Abstract
Background
This study was undertaken to determine the susceptibility profile and the mechanism of antibiotic resistance in Group B streptococcus (GBS) isolates detected in vaginal and rectal swabs from pregnant women attending Dr George Mukhari Academic Hospital, a University Teaching Hospital in Pretoria, South Africa.
Methods
The samples were collected over an 11-month period, cultured on selective media (colistin and nalidixic acid agar and Todd-Hewitt broth), and GBS positively identified by using different morphological and biochemical tests. The susceptibility testing was done using the Kirby–Bauer and E test methods according to CLSI guidelines 2012. The D test method was used for the detection of inducible clindamycin resistance. Multiplex PCR with specific primers was used to detect different genes coding for resistance.
Results
Out of 413 samples collected, 128 (30.9 %) were positive with GBS. The susceptibility testing revealed that 100 % of isolates were sensitive to penicillin, ampicillin, vancomycin and high level gentamicin. Erythromycin and clindamycin resistance was 21.1 and 17.2 %, respectively, in which 69 % had harboured constitutive macrolide, lincosamide and streptogramin B (MLSB), 17.4 % had inducible MLSB. The M and L phenotypes were present in 6.8 % each. The methylation of target encoded by ermB genes was the commonest mechanism of resistance observed in 55 % of isolates, 38 % of isolates had both ermB and linB genes and efflux pump mediated by mefA genes was also distributed among the isolates.
Conclusions
The study reaffirmed the appropriateness of penicillin as the antibiotic of choice for treating GBS infection. However it identified the challenges of resistance to macrolides and lincosamides used as alternative drugs for individuals allergic to penicillin. More GBS treatment options for penicillin allergic patients need to be researched on.
Keywords: Group B streptococcus, Antibiotic resistance, Africa
Background
Streptococcus agalactiae (Group B streptococcus, GBS) is the leading cause of neonatal infections in humans. It is an important cause of illness in pregnant women and the elderly with underlying illnesses such as diabetes mellitus or immunosuppression [1–3]. The organism is part of the normal flora of the gut and genital tract and is found colonizing 10–40 % of pregnant women [4].
In adults and pregnant women, GBS can cause urosepsis, chorioamnionitis, endometritis, pneumonia, skin and soft tissue infections [3, 4]. In newborns GBS is the cause of neonatal sepsis, pneumonia, and meningitis [5–7]. Mother to child transmission occurs via the ascending route from the maternal genital tract into the amniotic fluid or at delivery [8]. Infant GBS infection is classified as early-onset disease (EOD) when occurring from birth to 6 days (70–80 % of cases), and late-onset disease (LOD), when it occurs more than 7–90 days after birth; this is transmitted from mother or health care personnel to infants [9–11].
Penicillin and ampicillin are the antibiotics of choice, followed by the first-generation cephalosporins and vancomycin for the treatment of GBS infections [12, 13]. No resistance to penicillin has been reported except a few cases of isolates with intermediate sensitivity or reduced Minimum Inhibitory Concentrations (MICs) to penicillin [14–16]. Alternative antibiotics besides macrolides and lincosamide exist for penicillin allergic patients, although the use of vancomycin should be reserved for penicillin-allergic women with a high risk of anaphylaxis [17–19].
Erythromycin resistance mechanism in GBS is mostly due to ribosomal modification encoded by erm genes (ermB, erm A/TR) or through efflux pump mediated by mefA genes that cause resistance to 14- and 15-membered macrolides, which confers cross-resistance to all constitutive macrolide, lincosamide and streptogramin B (MLSB) antibiotics [20–22]. This resistance can either be inducible (iMLSB) or constitutive (cMLSB) [23, 24]. Moreover clindamycin resistance in GBS is less frequent and is due to ribosomal translocation encoded by linB genes [25]. Multiplex PCR can be used to detect the major erythromycin and clindamycin resistance genes in GBS strains [22, 26].
In South Africa there is a lack of sufficient data on antibiotic resistance in GBS isolates. The purpose of this study was to assess the susceptibility profile of GBS to different antibiotics, to determine genetic basis and the mechanisms of antibiotic resistance in GBS isolates from pregnant women at Dr George Mukhari Academic Hospital, in Pretoria.
