Abstract
Light and food are two major environmental factors that impact daily life. Light entrainment is centrally controlled by suprachiasmatic nuclei of the hypothalamus. Food entrainment might require cooperation between the intestine and dorsomedial hypothalamus. Clock genes that are essential for light entrainment also play a part in food entrainment. Understanding the role of clock genes in the entrainment of intestinal functions, as well as in gut–brain communication during food entrainment, will enhance our understanding of gastrointestinal and metabolic disorders. This review highlights recent studies examining light- and food-entrained regulation of plasma lipids and of various intestinal activities and offers insight into the role of the intestine in food entrainment.
Introduction
The rotation of the earth around the sun and on its own axis with remarkable periodicity, resulting in day and night, is a dominant environmental factor that affects each living organism. Adaptations by metazoans to these daily changes manifest as variations in physiological and behavioral activities that recur with approximately 24 h intervals. That mammals have retained circadian (see Glossary) rhythms that probably evolved billions of years ago in photosynthetic cyanobacteria underscores their fundamental importance in the propagation, survival and evolution of life. Food is another factor that affects daily physiological and behavioral activities because of the dependence of life on external sources of energy. Here, we briefly describe clock genes and discuss their role(s) in light- and food-entrained circadian variations. Our main focus is on the rhythmic variations of intestinal functions, the role of the intestine in food-entrained regulation and responses of intestinal proteins to changes in food availability. In addition, we discuss daily variations in plasma lipids and highlight issues that need exploration to understand the role of different proteins in the acclimatization of cells to recurrent environmental cues.
Light, clock genes and circadian rhythms
Daily changes in light are recognized by neurons in the retina, and this information is transmitted via a subpopulation of retinal ganglion cells to two suprachiasmatic nuclei (SCN) in the anterior hypothalamus of the brain (Figure 1). These nuclei are composed of numerous cells (~20 000 neurons) that are individually capable of eliciting endogenous, autonomous circadian rhythmicity and constitute the light-entrainable oscillator (LEO) (Figure 1a). The LEO converts the light-entrained information into neuronal and humoral output signals and influences various biological rhythms (e.g. locomotor activity, body temperature and hormone secretion) and, therefore, serves as a master pacemaker in the body [1–9]. The transmission of light signals leading to physiological and behavioral changes begins with the expression of certain transcription factors (i.e. clock genes) in the SCN. Clock genes show endogenous rhythms but need external cues for their entrainment. The most dominant factor that entrains clock genes is light.
Figure 1.

Light and food entrainment. (a) Light entrainment. Light is sensed by the retina, and this information is transmitted via the retino-hypothalamic tract to the suprachiasmatic nuclei (SCN) in the brain (LEO). This information elicits changes in the expression of various transcription factors that constitute the clock genes. Circadian regulation in the SCN is crucially dependent on two transcription factors, Clock and Bmal1. Clock and Bmal1 heterodimers activate three Per and two Cry genes, which heterodimerize to negatively regulate Clock/Bmal1 activity. Other peripheral tissues also express clock genes, and their circadian behavior is entrained by cues originating from the SCN. (b) Food entrainment. Availability of food at a given time with regular periodicity is also a strong cue to entrain different physiological and behavioral activities. It is likely that the intestine plays a crucial part in food entrainment. The intestine might send signals to entrain the dorsomedial hypothalamus about the availability of food (FEO). The dorsomedial hypothalamus, in turn, might send signals to other tissues to entrain various behavioral and physiologic functions associated with food anticipation, digestion and absorption.
Central to circadian regulatory mechanisms are two basic helix–loop–helix Period–Arnt–Single-minded transcription factors, Clock and Bmal1 [1–9] (Figure 2). They form heterodimers and bind to E-box sequences to enhance the transcription of Period (Per), Cryptochrome (Cry), Rev-erbα and retinoic-acid-related orphan receptor alpha (Rorα) genes; therefore, Clock and Bmal1 constitute a major positive feedback loop. Per and Cry transcription factors form heterodimers, move to the nucleus and repress the expression of clock genes, thereby constituting a negative feedback loop. The Per proteins are phosphorylated by casein kinases and targeted for degradation by proteasomes [10]. This degradation lowers their concentrations and helps initiate another positive circadian cycle. In addition, histone acetylation and deacetylation also play a part in controlling clock gene expression [11–13]. RORα, possibly along with PPARγ coactivator-1 alpha (PGC1α) [14], forms a secondary positive loop and activates Bmal1, whereas Rev-erbα inhibits Bmal1 expression and represents a secondary negative feedback loop [1–9]. The intricate timing of these molecular feedback events leads to the characteristic pattern of rhythmic expression of different phenotypes.
Figure 2.
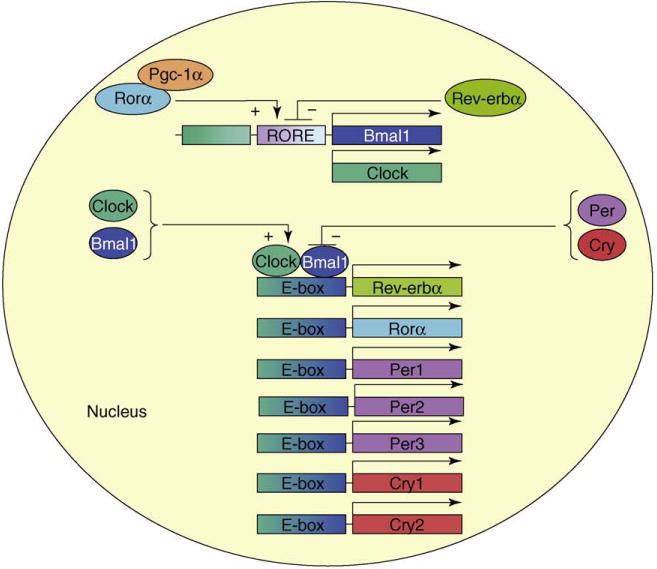
Clock genes and diurnal regulation. Clock, Bmal1, Per1, Per2, Per3, Cry1, Cry2, Rev-erbα and Rorα constitute core clock genes. Clock/Bmal1 heterodimers interact with promoter sequences in different genes to increase their transcription and represent a major feed-forward loop. Per and Cry protein heterodimers oppose their action to act as repressors and constitute a major negative feedback loop. Activator Rorα, possibly along with a co-activator (Pgc-1α), and repressor Rev-erbα regulate Bmal1 expression, constituting secondary positive and negative feedback loops, respectively. Different colors are used to represent individual transcription factors.
