogether, these data suggest that Smad1/5 are key regulators of sprouting angiogenesis and that excessive sprouting and reduced proliferation may contribute to the observed coalescence of vessels into large sinuses in mutant embryos.
Figure 2. Smad1/5 signaling regulates angiogenesis.
a, Dorsal vascular plexus (anti-CD31/anti-Nrp1) in flat-mounted hindbrain roofs (E9.5). Asterisks and boxed areas (right panels) show sprouts quantified in (b). b,c, Quantification of sprouts (b) and dorsal anastomoses (c). d, Dorsal aortae (anti-CD31) from control and dKOEC E9.5 embryos, arrow heads show ectopic sprouts. e,f, Quantification of proliferation (phospho H3-positive ECs) in the dorsal vascular plexus. g, Position of the Golgi apparatus (anti-GM130) in tip cells. h, Filopodia (anti-Nrp1) in endothelium (anti-CD31). Boxed region in g-h are magnified in right panels. Abbreviations: tc, tip cell. Quantifications are averages from several embryos (b,c: n ≥ 21; e: n ≥ 11). Scale bars: 200 μm (a, left panels), 20 μm (a, boxed area; g, left panels); 150 μm (d,f,h, left panels); 4 μm (g, boxed area) and 50 μm (h, boxed area). See also Figure S2 and S3.
Further analysis of dKOEC sprouts revealed severe defects in EC function and in tip cell and stalk cell formation. Tip cells were polarized in control embryos in a similar fashion as ECs grown in 2-dimensional cultures (Pi et al., 2007), with a compacted Golgi apparatus facing the leading edge of the cell (Figure 2g and Figure S3a). In contrast, the localization and compaction of the Golgi apparatus in tip cells of mutant embryos was random, suggesting that Smad1/5 are required for polarity and directed cell migration of tip cells. Similarly, Smad1/5KD HUVECs undergoing directed cell migration in Dunn chambers showed a random localization of the Golgi apparatus, while the Golgi apparatus predominantly localized at the leading edge of control cells (Figure S3b). Next, we performed transwell migration assays using complete growth medium containing VEGF as chemoattractant to evaluate the requirement of Smad1/5 for migration of ECs. Significantly fewer Smad1/5KD ECs migrated when compared to NT siRNA-transfected ECs (Figure S3c). Similarly, a decreased number of Smad1/5KD ECs underwent directed cell migration towards the source of complete growth medium in Dunn chambers (Figure S3d). Interestingly, these Smad1/5KD ECs presented cobblestone morphology with cytoskeletons composed of disorganized filaments of F-actin (Figure S3e). In contrast, the NT siRNA-transfected ECs were elongated with well organized parallel filaments of F-actin aligned towards the source of chemoattractant (Figure S3e). Altogether these results demonstrate that Smad1/5-mediated signaling is a crucial regulator of tip cell polarity and directed migration of ECs.
In addition to the polarity defect in mutant tip cells, all ECs facing the migration front had numerous ectopic and spiky filopodia whilst robust filopodia were restricted exclusively to tip cells in E9.5 control embryos (Figure 2h). Similarly, there was a significant increase in the number of filopodia formed by Smad1/5KD HUVECs when compared with control cells (Figure S3f-h). The increased filopodia formation was observed in ECs cultured with or without a chemoattractant gradient suggesting that Smad1/5 negatively regulate filopodia formation regardless of the migration status of the cell. The average length of these filopodia, however, did not differ between Smad1/5 and NT siRNA treated cells under these conditions (Figure S3f-h). The ectopic formation of filopodia suggested that the stalk cells acquire tip cell-like features in the absence of Smad1/5.
Smad1/5 mediated signaling via Id proteins is essential for stalk cell competence
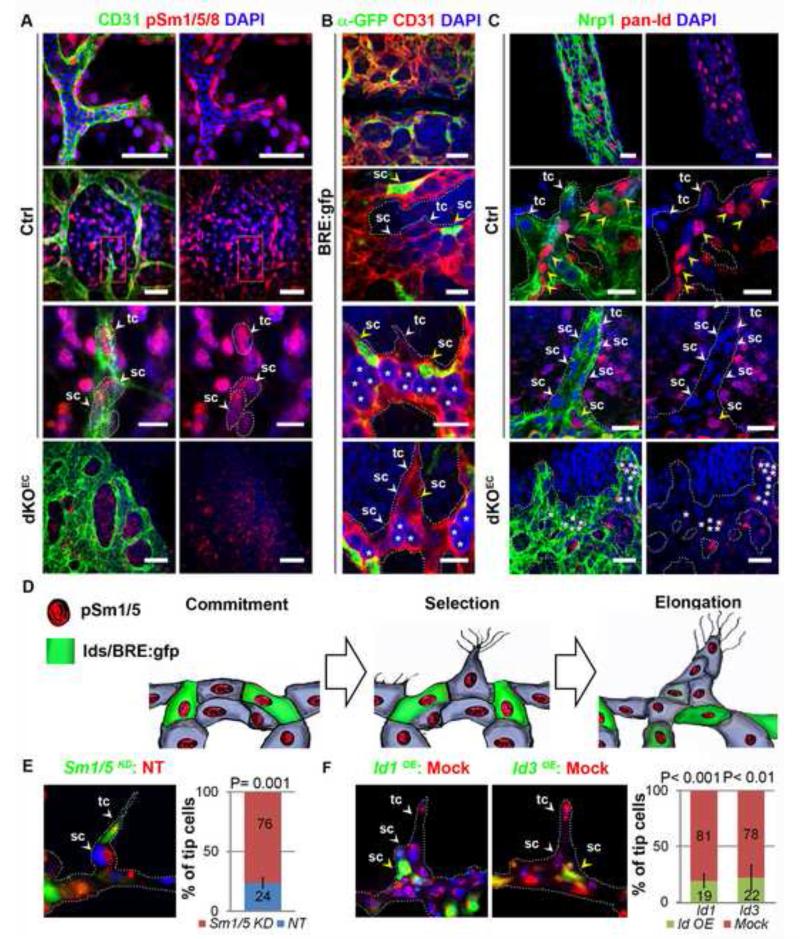
To gain a better understanding on the activities of Smad1/5 in the angiogenic endothelium, we analyzed the localization of the receptor-activated and C-terminally phosphorylated (p-) Smad1/5/8 proteins and compared it with the protein localization of known target genes for Smad1/5/8. The nuclear p-Smad1/5/8 staining in control E9.5 embryos revealed an ubiquitous localization and activity of BMP/Smad signaling throughout the venous and arterial endothelium, including tip and stalk cells (Figure 3a and Figure S4a). Nuclear p-Smad1/5/8 staining was absent in dKO endothelium, but normal elsewhere (Figure 3a), demonstrating that BMP-Smad signaling cascade is efficiently silenced in the endothelium of dKOEC embryos, and that Smad8 (if present) is not sufficient to compensate for the absence of the two other Smad proteins. Next, we monitored the activation of downstream target gene expression in reporter mouse embryos that are transgenic for a gene composed of a BMP response element (BRE) from the Id1 promotor driving green fluorescent protein synthesis (BRE:gfp)(Monteiro et al., 2008). Despite the ubiquitous localization of p-Smad1/5/8 in the endothelium, we found a scattered, non-tip cell distribution of GFP in the endothelium of E9.5 BRE:gfp embryos (Figure 3b). To confirm our observations, we analyzed the distribution of Id proteins using an antibody that detects all Id family members (Figure 3c). In control embryos, the Id proteins showed a scattered distribution in embryonic and extra-embryonic endothelium recapitulating the GFP localization in BRE:gfp embryos. This scattered distribution appeared to be highly dynamic and transient in the ECs, as it was found restricted to non-sprouting endothelium and the early stalk cells of emerging angiogenic sprouts, while ‘late’ stalk cells from elongated sprouts and all tip cells were always devoid of Id/gfp in E9.5 embryos (Figure 3b-d). Id proteins were undetectable in ECs of dKOEC E9.5 embryos, but preserved elsewhere (Figure 3c). This suggests that ubiquitous phosphorylation of Smad1/5 result in distinct activation of target genes in tip versus stalk cells at E9.5 and that, downstream of Smad1/5, a dynamic regulation of Id proteins may be required for proper stalk cell commitment/formation. In retinal angiogenesis, however, different or additional roles for Id proteins may be anticipated since Ids/gfp were more broadly distributed in the endothelium and were often scattered present in tip cells as well as in stalk cells (Figure S4b,c).
