Abstract
MicroRNA (miR) regulates hematopoiesis through targeting different genes post-transcriptionally. We have recently shown that Tip110 expression is downregulated during hematopoietic stem cell differentiation. However, the underlying mechanisms are not known. In this study, we identified a conserved miR-124-binding site on the Tip110 3′-untranslated region (3′-UTR) and showed that Tip110 was downregulated by miR-124 through its 3′-UTR. We then examined the relationship among miR-124 and Tip110 expression and differentiation of human cord blood CD34+ cells. We found that miR-124 was expressed in a low level in human cord blood CD34+ cells, but it was considerably upregulated during culturing and differentiation of these cells. Moreover, we demonstrated that miR-124 expression decreased Tip110 expression and promoted differentiation of human cord blood CD34+ cells, while miR-124 knockdown increased Tip110 expression, slowed down differentiation of human cord blood CD34+ cells, and caused an expansion of hematopoietic progenitor cells in vitro. Finally, we used mouse embryonic fibroblasts derived from Tip110 transgenic mice, performed the exon array analysis, and found that Tip110 altered a number of genes in the hematopoiesis pathways. Dnmt3a as de novo methyltransferase was also significantly upregulated. That miR-124 was markedly upregulated during human cord blood CD34+ cell differentiation could be the result of direct loss of its promoter methylation from Dnmt3a. Taken together, our study demonstrates that miR-124 regulates Tip110 expression and differentiation of human cord blood CD34+ cells and suggests important roles of miR-124/Tip110 in hematopoiesis.
Introduction
Hematopoiesis is tightly regulated by complex multidimensional mechanisms, including those mediated by transcription factors, microRNAs (miRNAs), and epigenetic modifiers [1]. miRNAs are naturally occurring, small noncoding RNA molecules, usually 21–25 nucleotides long, present in a wide variety of organisms, highly conserved during evolution, and regulate gene expression by targeting the 3′-untranslated regions (3′-UTR) of the mRNAs [2]. miRNAs are mostly potent negative regulators of gene expression leading to gene silencing. A few hundreds of miRNAs have been identified in the human genome. In the hematopoietic system, miRNAs are required and are essential for functional hematopoiesis and regulate hematopoietic stem cells (HSCs) and lineage-committed hematopoietic progenitor cells (HPCs) [3,4]. Deregulated expression of certain miRNAs in the hematopoietic system is linked to hematologic malignancies.
miR-124 was originally identified as one of the most abundantly expressed miRNAs in the central nervous system. It is highly conserved among diverse species. miR-124 is expressed during the terminal neuronal differentiation [5]. It is not expressed in neuronal stem cells and its expression begins during the transition from neuronal stem cells to neuronal progenitors [6]. miR-124 represses various types of cancer, such as breast cancer, gastric cancer, prostate cancer, colorectal cancer, glioma, and glioblastomas, through inhibiting cellular proliferation, invasion, and inducing apoptosis. Several target genes of miR-124 have been identified to suppress tumors, such as, Ets-1, ROCK1, SMC4, TGF-α, and EAE. In addition, miR-124 promotes microglia quiescence through its target gene, CCAAT/enhancer-binding protein-α (C/EBP-α) [7–11].
Human Tip110 was first identified as an RNA-binding nuclear protein to regulate HIV-1 gene expression and replication [12]. Our recent studies reported that Tip110 was expressed in HSCs and Tip110 expression levels decreased when HSCs differentiated [13]. Tip110 also regulates proliferation of HPCs [13]. Ectopic expression of Tip110 in quiescent CD34+ cord blood cells decreased apoptosis and enhanced numbers of cells entering into the cell cycle [13,14]. In the present study, we focused on the relationship among miR-124 and Tip110 expression and CD34+ cell differentiation. Using several molecular and cell biology strategies, we demonstrated that miR-124 targeted Tip110 expression and regulated CD34+ cell differentiation.
Materials and Methods
Cells
293T were purchased from the American Tissue Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Grand Island, NY) with 10% fetal bovine serum (FBS; Hyclone, Logan, UT) at 37°C and 5% CO2. Transfections were carried out using the standard calcium phosphate precipitation. Human cord blood was collected according to the institutional guidelines of Indiana University School of Medicine, Indianapolis, Indiana, and used to obtain CD34+ cells within 24 h of collection using an immunomagnetic selection (Miltenyi Biotech, San Diego, CA). CD34+ cells (>93% pure) were cultured in Iscove's modified Dulbecco's media (IMDM; Life Technologies) containing 10% FBS (Hyclone) and cytokines, human stem cell factor (100 ng/mL), human FLT3 ligand (100 ng/mL), and human thrombopoietin (100 ng/mL). All these three cytokines were from R&D Systems (Minneapolis, MN).
Plasmids and luciferase reporter gene assay
Human Tip110 3′-UTR-driven luciferase reporter plasmid pLightSwitch-Tip110.3′UTR was purchased from Switchgear Genomics (Carlsbad, CA), which includes the entire Tip110 3′-UTR (1,346 nt). Lentiviral vector expressing miR-124 (HmiR0427-MR03) and miR-124 inhibitor (HmiR-AN0074-AM03) and their paired respective controls were from GeneCopoeia, Rockville, MD. miR-124 mimic and its paired control were from Sigma, St. Louis, MO. For the luciferase reporter gene assay, 293T were transfected with indicated plasmids as well as pTK-βGal, which was used to normalize the transfection variations among all transfections. Forty-eight hours post-transfection, the cells were washed, lysed, and assayed for the luciferase activity using a luciferase assay kit (Promega, Madison, WI).
Western blot analysis
Western blot analysis of protein expression was performed as previously described [12]. Primary antibodies were mouse anti-human Tip110 antibody [12] and mouse anti-human β-actin antibody (Sigma Chem. Co., St. Louis, MO). β-Actin was included as a loading control.