The elaboration of rhythmic behavior by the SCN also involves ‘clock-controlled genes’ (Figure 3); these important intermediary transcription factors transmit circadian signals and control various metabolic pathways and physiologic functions. For example, D-site-binding protein (Dbp) affects drug and bile acid metabolism. Arginine vasopressin transmits the circadian response to regulate daily variations in drug metabolism and blood pressure. Rev-erbα and Rorα act as both clock genes and clock-controlled genes [15,16], transmitting circadian signals to other genes involved in physiologically important regulatory and metabolic pathways. Identification of new clock-controlled genes and mechanisms involved in the transmission of diurnal behavior of metabolic functions will elicit new information about regulatory circuitry between metabolism and circadian rhythms.
Figure 3.
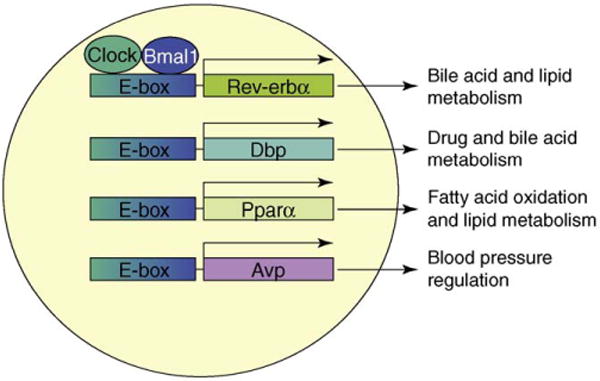
Clock-controlled genes and regulation of metabolic functions. Clock/Bmal1 heterodimers bind to E-boxes to increase expression of genes independent of the classical feedback loop of clock genes. These genes are referred to as ‘clock-controlled genes’. Induction of these clock-controlled genes helps propagate circadian signals via regulation of different metabolic genes leading to various physiological and behavioral outputs. For example, Pparα upregulates genes involved in fatty acid oxidation, and Rev-erbα suppresses genes involved in bile acid metabolism. Similarly, albumin D-site-binding protein (Dpb) and arginine vasopressin (Avp) have been shown to regulate genes involved in drug metabolism and blood pressure control, respectively. These clock-controlled genes modulate more than one physiologic activity; therefore, interactions among these clock-controlled genes might be important in regulating cellular and biochemical functions. This figure is intended to introduce the concept and is not comprehensive.
Expression of clock genes can be entrained by other nonphotic stimuli, such as temperature, food and drugs. Therefore, clock genes represent a basic set of genes, the expression of which can be modulated to exhibit a recurring pattern with 24 h intervals. Ubiquitous expression of clock genes in different tissues, however, indicates that circadian expression is not a specific property of the SCN; rather, all cells are capable of circadian expression. In fact, cells in culture exhibit circadian gene expression when subjected to external stimuli; however, they need repetitive cues to maintain rhythmicity. Hence, clock genes act like ‘servants’, ready to carry out recurring, rhythmic functions when commanded.
Food-entrainable oscillator
Food also acts as a synchronizer of various behavioral and physiological activities [17–21] (Figure 1b). As nocturnal animals, rodents consume food at night, and food consumption is accompanied by increases in locomotor activity with diurnal regularity [22,23] (Box 1). Feeding rodents during the day inverses the phase of these activities and uncouples SCN-regulated circadian rhythms in peripheral organs such as the liver, lung and heart [24–26]. SCN-lesioned rodents fail to respond to light but are entrained by restricted feeding [27]. These findings indicate that the food-entrainable oscillator (FEO) is distinct from the LEO. In fact, there is evidence that the LEO and FEO are anatomically distinct and are present in the SCN and dorsomedial hypothalamus (DMH), respectively [28,29]. Ablation studies to establish the importance of the DMH in food entrainment have been controversial and inconclusive [29,30]. Nevertheless, some biochemical and molecular studies indicate that the DMH might be involved in food-entrained regulation. For example, under ad libitum conditions in a 12 h light–dark cycle, the expression of c-fos, a measure of neuronal activity, in the DMH is in sync with that of the SCN [29]. During food entrainment, c-fos expression in the DMH dissociates from the SCN, showing maximum activity at mealtime. Similarly, an intense expression of Per1 and Per2 in the DMH occurs at mealtime in food-entrained mice [28,31,32]. Ablation of Per2 and Bmal1, but not Clock, abolishes food entrainment [28,33], and the loss of food entrainability in Bmal1−/− mice can be rescued by expressing Bmal1 in the DMH but not in the SCN. Therefore, the DMH and several of the clock genes might play a part in food entrainment.
Box 1. Activities associated with the FEO.
The FEO directs various behavioral and physiological changes. Animals subjected to food entrainment in a 12-h light–dark cycle exhibit new activities that differ significantly from those observed in animals fed ad libitum. These activities can be divided into those involved in preparation for meals (food-anticipatory activity) and in the ingestion, digestion and absorption of food (food-consuming activities). The anticipatory activities include changes in free wheel running, body temperature and bar tapping for food and increases in plasma cortical and free fatty acids. Food-consuming activities are accompanied by the upregulation of proteins involved in carbohydrate, protein and lipid uptake. A feature of food entrainment that has been overlooked is the cessation of these activities after the completion of a meal; failures in reducing these activities might cause metabolic disorders.