Figure 3. Smad1/5 signaling regulates stalk cell competence via Id proteins.
a, Anti-p-Smad1/5/8 staining in endothelium of E9.5 control and dKOEC littermates. Nuclei of tip and stalk cells are highlighted by dashed lines. b, GFP localization (BRE:gfp) in early stalk cells (middle panels) and elongated sprouts (bottom panel), c, Id protein localization in wild-type yolk sac artery (upper panels) and embryonic vasculature (middle panels). Absence of Id proteins in mutant sprouts (bottom panels). Yellow and white arrowheads indicate pan-Id/GFP positive or negative cells respectively. Asterisks represent red blood cells. d, Schematic representation of the dynamic p-Sm1/5/8 and Id/BRE:gfp localization in developmental angiogenic sprouts. e, Chimaeric tube formation assay and quantification of Smad1/5KD (green, calcein) or WT (red, DiI-AC-LDL) leading “tip” cells. f, Chimaeric tube formation assay and quantification of Id1OE/Id3OE (green, calcein) or WT (red, DiI-AC-LDL) leading “tip” cells. Abbreviations: sc, stalk cell; tc, tip cell. Quantifications are averages from several sprout/tubular structures (e: n ≥ 50; f: n ≥ 49). Scale bars from top to bottom: 15 μm, 50 μm, 15 μm and 40 μm in (a); 50 μm, 15 μm, 15 μm and 15 μm in (b); 50 μm, 15 μm, 15 μm and 50 μm in (c). See also Figure S4.
Competition chimaeric Matrigel tube formation assays were performed with control (NT siRNA) and Smad1/5KD HUVECs (1:1 ratio) to address Smad1/5 mediated signaling regulating EC function via Id proteins (Figure 3e-f and Figure S4d,e). Under such competitive conditions, ECs deficient for Smad1/5 signaling preferentially localized in the leading tip cell position of the tubular branches (Figure 3e). Furthermore, there was a significantly higher number of branching points formed in the chimaeric tubular networks when compared to the ones formed by control ECs only (Figure S4d). Conversely, ECs over expressing either Id1 or Id3 were excluded from the leading tip cell position in chimaeric tubular networks when co-cultured with mock transfected ECs (1:1 ratio) (Figure 3f). Moreover, there was a significantly lower number of branching points in non-chimaeric tubular networks composed of ECs overproducing Id1 or Id3 when compared with mock transfected control ECs (Figure S4e). This decreased number of branching points was rescued in chimaeric tubular networks composed of a mixture (1:1) of mock transfected ECs with Id1 or Id3 over expressing ECs, suggesting that Id1/3 expression inhibits branching/sprouting. Altogether, these data confirm that downstream of Smad1/5, Id proteins play a crucial and transient role in early stalk cell commitment/selection and suggest that the presence or absence of Id proteins in individual ECs may influence their potential to respond to the angiogenic stimuli to become tip or stalk cells.
Mutual interdependence of Notch and Smad1/5 mediated signaling
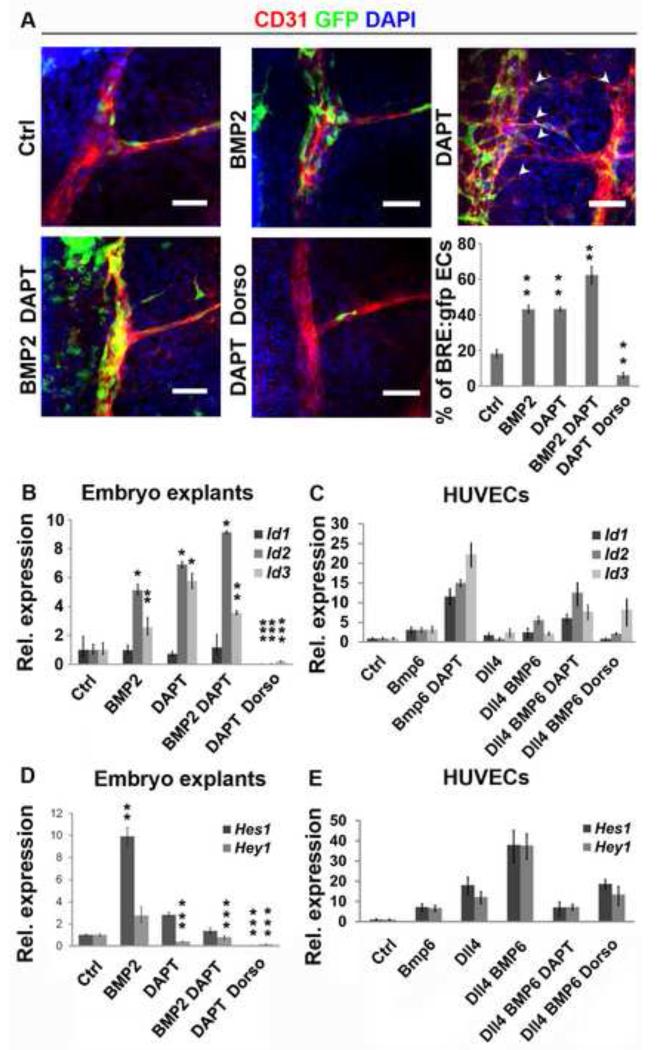
Stalk cells are transiently induced by pulses of Dll4/Notch signaling. It has been shown that a target gene of Notch, Hey1 (Herp2), negatively regulates Id protein production in cultured ECs (Itoh et al., 2004). The presence of of the Smad1/5-dependent Id proteins in the early stalk but not in stalk cells of elongated sprouts made us hypothesize that Id proteins may play an earlier role than Notch in regulating stalk cell selection. To evaluate a possible cross-signaling between BMP and Notch cascades, we developed an ex vivo mid-gestation angiogenic sprouting model and examined the effects of acknowledged pharmacological inhibitors of either pathway (Figure 4a). Dorsal aorta and intersomitic vessels in cultured explants of dorsal tissue of E9.5 BRE:gfp embryos preserved the scattered distribution of Id/gfp proteins observed in vivo in the embryos (Figure 4a versus Figure 3b,c). Interference with Notch signaling by the γ-secretase inhibitor DAPT resulted in increased numbers of cells producing gfp and sprouts from the dorsal aorta. Similarly, treatment with BMP2 increased the number of gfp-positive ECs (Figure 4a), but no ectopic sprouting was observed. The combined treatment with DAPT and BMP2 had an additive effect resulting in the majority of ECs producing gfp, but BMP2 counteracted the DAPT-induced excessive sprouting from the dorsal aorta. Combined treatment of Dorsomorphin, a BMP receptor inhibitor, and DAPT inhibited gfp overproduction below levels of non-stimulated controls (Figure 4a). An expression analysis was performed on these cultured embryo explants and ECs to address the interplay between both pathways. In HUVECs, Id1/2/3 transcripts increased only in response to BMP6 and not to BMP2 or BMP4 (Figure S5). Therefore, HUVECs were stimulated with BMP6 instead of BMP2. Treatment of explants with DAPT effectively down regulated Hey1 while up regulated Id2 and Id3 (Figure 4b-e), suggesting that the Notch-mediated inhibition of BMP signaling can be mediated by Hey1 (Itoh et al., 2004). Consistent with the expansion of the gfp-domain in BRE:gfp explants, the combined treatment with BMP2 and DAPT resulted in an additive induction of Id2 (Figure 4b). Inhibition of Notch signaling in HUVECs increased Id1-3 expression when combined with stimulation by BMP6 (Figure 4c). Next, we analyzed the effect of BMP signaling on the Notch target genes Hey1 and Hes1. Explants treated with BMP2, or HUVECs treated with BMP6, had increased Hes1 and Hey1 transcript levels, while treatment with Dorsomorphin dramatically downregulated Hes1 and Hey1 expression (Figure 4d,e). The combined treatment with BMP6 and Dll4 synergistically stimulated Hes1 and Hey1 expression in HUVECs (Figure 4e). These results further confirm a regulatory loop between the BMP and Notch signaling cascades in the endothelium, and suggest that in early stalk cells BMPs/Smad1/5 enforce Notch signaling to cooperatively activate target gene expression (Hes1 and Hey1), while Notch signaling negatively regulates Id expression.