Real time polymerase chain reaction
Total RNA was isolated from cells using TRIzol (Invitrogen, Grand Island, NY). Before RNA precipitation, RNA was extracted with acid phenol:chloroform:isoalcohol (125:24:1), pH 4.5, to prevent residual genomic DNA in the RNA preparations from amplification. cDNAs were synthesized using a ScriptII RT reagent kit (Promega) and oligo d(T)(18) as the primer. Tip110 sequences were amplified by using primers, 5′-CTT CAT CCA GGC CAC TGA TT-3′ and 5′-TTG AAA CGC TCT TCC ACC TC-3′. β-Actin was used as a reference for real time PCR and amplified using primers, 5′-AAC ACC CCA GCC ATG TAC GT-3′ and 5′-AGT ACT TGC GCT CAG GAG GA-3′.
Flow cytometric analysis of human cord blood CD34+ cells
Freshly purified and cultured human cord blood CD34+ cells from the different experimental groups were washed twice with PBS and stained for mouse anti-human CD34-PE/CD34-APC/CD34-FITC antibody, mouse anti-human CD38-PE antibody, mouse anti-human CD45RA-PECF594, mouse anti-human CD90-PECy7, and mouse anti-human CD49f-PerCPCy5.5 in the dark at room temperature for 30 min. All those antibodies were from BD Biosciences (San Jose, CA). Mouse IgG-FITC, mouse IgG-APC, and mouse IgG-PE were included as negative controls. The cells were subjected to flow cytometric analysis using FACS (Becton Dickinson, Franklin Lakes, NJ).
miR-124 analysis
Total RNA was isolated from human cord blood CD34+ cells using TRIzol (Invitrogen) according to the manufacturer's protocols and used (25 ng) for reverse transcription, followed by amplification using the TaqMan RNA-to-Ct 1-step Kit (Applied Biosystems, Foster City, CA). Specific miR-124 primers and miRNA TaqMan probes were designed by and obtained from Applied Biosystems. Reverse transcription was performed at 48°C for 15 min. PCR was performed for 40 cycles at 95°C for 15 s and 60°C for 60 s. miR-124 levels were calculated by the comparative ΔΔCt method and normalized to endogenous small nuclear U6 RNA.
Lentiviral preparation and transduction
293T were transfected with lentiviral plasmids expressing miR-124, miR-124 inhibitor, or their respective controls, along with pMD-gag-pol, pRSV-Rev, and pMDL-RRE (GeneCopoeia), using the calcium phosphate transfection. The cell culture supernatants were collected 48 h post-transfection, filtered through a 0.45-μm low-protein-binding filter (Millipore, Darmstadt, Germany), and used immediately for transduction, or stored in liquid nitrogen as lentivirus stock for later transduction. Human cord blood CD34+ cells were transduced with lentiviruses as previously described [15]. Briefly, freshly purified human cord blood CD34+ cells were cultured as described above for 24 h. The cells were then incubated with lentiviruses in the presence of 8 μg/mL polybrene for 8 h. After the end of incubation, the cells were washed extensively and cultured in IMDM medium. The transduction efficiency was estimated to be 20% [15].
In vitro hematopoietic colony-forming assay
After culturing for 4 days, lentivirus-transduced human cord blood CD34+ cells were seeded in triplicate onto a 35-mm dish at a density of 500 cells per dish in 1 mL 1% methylcellulose culture medium containing 100 mM 2-mercaptoethanol, 2 mM l-glutamine, and 30% FBS (Hyclone). Recombinant human erythropoietin (1 U/mL), stem cell factor (SCF) (50 ng/mL), human interleukin-3 (IL-3; 10 ng/mL), and granulocyte–macrophage colony-stimulating factor (GM-CSF; 10 ng/mL) (R&D System) were included in the culture medium containing EPO (Amgen, Inc., Thousand Oaks, CA) to stimulate colonies from multipotential [CFU-granulocyte–erythrocyte–macrophage–megakaryocyte (CFU-GEMM)], erythroid [burst-forming unit-erythroid (BFU-E)], and granulocyte–macrophage [CFU-GM)] progenitor cells. Colonies were scored after 14 days of incubation at 5% CO2 and lowered O2 (5%) under an inverted microscope.
Preparation of mouse embryonic fibroblast and exon microarray
Pregnant Tip110 transgenic (TG) and wild-type littermate (WT) control mice [13] were sacrificed at day 13. Each embryo was dissected from the placenta and embryonic sac. After finely mincing the tissue and treating it with trypsin, cells were washed with mouse embryonic fibroblast (MEF) culture medium (Life Technologies) and suspended in MEF medium. Total RNA was isolated using TRIzol (Invitrogen) and used for Affymetrix Exon Microarray (Affymetrix, Santa Clara, CA). Briefly, total RNA was converted into sense-strand cDNA using an Ambion WT Expression Kit (Invitrogen). The cDNA was then in vitro transcribed to create a complement RNA (cRNA) template. cRNA was combined with random primers and synthesized to produce cDNA. The cDNA was then fragmented and biotin labeled using an Affymetrix GeneChip® WT Terminal Labeling Kit (Affymetrix). The labeled cDNA was suspended in the hybridization cocktail and loaded onto GeneChip Mouse Exon 1.0 ST array (Affymetrix) for hybridization. After staining with streptavidin, signal amplification was assessed using biotinylated antistreptavidin. The images were analyzed with corresponding quality control metrics. Then, gene expression analysis was performed to identify genes of interest as follows: The first-pass analysis was performed using the Analysis Wizard of DAVID Bioinformatics Resources 6.7 from NIAID/NIH (http://david.abcc.ncifcrf.gov/). Normalized and filtered data were then entered into hematopoiesis pathway analysis. Genes identified in the pathway analysis were selected for analysis in KEGG pathways.