FEO activity is usually measured by recording wheel-running activity, which might represent a preparatory act to obtain food. This activity is probably controlled by sensory (taste) and nutrient responses. In contrast, changes in glucagon and free fatty acid levels might signify the fasting response. Increases in plasma triglycerides perhaps represent food consumption. The control of these different behavioral and physiological responses could be independently regulated. For example, restricted food availability increases locomotor activity and serum glucagon, free fatty acid and cortisol levels. However, entrainment for palatable food under ad libitum conditions entrains locomotor activity without affecting serum glucagon and free fatty acid levels [22].
It is unknown, however, how the DMH is entrained by food. It is possible that this entrainment involves gut–brain communication. We speculate that signals for the entrainment of the FEO originate from the intestine (Figure 1b). After coming in contact with food, the intestinal mucosa can elicit either humoral or vagal signals. Very little is known about the lumenal signals that entrain the intestine and about the humoral communications regarding food availability between the digestive system and the central nervous system. In addition, a mechanism or mechanisms entraining peripheral tissues by the FEO have not yet been elucidated. Although it was believed that glucocorticoids played a part [34], it is now thought that glucocorticoid rhythm driven by the SCN opposes the food-entrainment response [5]. Instead, it has been hypothesized that circulating macronutrients and consequent activation of signaling pathways might be involved in resetting peripheral clocks during food entrainment [2]. This hypothesis places enormous importance on the intestine and macronutrient absorption as a signal for the entrainment of peripheral clocks by food and warrants further research towards elucidating the role of the intestine in food-entrained regulation of behavioral activities, physiologic processes and molecular events.
Circadian regulation of intestinal functions
The main function of the intestine is to digest and absorb nutrients present in food. Macronutrients, carbohydrates, lipids and proteins are hydrolyzed in the lumen of the intestine, and products are retrieved by enterocytes involving various transporters. Several transporters involved in the uptake of hydrolyzed products of dietary fat have been identified [35]. However, little is known about their diurnal regulation. Carbohydrates are broken down to monosaccharides and disaccharides in the intestinal lumen and are taken up by two principle apical transporters: Na+-dependent glucose transporter 1 (Sglt1), which transports glucose as well as galactose, and a facilitated fructose transporter, Glut5. Intestinal epithelial cells express a basolateral Glut2 that transports all monosaccharides from the cytosol to the blood. Sglt1 exhibits circadian expression independent of dietary regulation [36–38]. Like Sglt1, Glut5 also shows diurnal expression and is transcriptionally regulated. Coordinate diurnal changes in Sglt1 and Glut5 indicate that carbohydrate metabolism is regulated to maximize absorption at mealtime.
Proteins are hydrolyzed to small peptides and free amino acids in the lumen of the intestine. H+-peptide cotransporter 1 (Pept1) expressed on the apical side of enterocytes is the major protein involved in the transport of small peptides [39]. Pept1 is expressed diurnally in the rat small intestinal epithelial cells with maximal mRNA and protein expression occurring at dark, coincident with Sglt1 expression [37]. Moreover, Pept1 expression is affected significantly by fasting and feeding schedules [36,40]. Recent studies indicate that circadian expression of Pept1 might be under the control of a clock-controlled gene, Dbp [41]. In addition to the peptide transporter, several amino acid transporters have been identified in intestinal epithelial cells [42,43]. However, little is known about their diurnal regulation. It is likely that some, if not all, of the amino acid transporters show diurnal rhythmicity.
In general, expression of these transporters is high at mealtime, and their expression pattern is altered significantly under conditions of food entrainment. Therefore, it seems that food has a prominent role in regulating intestinal transporters. In fact, the majority of transporters do not show dependence on vagal innervations [38]. There have been some speculations about the effect of humoral factors on enterocyte transporter expression. Lumenal signals associated with the passage of a meal through the intestinal tract might also be important. Tavakkolizadeh et al. [44] proposed two distinct and separate pathways regulating the expression and function of transporters in intestinal epithelial cells. One pathway involves gut lumenal signals (presumably food intake) to induce diurnal variations, whereas the second is a daily anticipatory mechanism that prepares the intestine for an expected increase in nutrients before exposure to lumenal contents. Therefore, several mechanisms might be operative in the entrainment of intestinal transporters.
In addition to nutrient absorption, several functions of the gastrointestinal tract exhibit circadian activities [45]. For example, peak DNA synthesis in the esophageal epithelial cells occurs towards the end of the dark cycle, and peak DNA synthesis in the rectum occurs much later [46]. Furthermore, intestinal epithelial cells frequently self-renew and exhibit circadian variations. The rhythmic property of intestinal cell proliferation has been the basis for timed chemotherapies and radiation therapies in the treatment of metastatic colon cancers [45]. Basal gastric acid output is high at night and low in the morning, which is indicative of circadian secretion. Vagotomy abolishes this rhythm, indicating a vagally controlled circadian rhythm. Gastric and colonic motilities show biological rhythms, and their disturbances contribute to constipation and irritable bowel syndrome. Interestingly, gastrointestinal symptoms (abdominal bloating and pain, alterations in bowel habits, constipation, and diarrhea) are commonly seen in people with abrupt routine shifts, shift workers and transcontinental travelers [47]. These examples indicate that some intestinal functions are rhythmically regulated and that their disruptions lead to health disorders. Therefore, understanding the mechanisms underlying diurnal variations in the digestion and absorption of food might be useful in the diagnosis and prevention of these disorders.
Rodent colon and stomach epithelial cells express clock genes that exhibit circadian behavior [48,49]. The circadian expression of clock genes in these tissues occurs in phase with that seen in the liver but is phase-delayed with respect to the SCN circadian clock and not affected by total darkness. Vagotomy has no effect on circadian expression of clock genes in the stomach, excluding the possibility that neuronal signals are required for entrainment. However, clock gene expression in the stomach is markedly altered when animals are subjected to food restriction, albeit with no effect on expression of these genes in the SCN [24]. These studies indicate that the colon and stomach express canonical clock genes in a circadian manner and are susceptible to attunement by food independent of the SCN. Further studies are required to determine the expression and regulation of clock genes in the small intestine and their regulation by food.