Figure 4. Cross-signaling between Notch and Smad1/5 pathways.
a, GFP localization in endothelium (anti-CD31) in E9.5 BRE:gfp dorsal explants exposed to different treatments. The arrows heads show ectopic intrasomitic vessels. Quantification of GFP positive ECs from dorsal aorta. b-e, mRNA expression analysis of Id1-3 in embryo explants (c) and HUVECs (d). Hes1 and Hey1 expression in embryo explants (c) and HUVECs (e). Scale bars: 50 μm. See also Figure S5.
Smad1/5 deficiency impairs Notch signaling in angiogenic endothelium
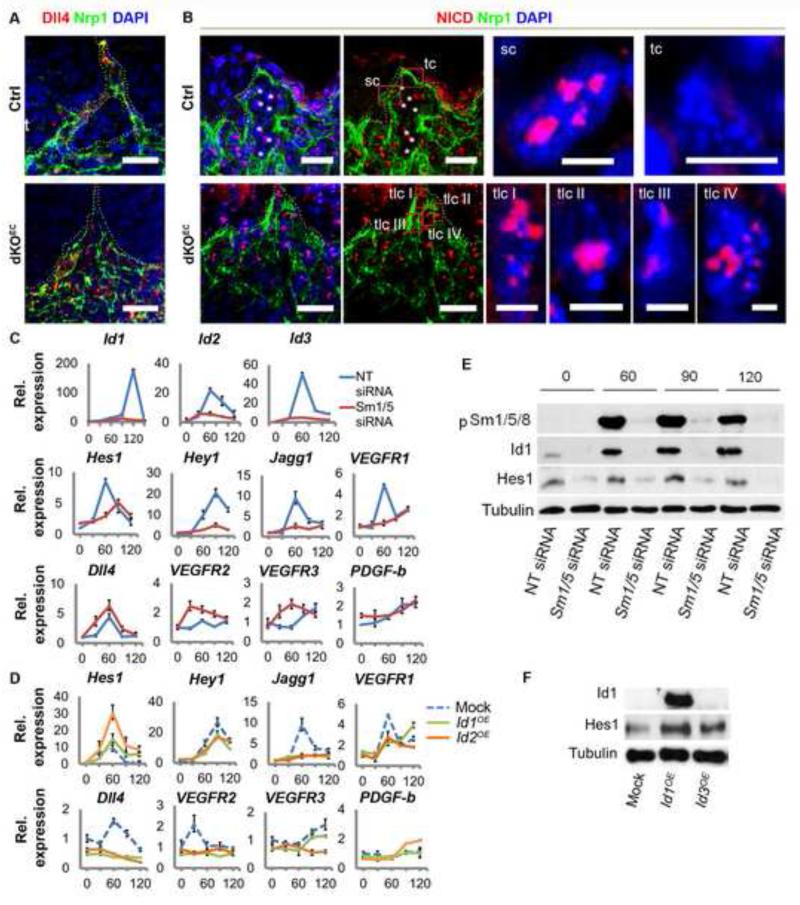
To evaluate whether Smad1/5 signaling reinforces Notch signaling during stalk cell formation during development, we evaluated the vascular distribution of Dll4 and NICD. In control embryos, tip cells showed a marked enrichment of Dll4 when compared with stalk cells. In contrast, Dll4 was not enriched in tip cells in mutants, but equally distributed in tip and stalk cells (Figure 5a). Despite this aberrant distribution of Dll4 in mutant ECs, Notch1 still became ligand-activated, proteolytically cleaved, and targeted to the nucleus as revealed by the staining of NICD in dKOEC embryos (Figure 5b). This nuclear staining, however, was equally present in presumptive tip and stalk cells, contrasting with the clear enrichment of NICD in stalk cells of control embryos. The widespread and overlapping distribution of Dll4 and NICD in mutant ECs suggested that the downregulation of Dll4 ultimately failed in mutant stalk cells. Hence, Smad1/5 are dispensable for Notch activation per se but critical for proper downstream expression and/or activity of target genes of Notch.
Figure 5. Smad1/5 mediated regulation of Dll4/Notch signaling in endothelial cells.
a,b Aberrant Dll4 (a) and NICD (b) localization in dKOEC angiogenic sprouts. In (b), left and middle panels are identical. Boxed areas of presumptive tip cells are magnified at the right. Asterisks represent red blood cells. c,d, Effect of Smad1/5 siRNA (c) or Id1/3 overexpression in the regulation of tip and stalk cell enriched transcripts in HUVECs. e,f, p-Smad1/5/8, Id1 and Hes1 proteins levels after Sm1/5 siRNA mediated downregulation (e) and Id1/3 overexpression (f) in HUVECs. Abbreviations: sc, stalk cell; tc, tip cell; tlc, tip cell-like cells. Scale bars from left to right: 25 μm in (a); 30 μm in (b) left and middle panels and 5 μm in right panels. See also Figures S6.