Statistical analysis
The data are mean±SD and representative of multiple independent experiments. All experiment data were analyzed by two-tailed student's t-test. A P value <0.05 was considered to be statistically significant, and a P value <0.005 was considered to be highly statistically significant.
Results
miR-124 targets Tip110 3′-UTR and downregulates Tip110 expression
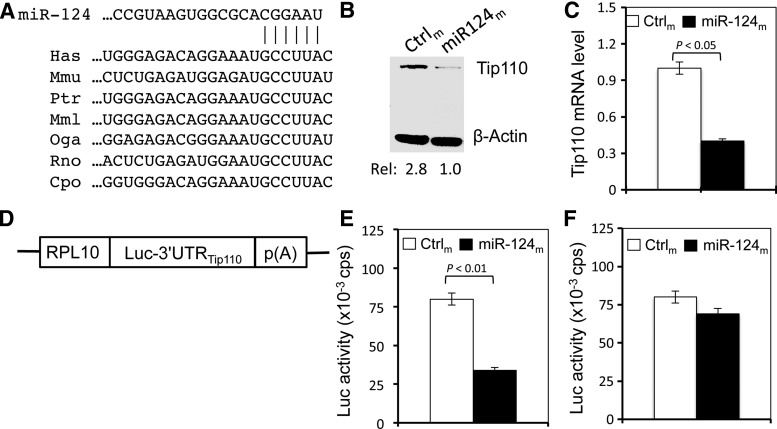
To determine the molecular mechanism for regulation of Tip110 expression during HSC differentiation, we focused on the 3′-UTR of the Tip110 gene. Using the TargetScan analysis (www.targetscan.org), we found that the 3′-UTR of human Tip110 mRNA contained a complementary site for the seed region of miR-124 (Fig. 1A). The aligned region was indeed conserved among Tip110 of various species, such as mouse, chimpanzee, rhesus, bushbaby, rat, and guinea pig. To determine whether Tip110 mRNA expression is regulated by miR-124, 293T were transfected with miR-124 mimics, which are chemically synthesized double-stranded RNAs that mimic mature endogenous miRNAs in transfected cells. Compared with the miRNA mimic control, the miR-124 mimic expression considerably decreased Tip110 protein expression (Fig. 1B) as well as Tip110 mRNA expression (Fig. 1C). To determine whether Tip110 3′-UTR is involved in miR-124-induced Tip110 mRNA and protein downregulation, we constructed a reporter plasmid by inserting the Tip110 mRNA 3′-UTR immediately next to the 3′ end of the luciferase gene (Fig. 1D). Similarly, coexpression of the reporter plasmid and miR-124 mimic led to significant decreases in the Tip110 3′-UTR-mediated luciferase activity (Fig. 1E). To further ascertain the specificity of the miR-124 interaction with Tip110 3′-UTR in Tip110 mRNA and protein downregulation, another reporter plasmid containing the deletion of the miR-124-binding site from the Tip110 3′-UTR were transfected with miR-124 mimics. Compared with the control, coexpression of the mutated reporter plasmid and miR-124 mimic showed no significant changes in the luciferase activity (Fig. 1F). Taken together, these results indicate that Tip110 is likely a target of miR-124 through its 3′-UTR.
FIG. 1.
Regulation of Tip110 expression by miR-124 through its 3′-UTR. (A) Sequence alignment between human miR-224 seed sequence and 3′-UTR of Tip110 mRNA from different species, including mouse, chimpanzee, rhesus, bushbaby, rat, and guinea pig. (B, C) 293T were transfected with miR-124 mimic (miR-124m) or its control (Ctrlm). Forty hours after transfection, cells were harvested for cell lysates and western blotting (B) or RNA isolation and real time polymerase chain reaction (qRT-PCR) (C). β-Actin was included as the loading control for western blotting (B) and as a reference for qRT-PCR (C). (D) Scheme of human Tip110 3′-UTR-containing luciferase reporter plasmid pLightSwitch-Tip110.3′UTR. (E, F) 293T were transfected with miR-124m and pLightSwitch-Tip110.3′UTR (E) or miR-124-binding site-deleted pSwitchgear-Tip110.3′UTR (F). Forty-eight hours after transfection, cells were harvested for cell lysates and luciferase reporter gene assay. pTK-βGal was included for transfection efficiency normalization (E, F). The data in (D, E) are mean±SEM and representative of three independent experiments. 3′-UTR, 3′-untranslated region.