Plasma lipid homeostasis and circadian rhythms
Thermogenesis and the sleep–wake cycle, plasma free fatty acids and triglyceride levels also exhibit circadian rhythmicity in humans and rodents [50]. Free fatty acids are transported bound to albumin in the plasma. By contrast, other lipids – such as triglycerides, phospholipids and cholesterol – are transported on special protein–lipid complexes called lipoproteins. Lipoproteins are generally classified based on their flotation properties. However, they can be also divided into two categories based on the presence or absence of apolipoprotein B (apoB). Triglycerides are transported in the plasma as part of apoB-containing lipoproteins (Box 2). These lipoproteins are mainly synthesized in the intestine and liver and require apoB, a structural protein, and microsomal triglyceride transfer protein (MTP), a chaperone present in the lumen of the endoplasmic reticulum [51,52]. Intestinal lipoproteins transport dietary and biliary lipids via the exogenous pathway (Box 2, Figure Ia). Hepatic lipoproteins remobilize lipids delivered by chylomicron remnants, as well as those synthesized in the liver via the endogenous pathway (Box 2, Figure Ib). We reported recently that diurnal variations in plasma triglyceride and cholesterol are mainly caused by changes in apoB lipoproteins [53]. In addition, intestinal and hepatic MTP gene expression show rhythmic expression in rats and mice [53]. Changes in MTP and plasma lipids are abrogated when mice are placed in total light or dark for five days, indicating regulation by the light-entrained SCN. Interestingly, circadian variations in both MTP and plasma lipids are altered when mice are subjected to food entrainment, implicating FEO involvement. Thus, both light- and food-entrainment mechanisms can regulate plasma lipids; however, key molecules involved in such regulation have not yet been identified. Another crucial question that has yet to be addressed is whether FEO acts independently or in cooperation with the LEO to elicit circadian variations in plasma lipids and in tissue MTP gene expression. It is possible that the FEO co-opts clock genes to regulate MTP expression and plasma lipids.
Box 2. Lipoprotein metabolism.
Lipoproteins, multi-molecular complexes that consist of lipids and proteins held together by non-covalent forces, transport hydrophobic lipids through the aqueous blood compartment. Lipoproteins are synthesized by the enterocytes to transport dietary fat and fat-soluble vitamins via the exogenous pathway (Figure Ia). Dietary lipids (triglycerides, phospholipids and cholesterol esters) are hydrolyzed in the lumen of the intestine and enterocytes absorb the products. (i) The lipids are re-synthesized by enterocytes and packaged into lipoproteins called ‘chylomicrons’. (ii) The assembly of chylomicrons occurs in the endoplasmic reticulum in a process requiring a structural protein, apolipoprotein B48 (apoB48), a dedicated chaperone, microsomal triglyceride transfer protein (MTP), triglycerides and phospholipids. The secreted particles are concentrated in lymphatics and delivered to the plasma at the thoracic duct. These particles acquire apolipoprotein CII from plasma and then interact with endothelial-cell-bound lipoprotein lipase (LPL). LPL hydrolyzes triglycerides, and various tissues (such as muscle, adipose and heart) take up generated free fatty acids. (iii) The remaining particles, chylomicron remnants, acquire apolipoprotein E (apoE) from plasma. ApoE is a high-affinity ligand for hepatic lipoprotein receptors, such as low-density lipoprotein receptors (LDL-R) and low-density lipoprotein-receptor-like protein (LRP). (iv) ApoE-containing chylomicron remnants are rapidly removed from the plasma by the liver via endocytosis.
The liver mobilizes the fat delivered by chylomicron remnants, as well as that synthesized in the liver by the endogenous pathway (Figure Ib). Lipoproteins taken up by the liver are degraded in lysosomes. (i) Some of the cholesterol is converted to bile acids and secreted into the lumen of the intestine. The mobilization of fat by the liver is similar to that in the intestine. (ii) Liver cells synthesize and secrete very-low-density lipoproteins (VLDLs) that contain apoB100 (in contrast to apoB48-containing chylomicrons synthesized by the intestine). Again, VLDL assembly is dependent on apoB, MTP, phospholipids and triglycerides. (iii) VLDLs are secreted into the blood. (iv) Like the catabolism of chylomicrons, the triglycerides present in VLDL are hydrolyzed by endothelial-cell-bound LPL, resulting in the generation of intermediate-density lipoproteins (IDL). There is one major distinction between the catabolism of chylomicrons and VLDLs. Whereas all chylomicrons are converted to chylomicron remnants, VLDLs are converted to two products: IDLs and (v) low-density lipoproteins (LDLs). (vi) IDLs acquire apoE and are cleared fast from plasma (i.e. they have a half-life of minutes) by hepatic receptors, LDL-R and LRP, via receptor-mediated endocytosis. (vii) However, LDLs are cleared slowly (i.e. they have a half life of approximately 24 h), relying on apoB100, which binds to the LDL-R in the liver and (viii) other tissues with low affinity.
Figure I.