To investigate how Smad1/5 signaling affects Notch signaling and stalk cell features, Smad1/5 loss and Id gain-of-function experiments were performed in HUVECs. Time course transcription analyses demonstrated that 1 hour after stimulation with serum-rich medium, the expression levels of Id2, Id3, Hes1, Jagged1 and VEGFR1 were highly upregulated in confluent control HUVECs transfected with NT siRNA. After 90 minutes, the level of this set of stalk cell-enriched transcripts decreased with a concomitant upregulation of Hey1 mRNA levels. The peak of Id1 expression appeared only 2 hours after the stimulus (Figure 5c). Smad1/5 siRNA-mediated downregulation effectively inhibited Id1/2/3 expression, as well as the expression of the Notch target genes Hes1 and Hey1, and the other stalk cell-enriched transcripts, Jagged1 and VEGFR1. In contrast, the expression of the tip cell enriched transcripts Dll4, VEGFR2 and VEGFR3 was upregulated in the Smad1/5KD HUVECs, whilst PDGFB expression remained unaffected. These results demonstrate that in the absence of Smad1/5, the levels of tip cell-enriched transcripts increase at the expense of the stalk cell-enriched transcripts. This confirms our in vivo observations, where the absence of Smad1/5 resulted in abrogated NICD signaling, and stalk cells acquired tip cell-like features. We next performed Id1 and Id3 gain-of-function experiments (Figure 5d). Interestingly, Id1 and, to a greater extend, Id3 overproduction, resulted in a dramatic increase of Hes1 expression. The expression of other stalk cell enriched transcripts, VEGFR1 and Hey1, was only mildly affected, except for Jagged1, which was drastically down regulated. Furthermore, Id1/3 overproduction resulted in downregulation of VEGFR2, VEGFR3 and Dll4, while PDGFB remained unaffected. Similar results were obtained using Id2 (not shown). Similarly to Id overproduction, Hes1 and NICD overproduction resulted in a marked downregulation of the tip cell enriched transcripts Dll4, VEGFR3 but also PDGFB (Figure S6). The endogenous Hes1 transcript levels also decreased in response to overproduction of Hes1 protein. Furthermore, Hes1 overproduction increased the levels of the stalk cell enriched transcripts Hey1, while NICD overproduction increased the levels of Hey1 and VEGFR1. Ids and Jagged1 expression was not affected either by Hes1 nor NICD overproduction. Altogether, these results confirm a crucial role for Smad1/5 and Id proteins in the regulation of stalk cell enriched transcripts, and suggest that this effect may be partially achieved by a positive regulation/stabilization of Hes1 expression.
Therefore, we next evaluated if Hes1 protein levels are similarly affected by pSmad1/5 and Id proteins in HUVECs. High levels of p-Sm1/5/8 and Id proteins were strongly detected 1 hour after stimulation with complete growth medium in NT siRNA-transfected ECs. Smad1/5KD led to a strong inhibition of p-Smad1/5, Id and Hes1 protein levels (Fig. 5e). Levels of Id and Hes1 proteins were already decreased in Smad1/5KD ECs from 0 minutes after stimulation, suggesting that it effectively inhibits basal levels of signaling. Conversely, Id1 or Id3 overproduction increased the levels of Hes1 prior to complete growth medium stimulation when compared to the mock transfected ECs (Figure 5f). Upon stimulation, however, the levels of Hes1 proteins in the mock transfected cells raised and became equally high as in the Id1/3 over expressing cells (not shown), likely reflecting that plateau levels of Hes1 have then been reached faster. Therefore, the presence of Id proteins allowed a fast increase of Hes1 transcription and protein production confirming that Smad1/5 signaling via Id proteins is required to stabilize Hes1 expression in ECs.
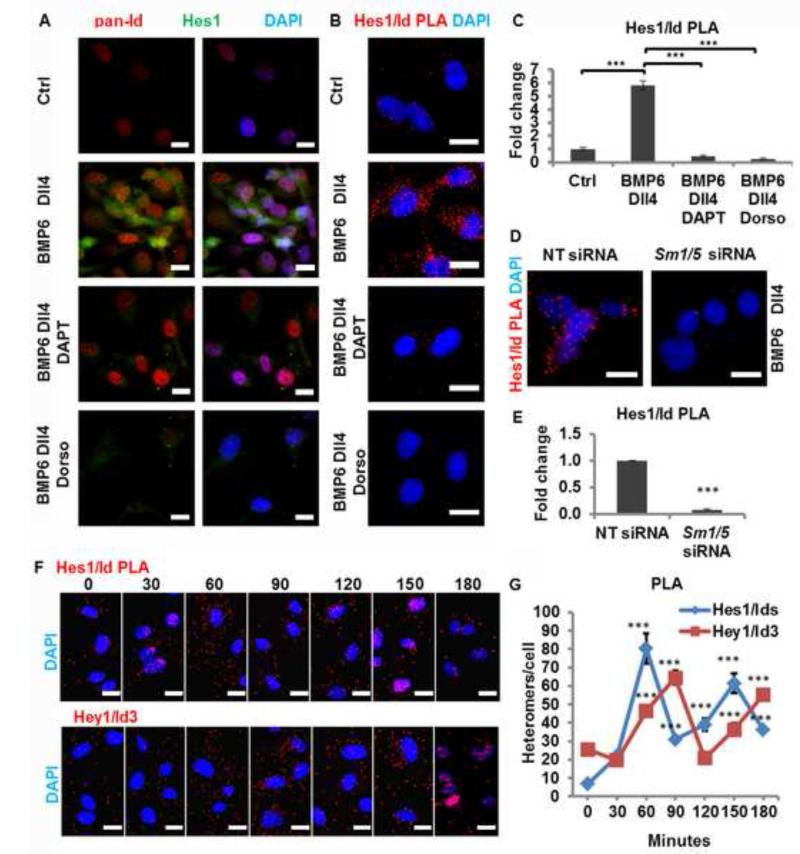
We next hypothesized that the Id-mediated Hes1 stabilization would depend on Id/Hes1 protein-protein interactions in ECs. Therefore, we used an in situ proximity ligation assay (PLA) for detection of Id/Hes1 heteromers. Notch activation by coated Dll4 effectively increased the levels of Hes1 protein, but had little effect on the levels of Id proteins (Figure S7a). Treatment with BMP6 increased Hes1 and Id protein levels. The stimulation with Dll4 or BMP6 induced the formation of Id/Hes1 heteromers above the levels of non-stimulated ECs (Figure S7b,c). The combined stimulation of BMP6 with Dll4 had an even greater effect on the levels of both Hes1 and Id-proteins and resulted in an additive increase (6-fold) in the number of heteromers formed when compared to basal levels (Figure 6a-c). Inhibition of either pathway with DAPT or Dorsomorphin significantly reduced the number of these heteromers (Figure 6b,c and Figure S7b,c). Similarly, siRNA-mediated downregulation of Smad1/5 also resulted in a significant reduction of the number of Id/Hes1 heteromers in HUVECs stimulated with Dll4 and BMP6 (Figure 6d,e), eliminating possible off-target effects of Dorsomorphin (Hao et al., 2010). Altogether, these results demonstrate that Hes1 and Id proteins functionally interact and form complexes in ECs.
Figure 6. Interaction of Hes1 and Id proteins in ECs.
a, Endogenous Hes1 and Id localization in HUVECs treated with Dll4, BMP6, DAPT or Dorsomorphin. b, Hes1/Ids heteromers detected by in situ PLA using pan-Id and Hes1 antibodies. c, PLA signal quantification of (b). d, e, Reduced levels of Hes1/Ids heteromers upon Sm1/5 siRNA transfection. f, g, Dynamic regulation of Hes1/Ids and Hey1/Id3 heteromers in function of time. P values were calculated using two-tailed Student’s t-test (c,d, n ≥ 71; g n, ≥ 105). Abbreviations: Dorso, dorsomorphin hydrochloride. Scale bars: 40 μm (a, f), 15 μm (b, d). See also Figures S7.