miR-124 expression is inversely correlated with CD34+ cell differentiation
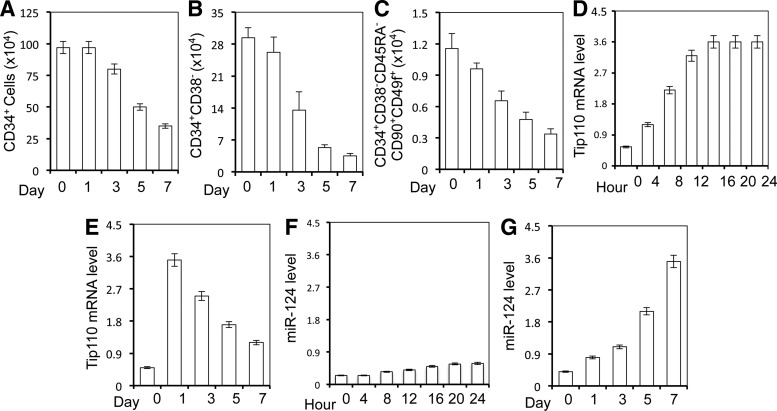
Several studies have demonstrated miRNA expression changes during hematopoiesis and hematopoietic differentiation of CD34+ cells [1,16,17]. However, it is not known whether miR-124 expression changes during hematopoietic differentiation of CD34+ cells. To investigate the expression kinetics of miR-124 in relation to CD34+ differentiation, we isolated fresh human cord blood CD34+ cells and cultured them in the medium containing 10% FBS and cytokines, SCF (100 ng/mL), FLT3 ligand (100 ng/mL), and thrombopoietin (100 ng/mL). Freshly isolated cells (day 0) and the cells that were cultured for various days were collected for immunostaining and flow cytometry or RNA isolation for qRT-PCR. CD34+ cells started to differentiate, as we reported previously [13], and as the number of CD34+ cells decreased (Fig. 2A), so did the number of CD34+CD38− cells (Fig. 2B), a marker for more immature cells compared with CD34+ cells, and the number of CD34+CD38−CD45RA−CD90+CD49f+ cells (Fig. 2C), a marker for purified definitive human HSCs [18]. In addition, as we demonstrated previously [13], Tip110 mRNA expression was significantly induced at the beginning of the differentiation, followed by a gradual decrease over time (Fig. 2D). In contrast to Tip110 mRNA expression, miR-124 was detected at a very low level in freshly isolated CD34+ cells (day 0) and gradually increased over CD34+ cell differentiation (Fig. 2E). The correlation coefficient (r) between the number of CD34+ cells and miR-124 expression, between the number of Cd34+CD38− cells and miR-124 expression, and the number of CD34+CD38−CD45RA−CD90+CD49f+ cells and miR-124 expression was −0.989, −0.899, and −0.915, respectively. These results suggest that miR-124 expression is inversely correlated with CD34+ cell differentiation.
FIG. 2.
Effects of CB CD34+ differentiation on miR-124 and Tip110 expression. Human core blood CD34+ cells were isolated and cultured in the presence of 10% fetal bovine serum and cytokines, 100 ng/mL SCF, 100 ng/mL FLT3 ligand, and 100 ng/mL thrombopoietin, for indicated days. The cells were collected. Separate aliquots were used for staining and flow cytometry analysis for CD34+ (A), CD34+CD38− (B), and CD34+CD38−CD45RA−CD90+CD49f+ (C) or RNA isolation and qRT-PCR for Tip110 mRNA level (D, E) and miR-124 level (F, G). β-Actin was included as a reference for Tip110 qRT-PCR (D), and U6 was included as a reference for miR-124 qRT-PCR (E). y-Axis in (A–C) represented the number of cells of the 1 million cells counted. The data are mean±SEM and representative of three independent experiments.
miR-124 expression alters CD34+ cell differentiation
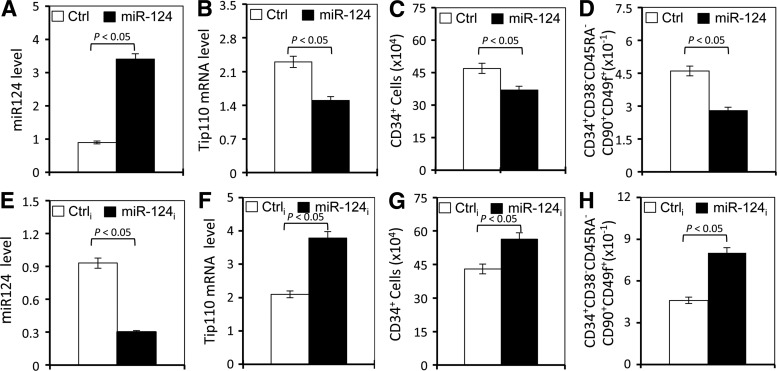
Next, we sought to determine whether manipulation of miR-124 expression would lead to changes in CD34+ cell differentiation. First, we overexpressed miR-124 in human cord blood CD34+ cells through lentivirus transduction, cultured the lentivirus-transduced cells for 4 days, and determined CD34+ cell differentiation. MiR-124 expression in the transduced human cord blood CD34+ cells was confirmed using qRT-PCR (Fig. 3A). As shown before (Fig. 1), miR-124 expression decreased Tip110 mRNA (Fig. 3B). Compared with the transduction control, miR-124 expression significantly decreased the number of CD34+ cells (Fig. 3C) and the number of CD34+CD38−CD45RA−CD90+CD49f+ cells (Fig. 3D) 4 days post-transduction. Then, we also expressed miR-124 inhibitor in human cord blood CD34+ cells and monitored CD34+ cell differentiation. As expected, miR-124 inhibitor expression decreased miR-124 in human cord blood CD34+ cells (Fig. 3E) and increased the Tip110 mRNA level (Fig. 3F). In contrast to miR-124 overexpression, miR-124 inhibitor expression significantly increased the number of CD34+ cells (Fig. 3G) and the number of CD34+CD38−CD45RA−CD90+CD49f+ cells (Fig. 3H) 4 days post-transduction.
FIG. 3.
Effects of miR-124 overexpression and knockdown on CD34 differentiation. Human cord blood CD34+ cells were isolated, cultured for 1 day, and transduced with lentiviruses expressing miR-124 (A–D) or expressing miR-124 inhibitor (miR-124i, E–H). Lentiviruses expressing the control miRNA (Ctrl) and control miRNA inhibitor (Ctrli) were included as the controls. After culturing for 4 days, the cells were collected. Separate aliquots were used for RNA isolation and qRT-PCR for the miR-124 level (A, E) and Tip110 mRNA level (B, F) or for staining and flow cytometry analysis for CD34+ (C, G) or CD34+CD38−CD45RA−CD90+CD49f+ (D, H). U6 was included as a reference for miR-124 qRT-PCR (A, E), while β-actin was included as a reference for Tip110 qRT-PCR (B, F). y-axis in (C–H) represented the number of cells of the 1 million cells counted. The data are mean±SEM and representative of three independent experiments.