Methods
Sample collection and culture
The procedure for collection was explained to each patient before specimens were taken. High vaginal swabs (HVS), low vaginal swabs (LVS) and rectal swabs (RS) were collected aseptically from pregnant women (age 18–45 years old); these were at the gestational period of 16–38 weeks, attending antenatal clinic at Dr George Mukhari Academic Hospital, in Garankuwa. This is a University Teaching Hospital associated with the University of Limpopo, Medical University of Southern Africa (Medunsa campus), located about 37 km north of Pretoria, Gauteng Province, in South Africa. It lies at an altitude of about 1350 m (4500 ft) above sea level; in a longitude of 25°37′14″S and latitude of 28°1′1. The women who indicated that they received antibiotic treatment 2 weeks prior to sample collection were excluded. However any women who needed antibiotic treatment after collection were referred for clinical management accordingly. Samples were collected from February 2012 until December 2012. Swabs were placed into Amies transport medium (Rochelle Chemicals and Lab Equipment, Pretoria, South Africa), properly labelled and put into a cooler box containing ice packs, and transported to the laboratory at the Department of Microbiological Pathology, University of Limpopo, Medunsa campus within 2–4 h of collection. Specimens, one per patient were cultured onto selective media, 5 % sheep blood, Columbia colistin and nalidixic acid (CNA) agar (DMP—National Health Laboratory Service, Pretoria, South Africa) and also inoculated into enriched selective GBS broth, Todd-Hewitt broth (DMP—NHLS, Pretoria, South Africa), with the same antibiotics concentration as in CNA (15 mg/l nalidixic acid and 8 mg/l gentamycin) and were incubated for 24–48 h in a 5 % CO2 atmosphere at 37 °C. Isolates were confirmed as GBS by using the following methods: morphology of bacteria, haemolytic activity, catalase test, microscopy (Gram’s stain), bile esculin, and CAMP reaction followed by latex agglutination test (Streptex—Slidex ® Strepto Plus—bioMérieux, Marcy l’Etoile, France) for antigen detection.
Antibiotic susceptibility testing
Purification of isolates was done before susceptibility testing. Susceptibility testing was done on one isolate per patient. For the three methods below, antibiotics were placed onto a Muller-Hinton agar added with 5 % sheep blood following bacterial inoculation (0.5 McFarland of bacterial suspension). The plates were incubated at 37 °C in a CO2 enriched environment for 20–24 h. For quality control, Streptococcus pneumoniae ATCC 49619 and Streptococcus agalactiae ATCC 12403 were used as control strains [16, 27].
Disc diffusion
All the 128 GBS positive isolates were tested by Kirby–Bauer method for susceptibility to ampicillin (10 μg), vancomycin (30 μg), high level gentamycin (120 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), and tetracycline (TE) (30 μg). The results were interpreted according the CLSI 2012 guidelines [27].
E test
The MICs of penicillin, erythromycin and clindamycin were determined by commercial paper strips or E test method (AB Biodisk, Davies-Diagnostics, Pretoria, South Africa), following the manufacturer’s instructions.
Double disc diffusion
The detection of inducible clindamycin resistance was done by using D test method as previously described [23, 24, 28]. Briefly, erythromycin (15 µg) and clindamycin (2 µg) disks (Oxoid, Davies-Diagnostics, Pretoria, South Africa) were placed 12 mm apart edge to edge [27]. Blunting was defined as growth within the clindamycin zone of inhibition proximal to the erythromycin disk, indicating MLSB-inducible methylation. Resistance to both erythromycin and clindamycin indicated MLSB-constitutive methylation. Resistance to erythromycin but susceptibility to clindamycin without blunting indicated an efflux mechanism (M phenotype). And finally resistance to clindamycin but susceptible to erythromycin was referred to as L phenotype as previously described [24, 28].
Molecular techniques
DNA extraction
DNA extraction was done using the Zymo Research—DNA MiniPrep-Kit (Zymo-Research—USA, Inqaba Biotechnical Industries, Pretoria, South Africa) and following the manufacturer instructions.
Multiplex PCR
All phenotypically resistant GBS isolates were tested to detect three genes for erythromycin resistance, ermB, ermTR, mefA and one gene for clindamycin resistance linB using a set of specific primers (Table 1) as previously described [22, 28–30]. The primers were synthesized at Inqaba—Biotechnical industries, Pretoria, South Africa. Briefly, a 50 µl PCR contained 2.5 mM Tris–HCl pH 8.6, 2.5 mM KCl, 2.5 mM MgCl2, 5 mM dNTP, 0.5 U Taq DNA polymerase (Thermo Scientific—Phusion Flash High-Fidelity PCR Master Mix, AB, Inqaba—Biotechnical Industry, Pretoria, South Africa), PCR water, and 1 µM primers pairs forwards and reverses. A total of 5 µl template DNA was used in the PCR. The cycling conditions on a My Cycler™ thermal cycler (BioRad Laboratories, London, UK) consisted of a single cycle of 95 °C for 3 min followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 57 °C for 1 min and extension at 72 °C for 1 min. A final extension step of 72 °C for 5 min was followed by a hold at 4 °C as previously described by Desjardins et al. [28].