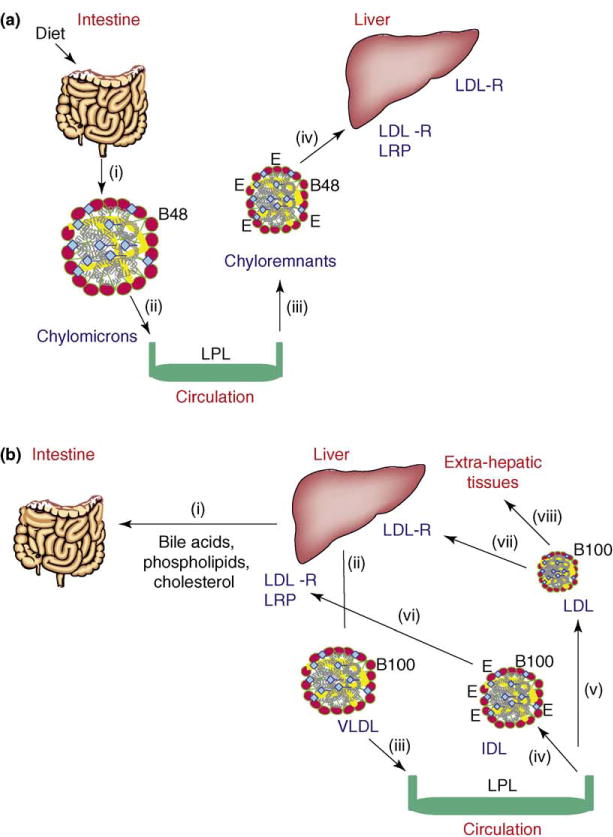
The endogenous and exogenous pathways ensure complete utilization of fat as an energy source. (a) Exogenous pathway. Absorption of dietary and biliary lipids by the intestines and their catabolism in the plasma. (b) Endogenous pathway. Liver mobilizes the delivered and newly synthesized fat.
Food entrainment enhances plasma triglycerides at mealtime. For example, even though chow is low in fat, chow-fed rodents show increased plasma triglycerides at mealtime [53]. Triglycerides also peak in humans after meals, indicating an eating response [50]. These studies indicate that surges in plasma triglyceride might be due to increased digestion and fat absorption. However, Escobar et al. [54] showed that plasma triglycerides and free fatty acids, but not glucose, show persistent circadian rhythmicity in rats fasted for 96 h. The increases in triglyceride during fasting, however, were lower than those observed in the fed state [54]. Similarly, Fukagawa et al. [55] showed plasma triglyceride circadian rhythms in fasted rats, indicating that an entrainable oscillator controls changes in plasma triglycerides. There is evidence to indicate that clock genes might have a role in circadian regulation of plasma lipids. Clock mutant C57Bl/6J mice are obese and exhibit characteristics of the metabolic syndrome: hypercholesterolemia, hypertriglyceridemia, hyperglycemia and hypoinsulinemia [56]. However, expression of a clock mutant protein in ICR mice has been shown to disrupt fat absorption, and these mice are resistant to diet-induced obesity [57]. It remains to be determined, then, how Clock modulates plasma lipids and why Clock mutant mice develop metabolic syndrome or resistance to obesity depending on their genetic background. Knowledge of the molecular and biological mechanisms regulating daily variations in plasma lipids might provide new insight regarding the pathobiology of common metabolic disorders.
Apart from plasma lipids and lipoproteins, biosynthesis of lipids in tissues also shows diurnal variations. Several in vitro and in vivo studies show that cholesterol synthesis exhibits circadian rhythm in the liver and intestine [58–63]. In fact, several proteins involved in lipid metabolism (such as hepatic cytochrome P450 cholesterol 7 α-hydroxylase, HMG CoA reductase, lipolytic enzymes, apolipoprotein AIV and Pparα) show diurnal variations in both humans and rodents. The expression of most of these enzymes does not show diurnal variations in Clock mutant mice, indicating that Clock is important for their diurnal expression in the liver [64,65]. Besides light-dependent regulation, the importance of feeding as a modulator of several metabolic enzymes has been revealed by numerous studies [61,66,67]. One hamster study showed that feeding patterns affect diurnal rhythmicity of cholesterol biosynthesis and HMG-CoA reductase activity in the intestine and liver [66]. A similar connection has been reported between food intake and the cyclic rise in HMG-CoA reductase activity [61]. Although it is generally believed that food intake is responsible for the major increase in intestine and liver cholesterol synthesis during the circadian cycle [67], there does not seem to be a general agreement on this point, and other theories point to hormonal regulation [68]. Thus, specific mechanisms responsible for changes in tissue cholesterol levels remain to be identified. As there is for cholesterol, there is evidence for the circadian expression of genes involved in triglyceride biosynthesis. For example, sterol-regulatory-element-binding protein-1c, acetyl-CoA carboxylase, fatty acid synthase and fatty-acid-binding protein 4 show diurnal variations in the liver and fat tissues of normal mice [69,70]. Mechanisms that control circadian expression of genes involved in cholesterol and triglyceride biosynthesis have not yet been explained.
As discussed previously, synthesis and transport of lipids show diurnal variation. Recent studies indicate that lipids might affect circadian expression of clock and clock-controlled genes in peripheral tissues. C57BL/6J mice on a high-fat diet, for example, show dampened diurnal rhythms of canonical clock and clock-controlled genes in the liver and adipose tissue [70]. In addition, diabetic mice show attenuated circadian expression of hepatic clock genes compared to non-obese control mice [71]. In contrast, high-fat or cholesterol and cholic acid diets in ICR mice do not affect circadian expression of hepatic clock genes [65,72]. These studies indicate that the effect of fat might be strain specific in mice. More experiments are necessary to explain the effect of fat accumulation on clock gene expression and other circadian activities.
Future perspectives
Alterations in sleeping habits owing to modern busy lifestyles affect circadian rhythms and could contribute to obesity [73]. Furthermore, easy accessibility to high-caloric ‘fast food’, changes in dietary content and inactivity can also contribute to the metabolic syndrome [74]. Indeed, high-fat diet disrupts circadian regulation of clock genes and other physiological rhythms [70], although how high fat alters circadian rhythms remains to be determined. It is possible that high fat disrupts signaling pathways that are important for exquisite elaboration of circadian rhythms. Alternatively, accumulation of fat might affect secondary metabolites and crucial molecules that might be intermediary in elucidating circadian rhythms. Conversely, restricted food availability or caloric restriction is thought to prolong life and reduce cancer risk; again, however, mechanisms for such effects are poorly defined [75]. Fasting is the primary approach self-imposed by many individuals to lose weight. How do these self-imposed food restrictions affect metabolic processes? Do they favor or avoid hyperlipidemia and the metabolic syndrome? Attempts to understand the molecular and biological mechanisms regulated by food entrainment might unravel new knowledge about the pathobiology of common metabolic disorders associated with fasting and feasting.