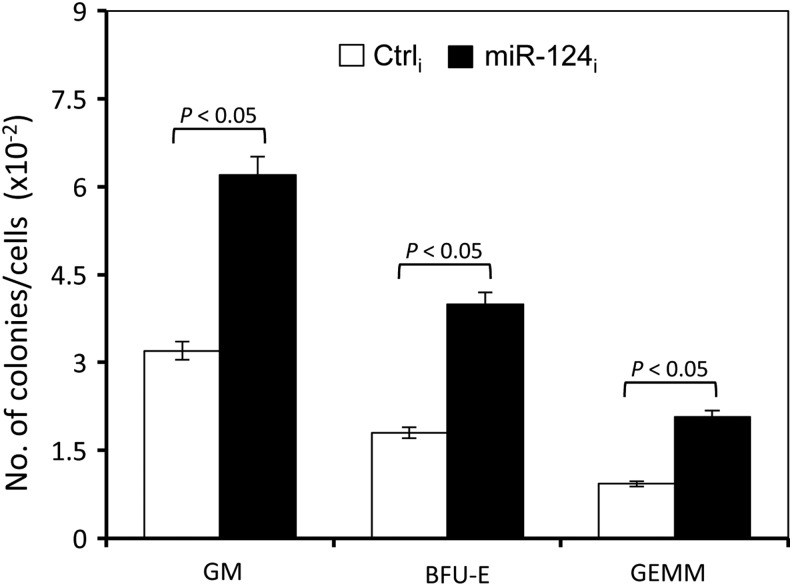
To further assess whether miR-124 affects functionally assayable HPCs, we transduced human cord blood CD34+ cells with lentiviruses expressing miR-124 inhibitor and performed the hematopoietic colony-forming assay. Compared with the control, miR-124 inhibitor expression in human cord blood CD34+ cells increased the number of granulocyte–macrophage (CFU-GM), erythroid (BFU-E), and multipotential (CFU-GEMM) progenitors (Fig. 4). Taken together, these results together suggest that miR-124 expression is a critical regulator of CD34+ cell differentiation and production of the different subsets of HPCs.
FIG. 4.
miR-124 knockdown increased hematopoietic colony formation of human CB CD34+ cells. Human cord blood CD34+ cells were isolated, cultured for 1 day, and transduced with lentiviruses expressing miR-124 inhibitor (miR-124i). Lentiviruses expressing the control miRNA inhibitor (Ctrli) were included as the control. After culturing for 4 days, the cells were seeded in methylcellulose medium at a density of 500 cells per mL for hematopoietic colony formation assay. The data are the ratio of the number of colonies to the number of input cells (ie, 500) and expressed as mean±SEM and representative of three independent experiments.
Link between Tip110 and hematopoiesis
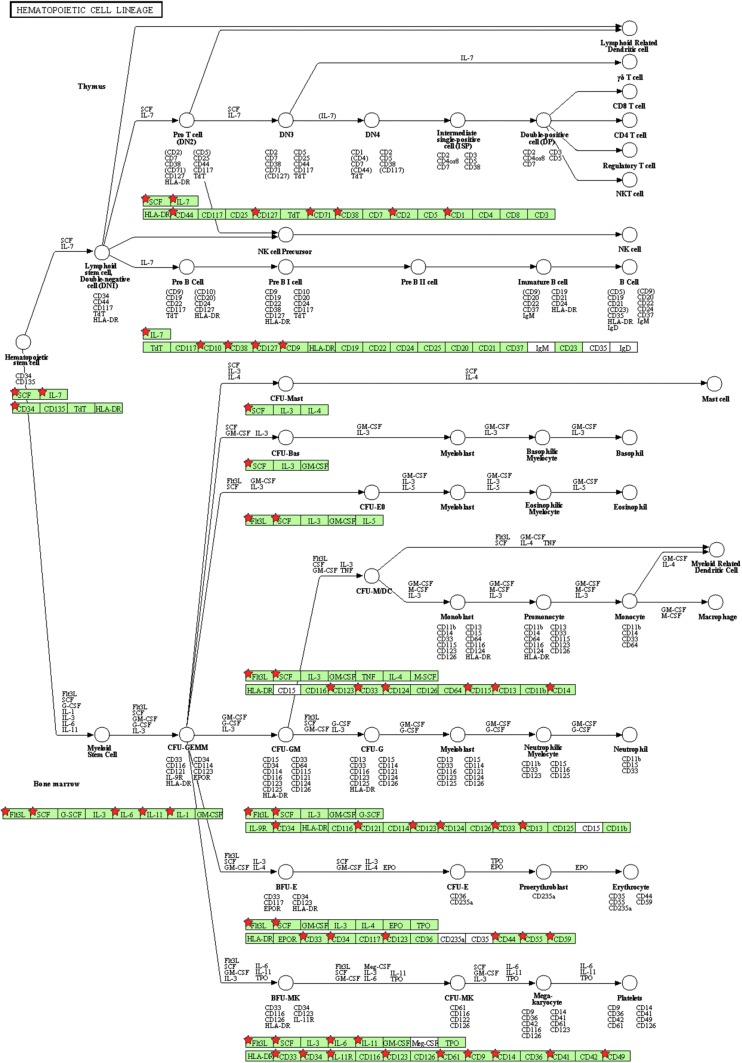
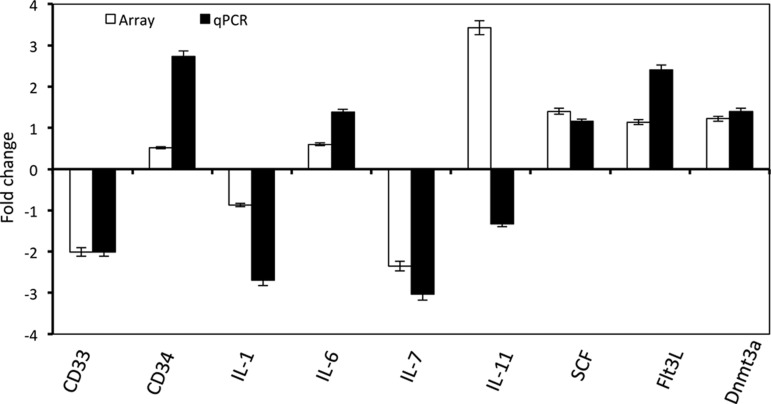
To explore the possible mechanisms that likely account for the roles of Tip110 in hematopoiesis, we performed a genome-wide expression analysis in the context of Tip110 overexpression using the Affymetrix exon array. We took advantage of the Tip110 transgenic mice [13,19]. Due to the scarcity of mouse HPCs, we used MEFs from Tip110 transgenic mice and cognate wild-type littermates and isolated total RNA for the genome-wide expression analysis. The difference of gene expression between Tip110 MEF and wild-type MEF was calculated as folds of changes using the GeneSpring software platform. The subsequent KEGG pathway analysis revealed a number of genes in the cellular pathways that were affected, including the hematopoiesis pathway (Fig. 5) that exhibited at least 2.0-fold changes by increased Tip110 expression (Table 2). We next selected eight genes of hematopoietic importance and performed qRT-PCR validation assay (Table 1). Among those were five upregulated genes, CD34, IL-6, IL-11, SCF, and Flt3L, and three downregulated genes, CD33, IL-1, and IL-7, in the Tip110 MEF (Fig. 6). All genes, except IL-11, showed consistent up- or downregulation in the Tip110 MEF between the array and the qRT-PCR, giving rise to about 87.5% validation rate.
FIG. 5.