Table 1.
PCR primers used for detection of resistance genes in GBS
| Gene | Primers | Primers sequence (5′–3′) | Products size (bp) | References |
|---|---|---|---|---|
| ermB | ermB1 | 5′_-GAA AAG GTA CTC AAC CAA ATA-3′_(F) | 640 | [22, 29] |
| ermB2 | 5′_-AGT AAC GGT ACT TAA ATT GTT TAC-3′_(R) | |||
| ermTR | ermTR1 | 5′_-GAA GTT TAG CTT TCC TAA-3′_(F) | 400 | [22, 28] |
| ermTR2 | 5′_-GCT TCA GCA CCT GTC TTA ATT GAT-3′_(R) | |||
| mefA | mefA1 | 5′_-AGT ATC ATT AAT CAC TAG TGC-3′_(F) | 348 | [22, 29] |
| mefA2 | 5′_-TTC TTC TGG TAC TAA AAG TGG-3′_(R) | |||
| linB | linB1 | 5′_-CCT ACC TAT TGT TTG TGG AA-3′_(F) | 944 | [22] |
| linB2 | 5′_-ATA ACG TTA CTC TCC TAT TC-3′_(R) |
Agarose gel
The different genes of resistance were analyzed based on presence or absence of bands in the agarose gel. Electrophoresis on 1 % agarose gels in 40 mM Tris acetate–2 mM EDTA buffer was used to distinguish PCR products as previously described [4], and bands were visualized using Gel Docs (BioRad Laboratories, London, UK). A culture of GBS ATCC 49447 strain was used as negative control.
Ethical considerations
The women recruited in the study gave informed and signed consent. The study was approved by the Medical Research and Ethics Committee of South Africa (MREC/P/02/2011: IR) and Directorate for Health and Social Affairs (Medical University of Southern Africa; MEDUNSA) and the College of Agriculture and Environmental Health Sciences, University of South Africa (UNISA).
Results
Of the 413 pregnant women recruited, 128 (30.9 %) were colonized with GBS in which 70 were recovered from Todd-Hewitt broth and 58 from CNA agar (22/58 RS, 9/58 LVS, 3/58 HVS and 24/58 all the sites). The susceptibility pattern was performed on 128 positive GBS isolates against 9 antimicrobial agents and results are presented in Tables 2 and 3 showing susceptibility profile of GBS isolates by disc diffusion method and by E test, respectively. All strains were 100 % susceptible to penicillin, ampicillin, vancomycin and high level gentamycin. However resistant strains to erythromycin and clindamycin were observed in 21.1 and 17.2 %, respectively. The strains that were resistant to tetracycline were 94.5 % of the isolates, 24.9 % were resistant to chloramphenicol and 18.6 % resistant to ciprofloxacin (all the intermediate values were considered as resistant). The MIC range for penicillin was found to be between 0.012 and 0.12 μg/ml and that for erythromycin and clindamycin both ranged between 0.02 and 0.25 μg/ml. All erythromycin and clindamycin resistant isolates were screened for resistance genes.
Table 2.
Susceptibility profile of GBS isolates (n = 128)
| Antibiotic/method | Susceptible | Intermediate* | Resistant |
|---|---|---|---|
| Disc diffusion | |||
| Ampicillin | 128 (100 %) | – | – |
| Vancomycin | 128 (100 %) | – | – |
| Gentamicin-high level | 128 (100 %) | – | – |
| Ciprofloxacin | 104 (81.2 %) | 17 (13.3 %) | 7 (5.5 %) |
| Chloramphenicol | 96 (75.0 %) | 11 (8.6 %) | 21 (16.4 %) |
| Tetracycline | 7 (5.5 %) | 10 (7.8 %) | 111 (86.7 %) |
* The intermediate values were assimilated to resistant
Table 3.