We have shown that Id proteins localize in early stalk cells, but not in stalk cells of elongated sprouts, that Id proteins stabilize Hes1 expression, and we have confirmed previous reports showing that the Notch pathway negatively regulates Id expression in ECs, probably via Hey1. We hypothesized that upon an angiogenic stimulus Id proteins would stabilize Hes1 expression and that afterwards the increasing Notch signalling/Hey1 levels will reduce Id protein levels in stalk cells. Therefore, we performed a PLA time course experiment to reveal the dynamics of Ids/Hes1 versus Id3/Hey1 complexes in ECs upon complete growth medium stimulation (Figure 6f,g). Indeed, Ids/Hes1 complexes peak at 1 hour after stimulation and were formed prior to the formation of Hey1/Id3 complexes. In contrast, the number of Hey1/Id3 complexes peaked with a delay of 30 minutes (90 minutes after stimulation) with a concomitant and rapid reduction of Id/Hes1 complexes. Later, at 2 hours after stimulation, the number of Hey1/Id3 complexes also decreased while those of Ids/Hes1 rose higher again. It has been reported that protein-protein interactions of Hey1 with Id proteins reduce Id protein levels by targeting them for degradation (Itoh et al., 2004). Therefore, these results suggest that Hey1 may indirectly destabilize Hes1 gene expression by negatively regulating the levels of Id proteins in the endothelium and, as such, probably contribute/generate Id/Hes1 oscillations that may influence how ECs interpret the angiogenic stimulus for becoming either a tip or stalk cell.
Discussion
Here we demonstrate that the EC-specific inactivation of Smad1/Smad5 primarily impairs tip cell polarity and Notch-mediated stalk cell selection; and we also report hyper sprouting from the dorsal aorta. Recently, it has been reported in zberafish that BMP signaling regulates the initial sprouting steps from axial vein but – unlike in the mouse model - not from the dorsal aorta (Wiley et al., 2011). Importantly, the different loss-of-function approaches used in both organisms target slightly different aspects of the BMP/TGFβ signaling cascades. The genetic deletion of Smad1/5 in mouse not only affects BMP signaling but can also interfere with Alk1/TGFβ signaling (Pardali et al., 2010); whereas noggin and dominant negative Bmpr1 overexpression or the use of pharmacological inhibitors like SL327 will affect in zebrafish also the non-Smad mediated signaling (MAP/Erk signaling), without interfering with Alk1-mediated BMP9-10/TGFβ signaling. The differences in loss-of-function defects in the zebrafish and mouse models may also reflect intrinsic variations in the spouting program used by these organisms.
Several BMPs have been implicated in EC migration in vitro, and Id proteins have been shown to be the mediators for this BMP-Smad1/5 induced EC migration (Itoh et al., 2004; Valdimarsdottir et al., 2002). Accordingly, we demonstrate that Smad1/5 are ubiquitously phosphorylated in tip cells, stalk cells and other ECs throughout the arterial and venous endothelium. However, the distribution of Id proteins is not ubiquitous in the embryonic vasculature but restricted to stalk cells flanking early tip cells and scattered ECs in non-sprouting endothelium, but never in tip cells. This localization pattern suggests that Smad1/5 may activate distinct target genes in tip and stalk cells, and that target genes for BMP-Smad1/5 different from Id genes are probably implicated in the regulation of tip cell polarity and directed cell migration. The molecular mechanism that drives the differential expression of target genes for activated Smad1/5 proteins in tip and stalk cells remains unclear. Yet, microarray analysis revealed that BMP2 and BMP6 induce Myosin10 (Myo10) in ECs (Pi et al., 2007). Furthermore, Myo10 is required for filopodial formation, cell alignment and directed cell migration. Myo10 translocates the BMP-receptor ALK6 into filopodia to reinforce the BMP-induced directed cell migration. Thus, Myo10 and other putative target genes for p-Smad1/5 in tip cells remain to be identified and/or confirmed in tip cells in vivo.
ECs are plastic and have the competence to become tip as well as stalk cells in a process largely controlled by Dll4/Notch signaling in response to precise doses of VEGF, which are on themselves induced by hypoxia (Gerhardt et al., 2003; Hellstrom et al., 2007). It has been proposed that tip and stalk cells are in flux in angiogenic vessels and pre-pattern the vessel continuously for efficient and robust tip cell selection upon stimulation with VEGF (Bentley et al., 2009; Hellstrom et al., 2007). Indeed, tip and stalk cells are highly transient. This results in cell shuffling and in a dynamic competition for the tip cell position, which involves differential regulation of VEGFR levels in a Notch-dependent manner (Jakobsson et al., 2010). Oscillatory expression of genes encoding Delta-like ligands and also of target genes for Notch can contribute to cellular competition and EC priming, as demonstrated in other cell types (Kobayashi et al., 2009; Shimojo et al., 2008), however up to date, oscillatory gene expression has not been demonstrated in endothelium.
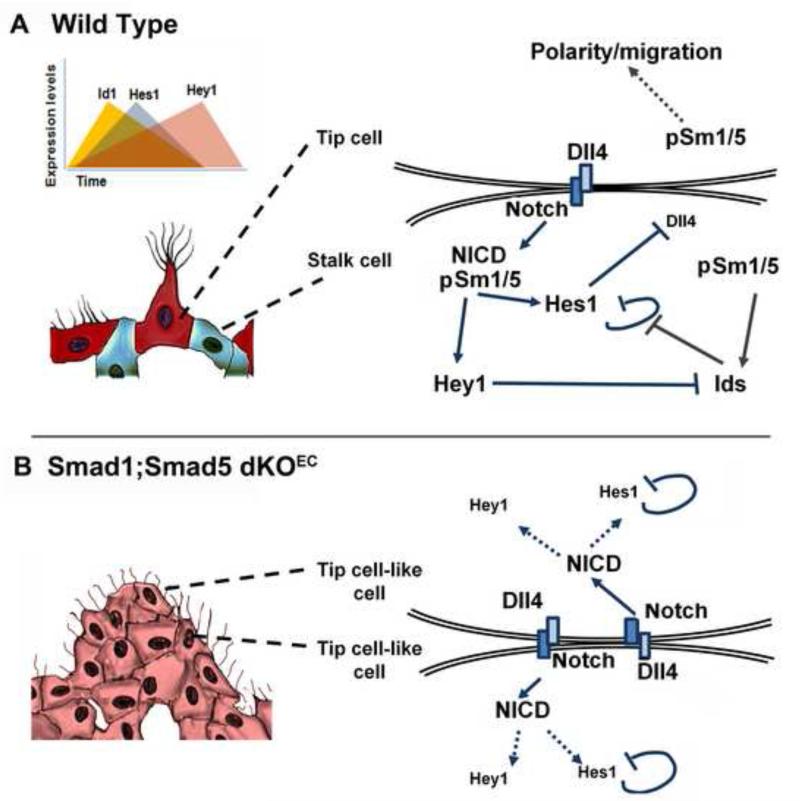
So far it remained unclear whether other pathways would assist in establishing oscillatory gene expression patterns of genes encoding Notch components in the angiogenic endothelium. We propose a model where cross-signaling between Notch and p-Smad1/5 orchestrates tip cell/stalk cell competence/selection and may generate oscillatory target gene expression of both pathways (Figure 7a). Id proteins localize scattered in ECs in E9.5 embryos and play a transient, but crucial role in stalk cell competence and selection. This scattered localization pattern of Id proteins in the endothelium suggests that single ECs may dynamically experience high over low to no Id protein levels with time and that dynamic pulses of Id protein synthesis occur. Interestingly, it has been shown that periodic waves of Smad1/5/8 phosphorylation can generate oscillatory target gene expression in mouse fibroblasts (Yoshiura et al., 2007). Furthermore, several Notch signaling pathway-related genes are EC-specific p-Smad1/5 target genes. In particular, Jag1 is regulated directly by Smad1/5 and its protein transactivates Notch signaling in the neighboring cells in culture (Morikawa et al., 2011). We propose that the mutual interdependence between Notch and Smad1/5 signaling demonstrated here can result in dynamic amplification but also subsequent inhibition of downstream targets of either cascade (Figure 7a). When present in sufficiently high levels, Id proteins would play a cell-autonomous and permissive role in ECs towards stalk cell competence, likely even prior to the perception of the first angiogenic stimulus. Upon perceiving this angiogenic