Regulatory pathways of hematopoiesis associated with Tip110 expression. Mouse embryonic fibroblasts (MEFs) isolated from Tip110 transgenic mice and congenic wild-type mice were used to isolate total RNA for the Affymetrix GeneChip® Mouse Exon 1.0 array. Genes with greater than twofold changes of expression between the Tip100 MEF and wild-type MEF were analyzed by the KEGG pathway analysis program. A number of genes that were affected by Tip110 expression, marked by a red star, were shown to be involved in hematopoiesis pathways.
Table 2.
Involvement of Tip110-Regulated Genes in the Cellular Pathway Identified by DAVID (the Database for Annotation, Visualization, and Integrated Discovery, Version 6.7) in KEGG Analysis (the Kyoto Encyclopedia of Genes and Genomes)
| Term | Gene count | P value |
|---|---|---|
| Chemokine signaling pathway | 25 | 8.1E-6 |
| Hematopoietic cell lineage | 29 | 2.1E-5 |
| Cytokine–cytokine receptor interaction | 15 | 5.1E-5 |
| Complement and coagulation cascades | 13 | 2.6E-4 |
| Systemic lupus erythematosus | 15 | 4.8E-4 |
| Natural killer cell-mediated cytotoxicity | 16 | 8.9E-4 |
| Toll-like receptor signaling pathway | 14 | 1.1E-3 |
| Cytosolic DNA-sensing pathway | 9 | 5.1E-3 |
| Cell adhesion molecules | 16 | 8.6E-3 |
| Axon guidance | 14 | 1.2E-2 |
| Glutathione metabolism | 8 | 1.3E-2 |
| Leukocyte transendothelial migration | 13 | 1.4E-2 |
Table 1.
Listed Genes and Primers Used in the Validation Study
| Gene | GeneBank no. | Region | Forward primer | Reverse primer | Expected size |
|---|---|---|---|---|---|
| CD33 | NM_001111058.1 | ORF:2321–2446 | CTAGCAATCTGTCTCTGTCTCATC | TCTCTCTCTCTCTCTCTCTCTCT | 104 |
| CD34 | NM_133654.3 | ORF:1493-1594 | GTCAAGTTGTGGTGGGAAGA | AGAGGCGAGAGAGGAGAAAT | 101 |
| IL-1 | NM_001146088.1 | ORF: 684-782 | CCATAAGCAGAGGCAGAGTAG | AGTAGGGAAGGAGAGGAGTAAG | 98 |
| IL-6 | J03783.1 | ORF:55-189 | CTTCCATCCAGTTGCCTTCT | CTCCGACTTGTGAAGTGGTATAG | 141 |
| IL-7 | AC125373.4 | ORP: 55-189 | GCAGGAGACTAGGACCCTATAA | CAATCCTAGCCTGCCTTAGATAC | 134 |
| IL-11 | NM_001290423.1 | ORF: 819-912 | GGGATCACCTGTGGCTTATT | GATCTCAGTTCCCTGCTCTTC | 93 |
| SCF | M59915.1 | ORF: 252-354 | CAGCTTGACTACTCTTCTGGAC | AACACGAGGTCATCCACTATTT | 102 |
| Flt3L | NM_013520.3 | ORF:811-935 | ACTCAGCCAGGGTCTTATCT | ATTACAGTGTCCATCGCCATAC | 124 |
| Dnmt3a | NM_007872. | ORF:778-880 | TCCATGAAAATGGAGGGCTC | TTGCTGATGTAGTAGGGGTC | 121 |
FIG. 6.
qRT-PCR validation of selected genes that were altered by Tip110 expression. Total RNA was prepared from Tip110 transgenic and wild-type MEFs. qRT-PCR was performed using the gene-specific primers. The fold changes between Tip110 transgenic and wild-type MEFs compared with those obtained by the above-mentioned exon array. β-Actin was included as a reference for qRT-PCR. The data are mean±SEM and representative of three independent experiments.