Susceptibility profile of GBS isolates by E test (n = 128)
| Antibiotic | MIC (µg/ml) | ||||
|---|---|---|---|---|---|
| Range | 50 % of isolates | 90 % of isolates | Break pointa (susceptible) | % Of isolates resistant | |
| Penicillin | 0.002–32 | 0.012 | 0.047 | ≤0.12 | – |
| Erythromycin | 0.016–256 | 0.16 | 8 | ≤0.25 | 21.1 |
| Clindamycin | 0.016–256 | 0.16 | 8 | ≤0.25 | 17.2 |
aCLSI guidelines 2012
The phenotype (by double-disc diffusion) and genotype results of resistant isolates are summarized in Table 4. Among the resistant isolates to both erythromycin and clindamycin, 69 % harboured constitutive MLSB, 17.4 % harboured inducible MLSB, the M phenotypes were present in 6.8 % and L phenotypes in 6.8 %.
Table 4.
Minimum inhibitory concentrations of erythromycin and clindamycin resistant isolates, D-shape and screened genes (n = 29)
| No | MIC (µg/ml) | D-shape | MLS | Genes | |
|---|---|---|---|---|---|
| Erythromycin | Clindamycin | ||||
| 1 | 1 (R) | 0.50 (I) | Negative | cMLSB | ermB |
| 2 | 3 (R) | 0.75 (I) | Negative | cMLSB | linB + ermB |
| 3 | 0.75 (I) | 0.75 (I) | Negative | cMLSB | linB + ermB |
| 4 | 0.50 (I) | 0.047 (S) | Negative | M phenotype | mefA |
| 5 | 0.75 (I) | 1 (R) | Negative | cMLSB | ermB |
| 6 | 4 (R) | 0.50 (I) | Negative | cMLSB | ermB |
| 7 | 1 (R) | 0.38 (I) | Negative | cMLSB | ermB |
| 8 | 0.75 (I) | 1 (R) | Negative | cMLSB | ermB |
| 9 | 0.75 (I) | 4 (R) | Negative | cMLSB | linB + ermB |
| 10 | 4 (R) | 7 (R) | Negative | cMLSB | ermB |
| 11 | 2 (R) | 0.047 (S) | Positive | iMLSB | ermTR |
| 12 | 1.5 (R) | 0.016 (S) | Positive | iMLSB | ermB |
| 13 | 0.75 (I) | 0.50 (I) | Negative | cMLSB | linB + ermB |
| 14 | 8 (R) | 0.38 (I) | Negative | cMLSB | ermB |
| 15 | 0.25 (S) | 2 (R) | Negative | L phenotype | linB + ermB |
| 16 | 8 (R) | 1 (R) | Negative | cMLSB | linB + ermB |
| 17 | 0.75 (I) | 1 (R) | Negative | cMLSB | ermB |
| 18 | 3 (R) | 0.38 (I) | Negative | cMLSB | ermB |
| 19 | 1 (R) | 0.38 (I) | Negative | cMLSB | ermB |
| 20 | 1.5 (R) | 0.25 (S) | Positive | iMLSB | ermB |
| 21 | 0.25 (S) | 1 (R) | Negative | L phenotype | linB + ermB |
| 22 | 3 (R) | 0.047 (S) | Positive | iMLSB | ermB |
| 23 | 0.50 (I) | 0.75 (I) | Negative | cMLSB | linB + ermB |
| 24 | 2 (R) | 8 (R) | Negative | cMLSB | ermB |
| 25 | 0.75 (I) | 0.50 (I) | Negative | cMLSB | linB + ermB |
| 26 | 0.75 (I) | 0.094 (S) | Negative | M phenotype | ermB |
| 27 | 4 (R) | 0.023 (S) | Positive | iMLSB | ermB |
| 28 | 0.50 (I) | 1 (R) | Negative | cMLSB | linB + ermB |
| 29 | 0.75 (I) | 0.50 (I) | Negative | cMLSB | linB + ermB |
S susceptible, R resistant, I intermediate, cMLS B MLSB-constitutive methylation [erythro (R), clinda (R)], iMLS B MLSB-inducible methylation [erythro (R), clinda (S) with blunting], M-phenotype efflux pump mechanism [erythro (R), clinda (S) without blunting], L-phenotype erythro (S), clinda (R)
The genotypic analysis of all isolates irrespective of whether they were resistant or sensitive phenotypically, was done using Multiplex PCR. This showed that the methylation of target encoded by ermB genes was the commonest mechanism of resistance observed in 55 % (16/29) of isolates, and efflux pump mediated by mefA genes were found in 3.4 % of isolates, ermTR genes were also found in 3.4 % of isolates and finally both ermB and linB were observed in 38 % (11/29) of isolates. The PCR products of isolates with resistant genes were distinguished by agarose gel electrophoresis as shown in Fig. 1.