One of the main purposes of this review is to inspire vigor in identifying the role of the intestine in food-entrained regulation of circadian rhythms. There are several fundamental questions that need to be addressed in this regard. How do intestinal cells communicate with the brain to elicit a food-entrainable response? How does the brain prepare organisms for mealtime? What molecules are crucial for food entrainment of the intestine and other peripheral tissues? Knowledge of these molecular and biological mechanisms, as well as the role of clock genes in food entrainment, might be useful in understanding etiologies of gastrointestinal, metabolic and behavioral abnormalities.
Obviously, several different approaches are required to answer these questions. Despite the evidence that vagal communication might not be necessary for food entrainment, there is a need to examine biochemical and molecular changes in the DMH and other areas of the brain after disruption of vagal innervations in food-entrained animals. Experiments are needed to precisely activate different regions of the brain and identify loci that are important in the elucidation of food-anticipatory and -consuming responses. It might be easier to expose food-entrained animals to different agonists and antagonists to recognize receptors and neurons important for food entrainment. Similarly, neuron-specific ablation might provide invaluable information about the role of candidate genes in food entrainment. Exposing brain cell cultures to various peptides and hormones elaborated by intestinal cells might highlight the role of humoral factors. The humoral communication from the DMH to the intestine and other peripheral tissues in food entrainment is another area that needs further experimentation. In this regard, newly identified molecules (e.g. C16:1n7-plamioleate [76], oleoylethanolamide [77], N-acylphosphatidylethanolamine [78] and adropin [79]) that have been shown to have a role in satiety and other physiologic functions might be interesting candidates for the entrainment of the DMH.
Another area of interest that needs immediate attention is the expression of intestinal clock genes. Are there variations along the jejunum–colon axis in expression of these genes? More importantly, why are clock genes expressed in the intestine? A simple explanation is that they are needed to control local circadian expression of several genes. Is it possible that clock genes might have roles other than their involvement in circadian regulation? Could clock genes be important in regulating genes that carry out normal physiologic functions? It is possible, for example, that clock genes play a part in the expression of genes involved in carbohydrate, protein and lipid absorption?
It is also essential to identify clock-controlled genes that are important in regulating various intestinal functions and to explain how clock genes transmit circadian signals to intestinal transporters in carrying out diurnal activities. It is likely that various clock-controlled genes, independently or in combination, control proteins involved in absorption of proteins, carbohydrates and lipids. Therefore, experiments are needed to understand molecular interactions, communications and coordination among clock genes and between other functional genes that regulate intestinal functions.
Gastrointestinal disturbances are one of the most common health problems reported in shift workers. Several theories have been posited to explain common maladies in shift workers, including gastrointestinal motility, acid–base imbalance, activation of stress response and immune depression. However, few molecular explanations for the high number of gastrointestinal complications in shift workers have been found. At the molecular level, it is possible that gastrointestinal functions are more dependent on clock genes than other tissues are. A critical evaluation of the dependence of intestinal functions, such as digestion and nutrient absorption, on clock genes should be performed. It is anticipated that future research will offer novel knowledge about the role of diet, the intestine and the FEO in regulating physiologic and behavioral activities, as well as provide new perceptions about the molecular mechanisms involved in the transmission and entrainment of biochemical and cellular events by food.
Acknowledgments
This work was supported in part by NIH grant DK-46700, as well as by a Grant-in-Aid (M.M.H.) and Postdoctoral Fellowship (X.P.) from the American Heart Association.
Glossary
- Circadian
a repeating act with an approximate interval of 24 h
- E-box
a DNA sequence (CACGTG) present in the promoter or enhancer regions of specific genes
- Enterocyte
differentiated intestinal epithelial cells that carry out nutrient absorption
- Entrainment
learning chores after exposure to the same stimuli at regular intervals
- Humoral factors
factors that travel through body fluids to affect function in other tissues
- Retino-hypothalamic tract
a set of neurons that originate in the eye and send signals to the hypothalamus
- Suprachiasmatic nuclei
a pair of tiny clusters of cells present in the hypothalamus above the chiasma
- Vagotomy
surgical disruption of the vagus nerve
References
- 1.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hastings MH, et al. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 3.Green CB, et al. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi JS, et al. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 6.Ramsey KM, et al. The clockwork of metabolism. Annu Rev Nutr. 2007;27:219–240. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- 7.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 8.Rutter J, et al. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 9.Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci. 2001;2:521–526. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- 10.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 11.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 12.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, et al. Transcriptional coactivator PGC-1a integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 15.Duez H, Staels B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab Vasc Dis Res. 2008;5:82–88. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- 16.Duez H, Staels B. Rev-erb alpha gives a time cue to metabolism. FEBS Lett. 2008;582:19–25. doi: 10.1016/j.febslet.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 17.Davidson AJ, et al. Is the food-entrainable circadian oscillator in the digestive system? Genes Brain Behav. 2003;2:32–39. doi: 10.1034/j.1601-183x.2003.00005.x. [DOI] [PubMed] [Google Scholar]