To investigate how miR-124 is induced in HSCs during the differentiation process, we searched the potential factors regulating miR-124 expression from Tip110-regulated genes in the splicing array. miR-124 was reported to be methylated on CpG islands on its promoter region by Dnmt3a [20]. We identified Dnmt3a to be upregulated by Tip110 expression through the array, which was further confirmed by qRT-PCR (Fig. 6). Loss of Dnmt3a expression might be caused by downregulation by Tip110 during HSC differentiation so as to result in less methylation on miR-124. This result suggested that miR-124 induction during HSC differentiation could be caused by alleviation of miR-124 methylation through the loss of Dnmt3a expression from Tip110 downregulation.
Discussion
miRNAs are essential for regulating hematopoiesis, including controlling self-renewal and differentiation [21]. Therefore, studies on the miRNA expression as well as their integrative analysis of affected genes are extremely important. The current study provides new insights into the role of miRNA in the function of normal human cord blood HSC self-renewal and differentiation. In this study, we have demonstrated that miR-124 directly suppresses expression of Tip110 (Fig. 1), which is an important factor for cord blood HSC self-renewal. miR-124 expression level increases during CB HSC differentiation (Fig. 2). Manipulation of the miR-124 expression level altered cord blood HSC self-renewal and differentiation (Fig. 3) and production of HPCs (Fig. 4). Furthermore, Tip110 expression leads to expression changes of a number of genes involved in hematopoiesis. Taken together, these results show for the first time that miR-124 is an important regulator of hematopoiesis and support further insights about the roles of Tip110 in hematopoiesis.
miR-124 has been viewed as a potential master regulator of differentiation of various types of cells in the central nervous system [22]. Overexpression of miRNA-124 in mouse embryonic stem cells induced differentiation of these cells [23]. miRNA-124 expression is repressed in various cancer cells, such as glioblastoma, gastric cancer, ovarian cancer, and bladder cancer [7–11]. In this study, we showed for the first time that miR-124 was markedly upregulated during human cord blood CD34+ cell differentiation. The similarity between normal stem cells and tumorigenic cancer cells is their proliferation capability [24]. The common function of miRNA-124 on stem cell differentiation, CNS development and adult neurogenesis, and cancer cell growth inhibition is linked to its ability to suppress cell proliferation toward the quiescent stage and induce cell differentiation.
In agreement with these findings, we showed in this study that miRNA-124 promoted human cord blood CD34+ cell differentiation. miR-124 effects on CD34+ cell differentiation may likely also involve the targets of miR-124, such as CDK4, CDK6, and cyclin D2 [25–28]. For example, miR-124 suppresses CDK6 expression and as a result, causes cell cycle arrest at the G1 phase and suppression of cell proliferation [29]. CDK6 itself has also been shown to regulate HSCs [30] and play a central role in hematopoietic malignancies [31]. Our previous findings that Tip110 is an important regulator of human cord blood hematopoiesis [13], along with our current findings that Tip110 is a novel target of miRNA-124, suggest that downregulation of Tip110 expression during human cord blood CD34+ cell differentiation is likely a result of the interaction between increased miR-124 expression during human cord blood CD34+ cell differentiation and Tip110 3′-UTR. In addition, miR-124 expression could be transcriptionally inactivated by CpG island hypermethylation in human tumors [25,28]. Therefore, it would be interesting to determine whether it is possible to manipulate human cord blood stem cell hematopoiesis through miR-124 expression in vivo.
Microarrays are used to determine genome-wide gene expressions and to define gene expression regulatory networks. To identify how Tip110 affects the hematopoiesis, we compared gene expression between the Tip110 transgenic MEF and its cognate wild-type MEF and identified the possible gene targets of Tip110 in hematopoiesis pathways. The results showed that Tip110 regulates a number of genes in the hematopoiesis pathway. Among them are CD34, SCF, and Flt3L upregulated by Tip110. SCF and Flt3L are potent costimulating cytokines for HSCs and HPCs [32]. On the other hand, CD33, downregulated by Tip110, is a myeloid differentiation antigen [33,34].
Dnmt3a acts as de novo methyltransferase for modification of unmethylated DNA [35]. During hematopoiesis, Dnmt3a expression was reported to be much higher in primitive long-term HSCs than differentiated cells [36]. Loss of Dnmt3a enables HSC differentiation. Microarray data between the Tip110 transgenic MEF and its cognate wild-type MEF identified Dnmt3a to be significantly upregulated. Oncoprotein EVI1 forms a complex with Dnmt3a for a specific target on the miR-124 regulatory region on its promoter [20]. That miR-124 was markedly upregulated during human cord blood CD34+ cell differentiation could be the result of direct loss of its promoter methylation from Dnmt3a.