Fig. 1.
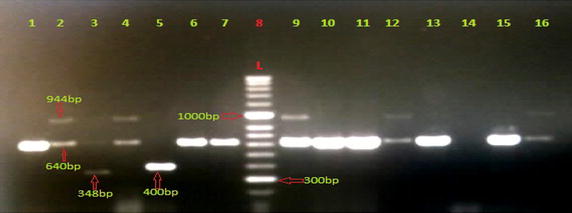
Result of Multiplex PCR of GBS isolates resistant to erythromycin and clindamycin. Lane 8 DNA Molecular Weight Marker Hyper Ladder™ 50 bp (BioLine). Lane 14 negative control (GBS ATCC 49447). Lane 1, 6, 7, 10, 11, 13, 15 presence of ermB genes, 640 bp, (sample no 15, 63, 65, 125, 148, 182, 183). Lane 2, 4, 9, 12, 16 presence of both ermB genes and linB genes, 944 bp (sample no 32, 57, 191,159, 184). Lane 5 presence of ermTR gene, 400 bp (sample no 83). Lane 3 presence of mefA gene, 348 bp (sample no 60)
Discussion
In our study the colonization rate was 30.9 %, the highest reported so far in South Africa. The susceptibility testing was performed on 128 GBS isolated against 9 antimicrobial agents. All strains were 100 % susceptible to penicillin, ampicillin, vancomycin and high level gentamycin. This is similar to a study conducted in Germany and to an Ethiopian study in which 100 % of isolates were found to be sensitive to penicillin, ampicillin, high-level gentamycin and vancomycin [12, 31]. Similar findings were also described in studies from the USA and Argentina where 100 % sensitivity to penicillin, ampicillin and vancomycin [4, 23].
Our findings slightly differ with those described in a study from the neighbouring country of Zimbabwean in which almost 100 % of isolates were sensitive to penicillin, but 2 % were intermediate susceptible to penicillin [8].
In this study 94.5 % of the isolates were resistant to tetracycline, 24.9 % resistant to chloramphenicol, 21.1 % resistant to erythromycin, 18.6 % resistant to ciprofloxacin and 17.2 % resistant to clindamycin. Considering erythromycin and clindamycin resistance rates, similar findings were reported in Canada, in which erythromycin and clindamycin resistant rates were found in 17 and 8 %, respectively [28]. This is again similar to a Tanzanian study that reported a GBS resistance rate of 17.6 and 13 % for erythromycin and clindamycin, respectively and to that described in the Malawian study where the erythromycin resistance rate was 21 % [32, 33]. This suggests that antibiotic resistance in GBS may be similar despite different geographic locations in sub-Saharan Africa. However phylogenetic studies are necessary to verify this. Erythromycin and clindamycin resistance rates in these studies were far lower when compared to the 50.7 % of erythromycin resistance and 38.4 % clindamycin resistance rates reported by Back et al. [18] in the USA, and far lower again than the 54 and 33 %, respectively reported by DiPersio et al. [34]. Erythromycin and clindamycin resistance rates in our findings were higher when compared to the Canadian study, where there was 8 % resistance for erythromycin and 4.5 % for clindamycin [30].
The 94.5 % tetracycline resistance rate found in our study is similar to the 96 % reported by Gray et al. [33], to the 86.8 % reported by De Azavedo et al. [30], and 100 % reported by Moyo et al. [8]. Resistance to tetracycline might be explained by wide and indiscriminate use of these antibiotics worldwide.
In our study, the phenotypic testing by double disk diffusion revealed that 29 isolates were resistant to either erythromycin alone or clindamycin alone or to both erythromycin and clindamycin in which 20 (69 %) isolates harboured cMLSB, 5 (17.2 %) harboured iMLSB, the M phenotypes were present in 2 (6.8 %) isolates and the L phenotypes in 2 (6.8 %). This finding was in agreement with a Canadian study in which 47.2 % had cMLSB resistance phenotype, 40 % had an iMLSB resistance phenotype, and 12.7 % of the isolates displayed M phenotypes [26]. In Ireland, a study found similar findings with 40 % of isolates that harboured iMLSB, 36 % had cMLSB, 24 % M phenotype and no L phenotype [24].