- 18.Stephan FK. The “other” circadian system: food as a Zeitgeber. J Biol Rhythms. 2002;17:284–292. doi: 10.1177/074873040201700402. [DOI] [PubMed] [Google Scholar]
- 19.Saper CB, Fuller PM. Inducible clocks: living in an unpredictable world. Cold Spring Harb Symp Quant Biol. 2007;72:543–550. doi: 10.1101/sqb.2007.72.008. [DOI] [PubMed] [Google Scholar]
- 20.Feillet CA, et al. “Feeding time” for the brain: a matter of clocks. J Physiol (Paris) 2006;100:252–260. doi: 10.1016/j.jphysparis.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Mendoza J. Circadian clocks: setting time by food. J Neuroendocrinol. 2007;19:127–137. doi: 10.1111/j.1365-2826.2006.01510.x. [DOI] [PubMed] [Google Scholar]
- 22.Mendoza J, et al. Entrainment by a palatable meal induces food-anticipatory activity and c-Fos expression in reward-related areas of the brain. Neuroscience. 2005;133:293–303. doi: 10.1016/j.neuroscience.2005.01.064. [DOI] [PubMed] [Google Scholar]
- 23.Pitts S, et al. Food-entrained circadian rhythms are sustained in arrhythmic Clk/Clk mutant mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R57–R67. doi: 10.1152/ajpregu.00023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokkan KA, et al. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 26.Hara R, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 27.Horikawa K, et al. Rapid damping of food-entrained circadian rhythm of clock gene expression in clock-defective peripheral tissues under fasting conditions. Neuroscience. 2005;134:335–343. doi: 10.1016/j.neuroscience.2005.03.057. [DOI] [PubMed] [Google Scholar]
- 28.Fuller PM, et al. Differential rescue of light- and food-entrainable circadian rhythms. Science. 2008;320:1074–1077. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gooley JJ, et al. The dorsomedial hypothalamic nucleus is critical for the expression of food-entrainable circadian rhythms. Nat Neurosci. 2006;9:398–407. doi: 10.1038/nn1651. [DOI] [PubMed] [Google Scholar]
- 30.Landry GJ, et al. The dorsomedial hypothalamic nucleus is not necessary for the expression of circadian food-anticipatory activity in rats. J Biol Rhythms. 2007;22:467–478. doi: 10.1177/0748730407307804. [DOI] [PubMed] [Google Scholar]
- 31.Mieda M, et al. The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc Natl Acad Sci U S A. 2006;103:12150–12155. doi: 10.1073/pnas.0604189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verwey M, et al. Region-specific modulation of PER2 expression in the limbic forebrain and hypothalamus by nighttime restricted feeding in rats. Neurosci Lett. 2008;440:54–58. doi: 10.1016/j.neulet.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 33.Feillet CA, et al. Lack of food anticipation in Per2 mutant mice. Curr Biol. 2006;16:2016–2022. doi: 10.1016/j.cub.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 34.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 35.Mansbach CM, Gorelick F. Development and physiological regulation of intestinal lipid absorption. II Dietary lipid absorption, complex lipid synthesis, and the intracellular packaging and secretion of chylomicrons. Am J Physiol Gastrointest Liver Physiol. 2007;293:G645–G650. doi: 10.1152/ajpgi.00299.2007. [DOI] [PubMed] [Google Scholar]
- 36.Pan X, et al. The diurnal rhythm of the intestinal transporters SGLT1 and PEPT1 is regulated by the feeding conditions in rats. J Nutr. 2004;134:2211–2215. doi: 10.1093/jn/134.9.2211. [DOI] [PubMed] [Google Scholar]
- 37.Pan X, et al. Diurnal rhythm of H+-peptide cotransporter in rat small intestine. Am J Physiol Gastrointest Liver Physiol. 2002;283:G57–G64. doi: 10.1152/ajpgi.00545.2001. [DOI] [PubMed] [Google Scholar]
- 38.Tavakkolizadeh A, et al. Differential role of vagus nerve in maintaining diurnal gene expression rhythms in the proximal small intestine. J Surg Res. 2005;129:73–78. doi: 10.1016/j.jss.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 39.Daniel H. Molecular and integrative physiology of intestinal peptide transport. Annu Rev Physiol. 2004;66:361–384. doi: 10.1146/annurev.physiol.66.032102.144149. [DOI] [PubMed] [Google Scholar]
- 40.Pan X, et al. Altered diurnal rhythm of intestinal peptide transporter by fasting and its effects on the pharmacokinetics of ceftibuten. J Pharmacol Exp Ther. 2003;307:626–632. doi: 10.1124/jpet.103.055939. [DOI] [PubMed] [Google Scholar]
- 41.Saito H, et al. Regulatory mechanism governing the diurnal rhythm of intestinal H+/peptide cotransporter 1 (PEPT1) Am J Physiol Gastrointest Liver Physiol. 2008;295:G395–G402. doi: 10.1152/ajpgi.90317.2008. [DOI] [PubMed] [Google Scholar]
- 42.Broer S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol Rev. 2008;88:249–286. doi: 10.1152/physrev.00018.2006. [DOI] [PubMed] [Google Scholar]
- 43.Terada T, et al. Expression profiles of various transporters for oligopeptides, amino acids and organic ions along the human digestive tract. Biochem Pharmacol. 2005;70:1756–1763. doi: 10.1016/j.bcp.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 44.Tavakkolizadeh A, et al. Diurnal rhythmicity in intestinal SGLT-1 function, Vmax, and mRNA expression topography. Am J Physiol Gastrointest Liver Physiol. 2001;280:G209–G215. doi: 10.1152/ajpgi.2001.280.2.G209. [DOI] [PubMed] [Google Scholar]
- 45.Hoogerwerf WA. Biologic clocks and the gut. Curr Gastroenterol Rep. 2006;8:353–359. doi: 10.1007/s11894-006-0019-3. [DOI] [PubMed] [Google Scholar]
- 46.Scheving LE, et al. Chronobiology of the intestinal tract of the mouse. Am J Anat. 1983;168:433–465. doi: 10.1002/aja.1001680405. [DOI] [PubMed] [Google Scholar]