stimulus, ECs will become enriched in VEGFR2/Dll4 levels, and this in turn will increase Notch signaling in neighboring cells. Neighbouring cell(s) with high levels of Id proteins will respond with a fast and robust amplification of (some of) the target genes of Notch signaling by the Id-mediated release of the negative autoregulatory loop of Hes1 (Bai et al., 2007). Direct binding of Hes1 to Dll1 and Dll4 promoters is reported to negatively regulate the expression of Dll1/Dll4 in stem cells (Kobayashi and Kageyama, 2010; Kobayashi et al., 2009). Accordingly, we showed that overexpression of Hes1 or of Ids in ECs results in a downregulation of Dll4 and of other different tip cell markers. As a consequence, these neighboring cell(s) with high levels of Id proteins will readily become the early stalk cells. ECs with lower levels of Id proteins will experience a moderate Notch/Hes1 response insufficient to trigger the stalk cell program. The observations in the Smad1/5 mutant embryos and the chimaeric competition experiment suggest that ECs are most sensitive to ‘tip’ cell selection when they have the lowest levels of Id-proteins, a cellular context in favor of a swift induction of Dll4 upon VEGFR2 signaling. The Id mediated stabilization of Hes1 in scattered ECs would result in a (direct) downregulation of Dll4 and other Hes1 target genes in the cells competent to become stalk cells, and as such ensuring a fast and robust initial selection of tip and stalk cells. The regulation of other target genes for Ids and Hes1 may also contribute with this process. Furthermore, p-Smad1/5 will concomitantly enforce the Notch pathway by forming complexes with NICD and synergistically activating target genes for Notch (Itoh et al., 2004), such as Hey and Hes in the cells that are now acquiring stalk cell features. This will further reduce the levels of Dll4 and the responsiveness to VEGF by down regulating VEGFR2 and VEGFR3, and upregulating VEGFR1 (Holderfield et al., 2006; Jakobsson et al., 2010). As the sprout elongates and Notch signaling levels augment in the stalk cells, the levels of Hey1 increase gradually resulting in the formation of Hey1-Id complexes and an accelerated degradation of Id proteins (Itoh et al., 2004). This will in turn decrease the number of Hes1-Id complexes and reconstitute the negative autoregulatory loop of Hes1 attenuating the Notch pathway in the stalk cell. This decreasing Notch activity can render stalk cells again permissive for cell shuffling and to acquire tip cell features.
Figure 7. Schematic model of the role of Smad1 and Smad5 in Dll4/Notch mediated stalk cell behavior.
a, In wild type embryos, the Dll4-rich leading tip cell (red) of the emerging sprout activates in adjacent cells higher levels of Notch signaling which directs them to acquire a stalk cell behavior (blue). Notch activation and target gene expression (Hey and Hes) downregulates the expression of VEGFR2, VEGFR3 and Dll4, and upregulates VEGFR1 resulting in stalk cell behavior. Downstream of Smad1/5 signaling, Id proteins release the negative autoregulatory loop of Hes1 in stalk cells. Conversely, increasing levels of the Notch target Hey1 will progressively inhibit Id steady-state levels, which will then indirectly cause attenuation of the Notch pathway by Hes1 downregulation. b, Absence of Id proteins in Smad1/5 deficient ECs will result in Hes1 downregulation. High levels of VEGFR2 and Dll4, and deficient Notch target gene expression in stalk cells turn them into a tip cell-like cell phenotype (pink) and prevents the downregulation of Notch1 signaling (nuclear NICD accumulation) in the leading ‘tip’ cells. The failure to balance tip versus stalk cell ratios and to transiently stabilize stalk cells, together with impaired tip cell polarity and directed migration, is likely to cause excessive sprouting and vessel coalescence in mutant embryos.
Inhibition of Notch signaling results in increased tip cell formation (Hellstrom et al., 2007), and we show that under such condition Id distribution gets expanded. This suggests that Id proteins alone do not induce stalk cells, but play a permissive role towards Notch/Hes1 signaling and stalk cell induction. Compound Id1;Id3 knockout mice are characterized by a local decrease in sprouting within the central nervous system (CNS)(Lyden et al., 1999). Despite that tip cell/stalk cell distribution was not reported, it is likely that Id2 functionally compensates for the absence of Id1 and Id3 in the vasculature of these compound mutants, except in the central nervous system where Id2 is not expressed. The angiogenic defect in these mutants is milder than in the embryos lacking Smad1 and Smad5 in the endothelium, which further supports our findings of other non-Id-mediated Smad1/5 functions, such as EC polarity and directed cell migration, in the developing vasculature.
The complex formation between p-Smad1/5 and NICD (Itoh et al., 2004) is expected to be required in stalk cells for a robust target gene expression of both pathways. In the absence of Smad1/5 in ECs (Figure 7b), Notch signaling is impaired and results in an increased number of filopodial protrusions and higher levels of several tip cell-enriched transcripts (VEGFR2/3, Dll4) at the expense of stalk cell-enriched transcripts (Id1-3, Hes1, Hey1, Jagged1 and VEGFR1). As a consequence, Dll4 does not become downregulated in Smad1/5 deficient stalk cells, a process reported to require a Notch-dependent differential regulation of VEGFR1/2 levels (Henderson et al., 2001; Holderfield et al., 2006; Jakobsson et al., 2010), or probably direct repression of Dll1 and Dll4 by Hes1 (Kobayashi et al., 2009). High and ubiquitous VEGFR2-Dll4 levels coupled to impaired Notch signaling (downstream of NICD) in mutant stalk cells drives them to acquire a tip cell-like cell phenotype and to accumulate nuclear NICD in the leading ‘tip’ cell like-cells (Figure 7B). Therefore, the complex phenotype that results from the absence of Smad1 and Smad5 in ECs results from the failure to balance the tip versus stalk cell ratio, excessive sprouting and defective directed cell migration. The sum of these defects is likely to cause the ‘local’ fusion of neighboring tip cell-like cells/sprouts that coalesce into sheets of sinusoid-like vessels in the mutant embryos. Our results put forward that BMP/TGFβ signaling via Smad1/5 is an integral co-player in priming vessel plasticity, initial stabilization of tip/stalk cell distribution and regulation of directed cell migration during mouse early-onset angiogenic sprouting.
Experimental Procedure
Mice
All animal procedures were performed according to the Animal Welfare Committee guidelines of KU Leuven, Belgium. Mice strains and genotyping of Smad1 and Smad5 floxed or recombined alleles, and the transgenic BRE:gfp allele are described in the Supplemental Experimental Procedures.