Considering the genotypic analysis by multiplex PCR, erythromycin and clindamycin resistance in GBS were mainly associated with ermB genes with 55 % of isolates, ermTR genes harboured 3.4 % and mefA genes 3.4 % of the isolates, both ermB and linB genes together were present in 38 % of the isolates, and none of the strains carried both ermB and ermTR nor both mefA and erm nor linB alone. This was similar to a French study in which ermB was found in 47 % of isolates, ermTR genes in 45 % of isolates and mefA gene in 6 % of the isolates and none of the strains carried both ermB and ermTR or both mefA and erm genes [17].
In this study, there were two isolates which were phenotypically sensitive to erythromycin, and their erm genes were also detected by molecular testing. This may be due to erm gene not being expressed, but will require further studies to confirm the interpretation. Also two isolates were resistant to clindamycin but no resistance mechanism was found. This situation could be explained by the fact that isolates may harbour mutations in genes coding for 23S rRNA. A similar situation was also reported in an Irish study where no recognized resistance mechanisms were found in nine isolates [24].
The limitations of the study were that the positive control strain (resistant to both erythromycin and clindamycin) was not available during the molecular stage of the study; thus only negative controls were used. Data from previous local studies in South Africa were not available to allow comparison of genetic mechanisms underlying resistance in GBS.
Conclusion
This study confirmed the appropriateness of penicillin as still being the antibiotic of choice for treating GBS infections in South Africa. The concern which still remains is the reported increase in the resistance to the macrolides and clindamycin used as alternative drugs for penicillin allergic patients, in other parts of the world. More GBS treatment options for penicillin allergic patients need to be explored. The methylation of targets encoded by ermB was the commonest mechanisms of resistance observed and efflux pump mediated by mefA genes was also distributed among the isolates. More research studies need to be done in various areas and populations of South Africa to determine GBS colonization.
Authors’ contributions
JYB, CMM, and MOC contributed to sample processing, data analysis and write-up. LSM, MST and TC set up the clinical component of the study including sample collection. MN and SRM conceived the idea for the project and supervised the work and contributed to write-up. MRBM and SLL supervised the work and contributed to write-up. RTM supervised the molecular work and contributed to write-up. All authors read and approved the final manuscript.
Acknowledgements
We appreciate the assistance of the staff and post-graduate students of the Department of Microbiological Pathology, University of Limpopo—MEDUNSA campus, especially Ms B. de Villiers and Ms N. Makhado for their contribution during molecular part of the study. We also thank the staff of National Health Laboratory Service (NHLS)–DGMAH–Tertiary Laboratory, especially Mr LD Nemuthavanani, for assisting with susceptibility testing. We finally thank The National Research Foundation (NRF), VLIR/UL Partnership Program and UNISA Research Directorate for financial support.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Abbreviations
- GBS
group B streptococcus
- MICs
minimum inhibitory concentrations
- cMLSB
constitutive macrolide, lincosamide and streptogramin B
- iMLSB
inducible macrolide, lincosamide and streptogramin B
- CNA
Columbia colistin and nalidixic acid agar
Contributor Information
John Y. Bolukaoto, Email: jbolukaoto@gmail.com
Charles M. Monyama, Email: monyamc@unisa.co.za
Martina O. Chukwu, Email: timummy2002@yahoo.com
Sebotse M. Lekala, Email: sebotselekala@yahoo.com
Maphoshane Nchabeleng, Email: Maphoshane.Nchabeleng@ul.ac.za.
Motlatji R. B. Maloba, Email: motlatji.maloba@nhls.ac.za
Rooyen T. Mavenyengwa, Email: rmavenyengwa@yahoo.com
Sogolo L. Lebelo, Email: lebelol@unisa.ac.za
Sam T. Monokoane, Email: Sam.Monokoane@ul.ac.za
Charles Tshepuwane, Email: charles4@yebo.co.za.