In conclusion, the data provide additional evidence to support the notion that Tip110 participates in the regulation of hematopoiesis and demonstrate for the first time a dynamic requirement of miR-124 for hematopoietic differentiation through downregulation of Tip110 expression.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Montagner S, Deho L. and Monticelli S. (2014). MicroRNAs in hematopoietic development. BMC Immunol 15:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangaraju VK. and Lin H. (2009). MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol 10:116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao R, Sun J, Zhang L, Lou G, Chen M, Zhou D, Chen Z. and Zhang S. (2008). MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem 104:805–817 [DOI] [PubMed] [Google Scholar]
- 5.Makeyev EV, Zhang J, Carrasco MA. and Maniatis T. (2007). The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27:435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng LC, Pastrana E, Tavazoie M. and Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 12:399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng XH, Huang HR, Lu J, Liu X, Zhao FP, Zhang B, Lin SX, Wang L, Chen HH, et al. (2014). MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol Cancer 13:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinushi T, Shibayama Y, Kinoshita I, Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H. and Iseki K. (2014). Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Med 3:1544–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Zang W, Liu P, Wang Y, Du Y, Chen X, Deng M, Sun W, Wang L, Zhao G. and Zhai B. (2014). MicroRNA-124 inhibits cellular proliferation and invasion by targeting Ets-1 in breast cancer. Tumour Biol 35:10897–904 [DOI] [PubMed] [Google Scholar]
- 10.Mucaj V, Lee SS, Skuli N, Giannoukos DN, Qiu B, Eisinger-Mathason TS, Nakazawa MS, Shay JE, Gopal PP, et al. (2014). MicroRNA-124 expression counteracts pro-survival stress responses in glioblastoma. Oncogene [Epub ahead of print]; doi: 10.1038/onc.2014.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Zheng L, Huang J, Gao F, Lin X, He L, Li D, Li Z, Ding Y. and Chen L. (2014). MiR-124 radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One 9:e93917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Li J, Kim BO, Pace BS. and He JJ. (2002). HIV-1 Tat protein-mediated transactivation of the HIV-1 long terminal repeat promoter is potentiated by a novel nuclear Tat-interacting protein of 110 kDa, Tip110. J Biol Chem 277:23854–23863 [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Timani K, Mantel C, Fan Y, Hangoc G, Cooper S, He JJ. and Broxmeyer HE. (2011). TIP110/p110nrb/SART3/p110 regulation of hematopoiesis through CMYC. Blood 117:5643–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Lee MR, Timani K, He JJ. and Broxmeyer HE. (2012). Tip110 maintains expression of pluripotent factors in and pluripotency of human embryonic stem cells. Stem Cells Dev 21:829–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Hangoc G, Campbell TB, Goodman M, Tao W, Pollok K, Srour EF. and Broxmeyer HE. (2008). Identification of parameters required for efficient lentiviral vector transduction and engraftment of human cord blood CD34(+) NOD/SCID-repopulating cells. Exp Hematol 36:947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissels U, Wild S, Tomiuk S, Hafner M, Scheel H, Mihailovic A, Choi YH, Tuschl T. and Bosio A. (2011). Combined characterization of microRNA and mRNA profiles delineates early differentiation pathways of CD133+ and CD34+ hematopoietic stem and progenitor cells. Stem Cells 29:847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgantas RW, 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM. and Civin CI. (2007). CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A 104:2750–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I. and Dick JE. (2011). Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333:218–221 [DOI] [PubMed] [Google Scholar]
- 19.Timani KA, Liu Y. and He JJ. (2013). Tip110 interacts with YB-1 and regulates each other's function. BMC Mol Biol 14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senyuk V, Premanand K, Xu P, Qian Z. and Nucifora G. (2011). The oncoprotein EVI1 and the DNA methyltransferase Dnmt3 co-operate in binding and de novo methylation of target DNA. PLoS One 6:e20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazare SS, Wojtowicz EE, Bystrykh LV. and de Haan G. (2014). microRNAs in hematopoiesis. Exp Cell Res 329:234–238 [DOI] [PubMed] [Google Scholar]
- 22.Ponomarev ED, Veremeyko T. and Weiner HL. (2013). MicroRNAs are universal regulators of differentiation, activation, and polarization of microglia and macrophages in normal and diseased CNS. Glia 61:91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosik KS. (2006). The neuronal microRNA system. Nat Rev Neurosci 7:911–920 [DOI] [PubMed] [Google Scholar]
- 24.Shackleton M. (2010). Normal stem cells and cancer stem cells: similar and different. Semin Cancer Biol 20:85–92 [DOI] [PubMed] [Google Scholar]
- 25.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, et al. (2007). Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67:1424–1429 [DOI] [PubMed] [Google Scholar]
- 26.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, et al. (2008). miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, Martin-Subero JI, Cordeu L, Garate L, San Jose-Eneriz E, Abizanda G, Rodriguez-Otero P, et al. (2009). Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res 69:4443–4453 [DOI] [PubMed] [Google Scholar]
- 28.Chen X, He D, Dong XD, Dong F, Wang J, Wang L, Tang J, Hu DN, Yan D. and Tu L. (2013). MicroRNA-124a is epigenetically regulated and acts as a tumor suppressor by controlling multiple targets in uveal melanoma. Invest Ophthalmol Vis Sci 54:2248–2256 [DOI] [PubMed] [Google Scholar]
- 29.Pierson J, Hostager B, Fan R. and Vibhakar R. (2008). Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 90:1–7 [DOI] [PubMed] [Google Scholar]
- 30.Scheicher R, Hoelbl-Kovacic A, Bellutti F, Tigan AS, Prchal-Murphy M, Heller G, Schneckenleithner C, Salazar-Roa M, Zochbauer-Muller S, et al. (2014). CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood 125:90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, Schafer M, Fajmann S, Schlederer M, et al. (2013). A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 24:167–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fadilah SA, Vuckovic S, Khalil D. and Hart DN. (2007). Cord blood CD34+ cells cultured with FLT3L, stem cell factor, interleukin-6, and IL-3 produce CD11c+CD1a-/c- myeloid dendritic cells. Stem Cells Dev 16:849–855 [DOI] [PubMed] [Google Scholar]
- 33.Laszlo GS, Estey EH. and Walter RB. (2014). The past and future of CD33 as therapeutic target in acute myeloid leukemia. Blood Rev 28:143–153 [DOI] [PubMed] [Google Scholar]
- 34.Walter RB. (2014). The role of CD33 as therapeutic target in acute myeloid leukemia. Expert Opin Ther Targets 18:715–718 [DOI] [PubMed] [Google Scholar]
- 35.Okano M, Bell DW, Haber DA. and Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257 [DOI] [PubMed] [Google Scholar]
- 36.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, et al. (2012). Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]