Whole-mount detection of β-galactosidase activity, in situ hybridization and TUNEL assay
Whole-mount staining with 5-bromo-4-chloro-3-indolyl-beta-D-galacto-pyranoside (X-gal) (Fermentas, R0941), histology on paraffin-embedded tissue sections and TUNEL assay (ROCHE, 11684795910) were performed according to standard procedures. RNA in situ hybridization and Nkx2.5 riboprobe synthesis was performed as described in (Umans et al., 2003). Quantifications In vivo quantifications were done on high resolution confocal images for the discrimination between endothelial and non-endothelial cells. All images (field size 998×998 μm) were number coded and evaluated in a blinded way. The number of HUVECs or filopodia length and number were identified by nuclear staining (DAPI, Invitrogen) and/or cytoskeleton staining (phalloidin, Invitrogen). More details are provided in the Supplemental Experimental Procedures. All experiments were repeated at least 3 times.
Immunofluorescence
Dissected embryos and retinas were fixed for 2 hours in MEMFA (2 μM EGTA, 1 μM MgSO4, 0.1 M MOPS, pH 7.4, 3.7% formaldehyde) at RT or overnight in Dent’s Fixative (8:2 methanol:DMSO) at −20°C, and stored in methanol at −20°C. Embryos were incubated overnight with primary or secondary antibodies (22–25°C). Primary antibodies against CD31 (clone MEC13.3) and GM130 (610822) were from BD Biosciences; Nrp1 (AF566) and Dll4 (AF1389) from R&D Systems; C-terminally phophorylated Smad1/5/8 (9511S) from Cell Signaling; Id1z8 (sc-427), and GFP (sc-9996) from Santa Cruz; Alpha Smooth Muscle Actin (M0851) from Dako, and NICD (ab8925), Desmin (ab8592), Hes1 (ab87395) and pH3 (ab5176) from Abcam. Alexa Fluor 488 and 568 donkey secondary antibodies were from Invitrogen. Confocal images were acquired with a BioRad Radiance 2100 microscope.
Culture of dorsal midgestation embryo explants
E9.5 embryos were dissected in ice-cold 10% FCS in DMEM (Gibco). The roof of the hindbrain and neural tube were explanted and cultured. These dorsal explants were cultured on Millicell cell culture inserts (Millipore, PICM0RG50) in DMEM supplemented with 20% FBS and 50 μg/ml Endothelial Cell Growth Supplement (Millipore) in a humidified incubator at 37°C 5% CO2 for 6 hours. Growth factors (100 ng BMP2/ml, gift from W. Sebald and J. Nickel) and/or N-[N-(3,5-Difluorophenacetyl-L-alanyl)]-S-phenylglycine t-Butyl Ester, DAPT (100 μM, Calbiochem) or Dorsomorphin (10 μM, Tolcris) were applied directly to the medium. Details on EC medium and culture protocol are in Supplemental Experimental Procedures.
Gene expression analysis
RNA was extracted and purified with RNeasy purification columns (Qiagen). Reverse transcription was performed using MuMLV reverse transcriptase (Fermentas), oligo-dT and random primers (Invitrogen). Real-time qPCR on mouse dorsal explants was performed on ABI7000 using the SYBRgreen amplification reagent (Eurogentec). The expression analysis on HUVECs was performed using a LightCycler® 480 Real-Time PCR System and LightCycler® 480 SYBR Green I Master (ROCHE). Primer sequences are listed in the Supplemental Experimental Procedures.
Western blot analysis
Cells were lysed in Tropix lysis buffer. Total protein content was measured using a Bradford protein assay kit (Bio-Rad Laboratories, Hercules, CA). Samples were run on SDS-polyacrylamide gels, transferred onto PVDF membranes and incubated with appropriate antibodies overnight at 4 °C. Incubation with HRP-conjugated secondary antibodies was for 30 min at RT. Primary antibody against Hes1 (ABIN307170) was from LiveSpam technologies, Id1z8 (sc-427) from Santa Cruz, phospho-Smad1/5/8 (9511S) from Cell Signaling and Tubulin (ProBio) and HRP-conjugated secondary antibodies (Jackson ImmunoResearch). Detection was with ECL reagent (Perkin Helmer).
Cell culture and transfection
HUVECs (Lonza) were cultured in EGM®-2MV Microvascular Endothelial Cell Growth Medium-2 (fullEC medium, Lonza) and used between passages 6-9 from purchase. Silencing of endogenous Smad1 and Smad5 was performed by co-transfection of ON-TARGET plus SMARTpools of Smad1 and Smad5 siRNAs (Dharmacon) or non-targeting control siRNA at a final concentration of 5 nM in starvation medium (EBM medium with 0,1% FBS, no growth factors), using Lipofectamine 2000 (Invitrogen). The following expression plasmids were kindly provided by P. ten Dijke (pCDNA3-Id1 and pBlueScript II-Ks-Id3), R. Kageyama (Hes1) and R. Kopan (pCD2-NICD). Details on Dunn and Boyden chamber migration assays, time course experiments and Matrigel tube formation assays are provided in the Supplemental Experimental Procedures.
In situ proximity ligation assay (PLA)
Confluent HUVECs were serum starved for 12 hours, trypsinized and plated on 14 mm coverslips that were first coated overnight with 0.1% gelatin at 4°C. When appropriate, 1 μg/ml Dll4 (R&D) was included in the gelatin solution. DAPT (10 mM, Calbiochem) and Dorsomorphin (10 μM, Tolcris) were added directly to starvation medium containing VEGF (10 ng/ml, R&D) at cell seeding. After 8 hours of culture, HUVECs were fixed in 2% paraformaldehyde in PBS for 20 min and subjected to in situ PLA using Duolink Detection kit (Olink Bioscience). Cells were serum starved for 12 hours, and stimulated with fullEC medium for PLA time course experiments. Samples were fixed every 30 min and treated as described above. Antibodies and PLA protocol can be found in the Supplemental Experimental Procedures.
Statistical analysis
Statistical evaluation was done by student t-test. P values <0.05 were considered significant: *P < 0.05, **P < 0.01, or ***P < 0.001. Error bars represent s.e.m in all figures.
Supplementary Material
Highlights.
-
■
BMP signaling is integral to priming blood vessel plasticity
-
■
Smad1/5 signaling in endothelium affects endothelial cell polarity and migration
-
■
Reciprocal Smad1/5 and Notch signaling is crucial for stalk cell selection
-
■
Hes1 and Id protein interaction strengthens Notch signaling in endothelium
Acknowledgements
All members of the Zwijsen and Huylebroeck teams are thanked for continuous support, M. Missoul for mouse husbandry, and P. Zimmermann for imaging help. We thank A. Noël (Ulg, Liège, Belgium) and J. Haigh (VIB, Ghent, Belgium) for discussions, W. Sebald and J. Nickel (Würzburg, Germany) for recombinant BMP2, S. Chuva de Sousa Lopes (LUMC, Leiden, Netherlands) for Bre:gfp mice, M. Yanagisawa (University of Texas Southwestern Medical Center, Dallas, US) for Tie-2-Cre-mice, R. Kageyama (Kyoto University, Kyoto, Japan), R. Kopan (Washington University, US), and P. ten Dijke (LUMC, Leiden, Netherlands) for Hes1, NICD and Id1/3 expression plasmids respectively.
IMM is a VIB-predoctoral fellow. This work is supported by VIB, the Interuniversity Attraction Poles Program IUAP-6/20, grants from the Research Council of the KU Leuven (OT/09/053 and GOA/11/012) and Hercules Foundation type 3 large infrastructure (ZW09-03 InfraMouse).