Sylvester R. Moyo, Email: srmoyo@polytechnic.edu.na
References
- 1.Arana DM, Rojo-Bezares B, Torres C, Alos JI. First clinical isolate in Europe of clindamycin-resistant group B Streptococcus mediated by the lin(B) gene. Rev Esp Quimioter. 2014;27(2):106–109. [PubMed] [Google Scholar]
- 2.Lammler CH, Scwarz S, Wibawan IWT, Otte E, Bopp V, Martinez-Tagle A. Comparison of streptococci of serological group B isolated from healthy carriers and active disease in Chile. J Med Microbiol. 1995;42:161–164. doi: 10.1099/00222615-42-3-161. [DOI] [PubMed] [Google Scholar]
- 3.Murayama SY, Chizuko S, Hiroshi S, Katsuhiko S, Eiichi N, Satoshi I, Keisuke S, Kimino U. Capsular type and antibiotic resistance in Streptococcus agalactiae isolates from patients, ranging from newborns to the elderly, with invasive infections. Anti Ag Chem. 2009;53:2650–2653. doi: 10.1128/AAC.01716-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning SD. Molecular epidemiology of Streptococcus agalactiae (Group B streptococcus) Front Bio. 2003;8:1–18. doi: 10.2741/985. [DOI] [PubMed] [Google Scholar]
- 5.Larsson C, Lindroth M, Nordin P, Stalhammar-Carlemalm M, Lindahl G, Krantz I. Association between low concentrations of antibodies to protein α and Rib and invasive neonatal GBS infection. Arch Dis Child Fetal Neona. 2006;91:403–408. doi: 10.1136/adc.2005.090472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavenyengwa RT, Moyo SR, Nordbo SA. Streptococcus agalactiae colonization and correlation with HIV-1 and HBV seroprevalence in pregnant women from Zimbabwe. Euro J Obst Gynaecol Reprod Biol. 2010;150:34–38. doi: 10.1016/j.ejogrb.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Edmond KM, Kortsalioudaki C, Scott S, Schrag SJ, Zaidi AKM, Cousens S, Heath PT. Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet. 2012;379:547–556. doi: 10.1016/S0140-6736(11)61651-6. [DOI] [PubMed] [Google Scholar]
- 8.Moyo SR, Maeland JA, Munemo ES. Susceptibility of Zimbabwean Streptococcus agalactiae (group B streptococcus: GBS) isolates to four different antibiotics. Cent Afr J Med. 2001;47:226–229. [PubMed] [Google Scholar]
- 9.Madhi SA, Radebe K, Crewe-Brown H, Frasch CE, Arakere G, Mokhachane M, Kimura A. High burden of invasive GBS in South African infants. Ann Trop Paediatr. 2003;23:15–23. doi: 10.1179/000349803125002814. [DOI] [PubMed] [Google Scholar]
- 10.Martins ER, Andreu A, Correia P, Juncosa T, Bosch J, Ramirez M, Melo-Cristino J. Group B streptococci causing neonatal infections in Barcelona are a stable clonal population: 18-year surveillance. J Clin Microbiol. 2011;49:2911–2918. doi: 10.1128/JCM.00271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madzivhandila M, Adrian PV, Cutland CL, Kuwanda L, Schrag SJ, Madhi SA. Serotype distribution and invasive potential of group B streptococcus isolates causing disease in infants and colonizing maternal-newborn dyads. PLoS One. 2011;6:1–6. doi: 10.1371/journal.pone.0017861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musa M, Asrat D, Woldeamanuel Y, Demissie A. Prevalence of group B streptococcus colonization among pregnant women attending antenatal clinic of Hawassa Health Centre, Hawassa, Ethiopia. Ethiop J Health Dev. 2012;26:36–42. [Google Scholar]
- 13.Centers for Disease Control and Prevention (CDC). Morbidity and mortality weekly report. Prevention of perinatal group b streptococcal disease revised guidelines from CDC. 2010. http://www.cdc.gov/mmwr/groupbstrep. Accessed 12 Feb 2014. [PubMed]
- 14.Rouse DJ, Andrews WW, Lin FC, Mott CW, Ware JC, Philips JB. Antibiotic susceptibility profile of group B streptococcus acquired vertically. Obstet Gynecol. 1998;92:931–934. doi: 10.1016/S0029-7844(98)00263-4. [DOI] [PubMed] [Google Scholar]