- 47.Vener KJ, et al. The effect of shift work on gastrointestinal (GI) function: a review. Chronobiologia. 1989;16:421–439. [PubMed] [Google Scholar]
- 48.Hoogerwerf WA, et al. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133:1250–1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Sladek M, et al. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. 2007;133:1240–1249. doi: 10.1053/j.gastro.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 50.Maillot F, et al. Changes in plasma triacylglycerol concentrations after sequential lunch and dinner in healthy subjects. Diabetes Metab. 2005;31:69–77. doi: 10.1016/s1262-3636(07)70169-6. [DOI] [PubMed] [Google Scholar]
- 51.Hussain MM, et al. Microsomal triglyceride transfer protein and its role in apolipoprotein B-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 52.Hussain MM, et al. Microsomal triglyceride transfer protein in plasma and cellular lipid metabolism. Curr Opin Lipidol. 2008;19:277–284. doi: 10.1097/MOL.0b013e3282feea85. [DOI] [PubMed] [Google Scholar]
- 53.Pan X, Hussain MM. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J Biol Chem. 2007;282:24707–24719. doi: 10.1074/jbc.M701305200. [DOI] [PubMed] [Google Scholar]
- 54.Escobar C, et al. Persistence of metabolic rhythmicity during fasting and its entrainment by restricted feeding schedules in rats. Am J Physiol. 1998;274:R1309–R1316. doi: 10.1152/ajpregu.1998.274.5.R1309. [DOI] [PubMed] [Google Scholar]
- 55.Fukagawa K, et al. Circadian rhythm of serum and lymph apolipoprotein AIV in ad libitum-fed and fasted rats. Am J Physiol. 1994;267:R1385–R1390. doi: 10.1152/ajpregu.1994.267.5.R1385. [DOI] [PubMed] [Google Scholar]
- 56.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oishi K, et al. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 58.Mortimer BC, et al. The diurnal rhythms of cholesterol metabolism and plasma clearance of model chylomicrons: comparison of normal and genetically hypercholesterolemic rats (RICO) Comp Biochem Physiol A Mol Integr Physiol. 1998;120:671–680. doi: 10.1016/s1095-6433(98)10085-5. [DOI] [PubMed] [Google Scholar]
- 59.Edwards PA, et al. In vivo demonstration of the circadian rhythm of cholesterol biosynthesis in the liver and intestine of the rat. J Lipid Res. 1972;13:396–401. [PubMed] [Google Scholar]
- 60.Back P, et al. Regulation of cholesterol biosynthesis in rat liver: diurnal changes of activity and influence of bile acids. Arch Biochem Biophys. 1969;133:11–21. doi: 10.1016/0003-9861(69)90482-2. [DOI] [PubMed] [Google Scholar]
- 61.Shapiro DJ, Rodwell VW. Diurnal variation and cholesterol regulation of hepatic HMG-CoA reductase activity. Biochem Biophys Res Commun. 1969;37:867–872. doi: 10.1016/0006-291x(69)90972-3. [DOI] [PubMed] [Google Scholar]
- 62.Hamprecht B, et al. Rhythmic changes of hydroxymethylglutaryl coenzyme a reductase activity in livers of fed and fasted rats. FEBS Lett. 1969;4:117–121. doi: 10.1016/0014-5793(69)80210-3. [DOI] [PubMed] [Google Scholar]
- 63.Mayer D. The circadian rhythm of synthesis and catabolism of cholesterol. Arch Toxicol. 1976;36:267–276. doi: 10.1007/BF00340534. [DOI] [PubMed] [Google Scholar]
- 64.Oishi K, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 65.Kudo T, et al. Clock mutation facilitates accumulation of cholesterol in the liver of mice fed a cholesterol and/or cholic acid diet. Am J Physiol Endocrinol Metab. 2008;294:E120–E130. doi: 10.1152/ajpendo.00061.2007. [DOI] [PubMed] [Google Scholar]
- 66.Ho KJ. Circadian rhythm of cholesterol biosynthesis: dietary regulation in the liver and small intestine of hamsters. Int J Chronobiol. 1979;6:39–50. [PubMed] [Google Scholar]
- 67.Jurevics H, et al. Diurnal and dietary-induced changes in cholesterol synthesis correlate with levels of mRNA for HMG-CoA reductase. J Lipid Res. 2000;41:1048–1054. [PubMed] [Google Scholar]
- 68.Mirani-Oostdijk CP, et al. Diurnal changes in serum triglycerides as related to changes in lipolytic enzymes, lipoproteins and hormones in patients with primary endogenous hypertriglyceridaemia on a carbohydrate-rich diet. Atherosclerosis. 1985;57:129–137. doi: 10.1016/0021-9150(85)90026-7. [DOI] [PubMed] [Google Scholar]
- 69.Kudo T, et al. Attenuating effect of clock mutation on triglyceride contents in the ICR mouse liver under a high-fat diet. J Biol Rhythms. 2007;22:312–323. doi: 10.1177/0748730407302625. [DOI] [PubMed] [Google Scholar]
- 70.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Ando H, et al. Profile of rhythmic gene expression in the livers of obese diabetic KK-Aψ mice. Biochem Biophys Res Commun. 2006;346:1297–1302. doi: 10.1016/j.bbrc.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 72.Yanagihara H, et al. High-fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol Int. 2006;23:905–914. doi: 10.1080/07420520600827103. [DOI] [PubMed] [Google Scholar]
- 73.Bray MS, Young ME. Circadian rhythms in the development of obesity: potential role for the circadian clock within the adipocyte. Obes Rev. 2007;8:169–181. doi: 10.1111/j.1467-789X.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- 74.Zimmet P, et al. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 75.Froy O, Miskin R. The interrelations among feeding, circadian rhythms and ageing. Prog Neurobiol. 2007;82:142–150. doi: 10.1016/j.pneurobio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 76.Cao H, et al. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwartz GJ, et al. The lipid messenger OEA links dietary fat intake to satiety. Cell Metab. 2008;8:281–288. doi: 10.1016/j.cmet.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gillum MP, et al. N-acylphosphatidylethanolamine, a gut-derived circulating factor induced by fat ingestion, inhibits food intake. Cell. 2008;135:813–824. doi: 10.1016/j.cell.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar KG, et al. Identification of adropin as a secreted factor linking dietary macronutrient intake with energy homeostasis and lipid metabolism. Cell Metab. 2008;8:468–481. doi: 10.1016/j.cmet.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