References
- Bai G, Sheng N, Xie Z, Bian W, Yokota Y, Benezra R, Kageyama R, Guillemot F, Jing N. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Bentley K, Mariggi G, Gerhardt H, Bates PA. Tipping the balance: robustness of tip cell selection, migration and fusion in angiogenesis. PLoS Comput Biol. 2009;5:e1000549. doi: 10.1371/journal.pcbi.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Huylebroeck D, Verschueren K, Guo Q, Matzuk MM, Zwijsen A. Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development. 1999;126:1631–1642. doi: 10.1242/dev.126.8.1631. [DOI] [PubMed] [Google Scholar]
- Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K, Ibanez CF, Lendahl U. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development. 2003;130:6089–6099. doi: 10.1242/dev.00834. [DOI] [PubMed] [Google Scholar]
- David L, Feige JJ, Bailly S. Emerging role of bone morphogenetic proteins in angiogenesis. Cytokine Growth Factor Rev. 2009;20:203–212. doi: 10.1016/j.cytogfr.2009.05.001. [DOI] [PubMed] [Google Scholar]
- De Smet F, Segura I, De Bock K, Hohensinner PJ, Carmeliet P. Mechanisms of vessel branching: filopodia on endothelial tip cells lead the way. Arterioscler Thromb Vasc Biol. 2009;29:639–649. doi: 10.1161/ATVBAHA.109.185165. [DOI] [PubMed] [Google Scholar]
- Eilken HM, Adams RH. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LS, Sainson RC, Williams CK, Taylor JM, Shi W, Li JL, Harris AL. Regulation of multiple angiogenic pathways by Dll4 and Notch in human umbilical vein endothelial cells. Microvasc Res. 2008;75:144–154. doi: 10.1016/j.mvr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Hellstrom M, Phng L-K, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson A-K, Karlsson L, Gaiano N, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- Henderson AM, Wang SJ, Taylor AC, Aitkenhead M, Hughes CC. The basic helix-loop-helix transcription factor HESR1 regulates endothelial cell tube formation. J Biol Chem. 2001;276:6169–6176. doi: 10.1074/jbc.M008506200. [DOI] [PubMed] [Google Scholar]
- Holderfield MT, Henderson Anderson AM, Kokubo H, Chin MT, Johnson RL, Hughes CC. HESR1/CHF2 suppresses VEGFR2 transcription independent of binding to E-boxes. Biochem Biophys Res Commun. 2006;346:637–648. doi: 10.1016/j.bbrc.2006.05.177. [DOI] [PubMed] [Google Scholar]
- Hu-Lowe DD, Chen E, Zhang L, Watson KD, Mancuso P, Lappin P, Wickman G, Chen JH, Wang J, Jiang X, et al. Targeting activin receptor-like kinase 1 inhibits angiogenesis and tumorigenesis through a mechanism of action complementary to anti-VEGF therapies. Cancer Res. 2011;71:1362–1373. doi: 10.1158/0008-5472.CAN-10-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh F, Itoh S, Goumans MJ, Valdimarsdottir G, Iso T, Dotto GP, Hamamori Y, Kedes L, Kato M, ten Dijke Pt P. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 2004;23:541–551. doi: 10.1038/sj.emboj.7600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson L, Franco CA, Bentley K, Collins RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G, Medvinsky A, et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Kageyama R. Hes1 regulates embryonic stem cell differentiation by suppressing Notch signaling. Genes Cells. 2010;15:689–698. doi: 10.1111/j.1365-2443.2010.01413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Mizuno H, Imayoshi I, Furusawa C, Shirahige K, Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–1875. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechleider RJ, Ryan JL, Garrett L, Eng C, Deng C, Wynshaw-Boris A, Roberts AB. Targeted mutagenesis of Smad1 reveals an essential role in chorioallantoic fusion. Dev Biol. 2001;240:157–167. doi: 10.1006/dbio.2001.0469. [DOI] [PubMed] [Google Scholar]
- Leslie JD, Ariza-McNaughton L, Bermange AL, McAdow R, Johnson SL, Lewis J. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- Lobov IB, Renard RA, Papadopoulos N, Gale NW, Thurston G, Yancopoulos GD, Wiegand SJ. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proceedings of the National Academy of Sciences. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O’Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- Monteiro RM, de Sousa Lopes SM, Bialecka M, de Boer S, Zwijsen A, Mummery CL. Real time monitoring of BMP Smads transcriptional activity during mouse development. Genesis. 2008;46:335–346. doi: 10.1002/dvg.20402. [DOI] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin CH, Aburatani H, Miyazono K. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- Norton JD, Atherton GT. Coupling of cell growth control and apoptosis functions of Id proteins. Mol Cell Biol. 1998;18:2371–2381. doi: 10.1128/mcb.18.4.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali E, Goumans MJ, Ten Dijke P. Signaling by members of the TGF-beta family in vascular morphogenesis and disease. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Pi X, Ren R, Kelley R, Zhang C, Moser M, Bohil AB, Divito M, Cheney RE, Patterson C. Sequential roles for myosin-X in BMP6-dependent filopodial extension, migration, and activation of BMP receptors. J Cell Biol. 2007;179:1569–1582. doi: 10.1083/jcb.200704010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillien A, Blanco-Sanchez B, Halluin C, Moore JC, Lawson ND, Blader P, Cau E. BMP signaling orchestrates photoreceptor specification in the zebrafish pineal gland in collaboration with Notch. Development. 2011;138:2293–2302. doi: 10.1242/dev.060988. [DOI] [PubMed] [Google Scholar]
- Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- Ruhrberg C, Gerhardt H, Golding M, Watson R, Ioannidou S, Fujisawa H, Betsholtz C, Shima DT. Spatially restricted patterning cues provided by heparin-binding VEGF-A control blood vessel branching morphogenesis. Genes Dev. 2002;16:2684–2698. doi: 10.1101/gad.242002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–784. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Dunn NR, Robertson EJ. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- Umans L, Vermeire L, Francis A, Chang H, Huylebroeck D, Zwijsen A. Generation of a floxed allele of Smad5 for cre-mediated conditional knockout in the mouse. Genesis. 2003;37:5–11. doi: 10.1002/gene.10219. [DOI] [PubMed] [Google Scholar]
- Valdimarsdottir G, Goumans MJ, Rosendahl A, Brugman M, Itoh S, Lebrin F, Sideras P, ten Dijke P. Stimulation of Id1 expression by bone morphogenetic protein is sufficient and necessary for bone morphogenetic protein-induced activation of endothelial cells. Circulation. 2002;106:2263–2270. doi: 10.1161/01.cir.0000033830.36431.46. [DOI] [PubMed] [Google Scholar]
- Wiley DM, Kim JD, Hao J, Hong CC, Bautch VL, Jin SW. Distinct signalling pathways regulate sprouting angiogenesis from the dorsal aorta and the axial vein. Nat Cell Biol. 2011;13:687–693. doi: 10.1038/ncb2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Castilla LH, Xu X, Li C, Gotay J, Weinstein M, Liu PP, Deng CX. Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development. 1999;126:1571–1580. doi: 10.1242/dev.126.8.1571. [DOI] [PubMed] [Google Scholar]
- Yoshiura S, Ohtsuka T, Takenaka Y, Nagahara H, Yoshikawa K, Kageyama R. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc Natl Acad Sci U S A. 2007;104:11292–11297. doi: 10.1073/pnas.0701837104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene. 2001;20:8317–8325. doi: 10.1038/sj.onc.1205092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.