- 15.Simoes JA, Aroutcheva AA, Heimler I, Faro S. Antibiotic resistance patterns of group B streptococcal clinical isolates. Infect Dis Obstet Gynecol. 2004;12:1–8. doi: 10.1080/10647440410001722269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura K, Suzuki S, Jun-ichi W, Hiroshi K, Kunikazu Y, Naohiro S, Nagano N, Haru K, Shibayama K, Yoshichika A. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Ant Ag Chem. 2008;52:2890–2897. doi: 10.1128/AAC.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitoussi F, Loukil C, Gros I, Clermont O, Mariani P, Bonaonacorsi S, Thomas I, Deforched D, Bingen E. Mechanisms of macrolide resistance in clinical Group B streptococci isolated in France. Ant Ag Chem. 2001;45:1889–1891. doi: 10.1128/AAC.45.6.1889-1891.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Back E, O’Grady EJ, Back JD. High rates of perinatal GBS clindamycin and erythromycin resistance in an Upstate New York Hospital. Ant Ag Chem. 2012;56:739–742. doi: 10.1128/AAC.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verani JR, McGee L, Schrag SJ. Prevention of perinatal group B streptococcal disease-revised guidelines from CDC, 2010. MMWR Recomm Rep. 2010;59:1–36. [PubMed] [Google Scholar]
- 20.Arpin C, Daube H, Tessier F, Quentin C. Presence of mefA and mefE genes in Streptococcus agalactiae. Ant Ag Chem. 1999;43:944–946. doi: 10.1128/aac.43.4.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng X, Kong F, Hui W, Achie D, Gilbert LG. Simultaneous detection of nine antibiotic resistance-related genes in Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization assay. Ant Ag Chem. 2006;50:204–209. doi: 10.1128/AAC.50.1.204-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gygax SE, Schuyler JA, Trama JP, Mordechai E, Adelson ME. Detection of erythromycin and clindamycin resistance genes in Group B streptococcal clinical isolates and cervicovaginal-rectal swabs. Microb Drug Resist. 2007;13:119–123. doi: 10.1089/mdr.2007.732. [DOI] [PubMed] [Google Scholar]
- 23.Quiroga M, Pegels E, Oviedo P, Pereyra E, Vergara M. Susceptibility patterns and prevalence of GBS isolated from pregnant women in Misiones, Argentina. Braz J Microbiol. 2008;39:245–250. doi: 10.1590/S1517-83822008000200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan AS, Walsh A, Crowleyet B. Role of efflux in macrolide resistance in β-hemolytic streptococci of groups A, B, C and G collected in an Irish teaching hospital. J Med Microbiol. 2011;60:262–264. doi: 10.1099/jmm.0.023788-0. [DOI] [PubMed] [Google Scholar]
- 25.Heelan JS, Hasenbein ME, McAdam AJ. Resistance of group B streptococcus to selected antibiotics, including erythromycin and clindamycin. J Clin Microbiol. 2004;42:1263–1264. doi: 10.1128/JCM.42.3.1263-1264.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gosiewski T, Brzychczy-Włoch M, Heczko PB. The application of multiplex PCR to detect seven different DNA targets in group B streptococci. Folia Microbial (Praha) 2012;57:163–167. doi: 10.1007/s12223-012-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. M100-S22, M2–7 and M–7, Wayne, PA. 2012. Guidelines for streptococcus spp, β-hemolytic group 2012.
- 28.Desjardins M, Delgaty KL, Ramotar K, Seetaram C, Toye B. Prevalence and mechanisms of erythromycin resistance in group A and group B streptococcus: implications for reporting susceptibility results. J Clin Microbiol. 2004;42:5620–5623. doi: 10.1128/JCM.42.12.5620-5623.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin resistant by PCR. Ant Ag Chem. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Azavedo JCS, McGavin M, Duncan C, Low DE, McGeer A. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Ant Ag Chem. 2001;45:3504–3508. doi: 10.1128/AAC.45.12.3504-3508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fluegge K, Supper S, Siedler A, Berner R. Antibiotic susceptibility in neonatal invasive isolates of GBS in a 2-year nationwide surveillance study in Germany. Ant Ag Chem. 2004;48:4444–4446. doi: 10.1128/AAC.48.11.4444-4446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joachim A, Matee MI, Massawe FA, Lyamuya EF. Maternal and neonatal colonization of GBS at Muhimbili National Hospital in Dar es Salaam: prevalence, risk factors and antimicrobial resistance. BMC Public Health. 2009;9:437. doi: 10.1186/1471-2458-9-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray KJ, Bennett SL, French N, Phiri AJ, Grahan SM. Invasive group B Streptococcal infection in infants, Malawi. Emerg Infect Dis. 2007;13:223–229. doi: 10.3201/eid1302.060680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiPersio LP, DiPersio JR. High rates of erythromycin and clindamycin resistance among OBGYN isolates of group B streptococcus. Diagn Microbiol Infect Dis. 2006;54:79–82. doi: 10.1016/j.diagmicrobio.2005.07.003. [DOI] [PubMed] [Google Scholar]


