Summary
The structure and amino acid diversity of the T-cell receptor (TCR), similar in nature to that of Fab portions of antibodies, would suggest these proteins have a nearly infinite capacity to recognize antigen. Yet all currently defined native T cells expressing an α and β chain in their TCR can only sense antigen when presented in the context of a major histocompatibility complex (MHC) molecule. This MHC molecule can be one of many that exist in vertebrates, presenting small peptide fragments, lipid molecules, or small molecule metabolites. Here we review the pattern of TCR recognition of MHC molecules throughout a broad sampling of species and T-cell lineages and also touch upon T cells that do not appear to require MHC presentation for their surveillance function. We review the diversity of MHC molecules and information on the corresponding T-cell lineages identified in divergent species. We also discuss TCRs with structural domains unlike that of conventional TCRs of mouse and human. By presenting this broad view of TCR sequence, structure, domain organization, and function, we seek to explore how this receptor has evolved across time and been selected for alternative antigen-recognition capabilities in divergent lineages.
Keywords: TCR recognition, MHC, MHC-like class I, non-classical class I, structure, evolution
Introduction
The T-cell receptor (TCR) structural scaffold is remarkably similar to that of the antigen recognition domain of antibodies (Fab), with a heterodimeric assembly of two polypeptide chains that form a highly diverse antigen recognition domain composed of six to eight complementary determining region (CDR) loops (Fig. 1A). The diversity inherent in these loops has two origins. The CDR1 and CDR2 (and sometimes framework region 4 (FR4), included in this discussion because it occasionally contributes to antigen contact) loops are ‘germline-encoded’, in other words their diversity comes from differences in the V gene segments that are used within a species during rearrangement of receptors for a particular T-cell lineage. Amino acid diversity in the two CDR3 loops comes from the recombination-activating genes (RAG)-dependent process of gene segment rearrangement (V-D-J or V-J); the random joining of these gene segments alone generates significant diversity that is amplified by both templated and non-templated nucleotide addition or subtraction in these junctions.
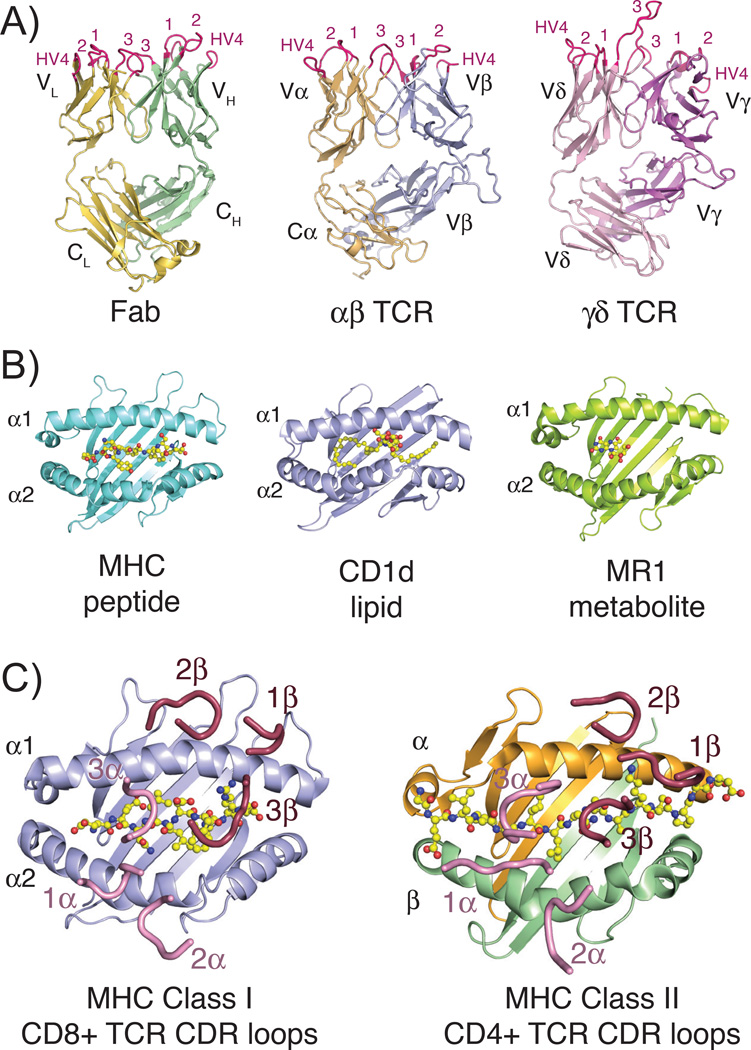
Fig. 1. Structures of TCR, Fab, and MHC molecules and the classical docking paradigm.
(A) Three-dimensional structures of the three classical rearranged receptors in jawed vertebrates: Fab, αβ TCR and γδ TCR (PDB IDs: 1MLC, 2CKB and 1YPZ). Domains are labeled according to their chain designations and CDR loops are colored in hot pink and labeled accordingly. (B) Representative three-dimensional structures of the three antigen-presenting MHC molecules: classical class I MHC with peptide, CD1d with lipid and MR1 with small molecule metabolite (PDB IDs: 2CKB, 1ZT4 and 4LCC). Ligands are shown in stick representations and colored yellow, red and blue indicating carbon, oxygen and nitrogen atoms, respectively. (C) Complex crystal structures of a CD8+ αβ TCR with MHC class I molecule (PDB ID: 3DXA) and CD4+ αβ TCR with MHC class II molecule (PDB ID: 4E41), with just CDR loops shown as they are positioned in the complex structure. Peptide ligands are shown as described above for 1B. In both complexes, CDR loops of the α chain are colored pink, of the β chain are colored raspberry and each are numbered accordingly. All three-dimensional structure figures were made with the program Pymol (Schrödinger).
For antibodies, their CDR diversity is further honed by the process of somatic hypermutation, generating an affinity-optimized scaffold that is well-equipped to bind almost any antigenic target. TCRs, lacking somatic hypermutation, have significant potential diversity within their CDR loops; however, in the species where they are most well-studied, mouse and human, the αβ T-cell lineage appears to recognize antigens only when presented by major histocompatibility complex (MHC) molecules (Fig. 1B). This phenomenon was first described for αβ T-cell recognition of the classical, peptide-presenting MHC (MHCp) class I and class II molecules, whereby the TCR probes a composite surface of the MHC surface and the presented peptide. Numerous structural studies have dissected the atomic underpinnings of the TCR/MHCp recognition process, and remarkably, the vast majority of these structures demonstrate that the TCR docks onto both MHC class I and class II surfaces using a conserved diagonal orientation, whereby the TCR β chain is mainly oriented over the α1 (called such in both class I and II molecules), and the TCR α chain is oriented over the α2 or β1 helix (for class I and class II molecules, respectively) (1, 2) (Fig. 1C).
Debate continues as to the origin of this MHC restriction and docking orientation bias: whether there is direct coevolution that has occurred between TCR variable (V) genes and their respective MHC ligands that would enforce both restriction and a particular docking orientation (3–6) or whether this instead is a product of selection in the thymus, enforced by co-receptor (CD4 or CD8) requirement for engagement of MHC in a particular orientation (7–10). This controversy has been covered in other reviews and reports and is not the focus of this review. Instead, we review the current state of TCR recognition of nonclassical and MHC-like proteins and place these in the context of conventional TCR recognition of MHCp. Furthermore, we review T-cell lineages, such as γδ T cells, where there is precedent for both MHC restriction and TCR recognition of antigens without requirement for MHC involvement. To place this broadly in the context of vertebrate evolution, we also survey the current understanding of the breadth of nonclassical or MHC-like loci, TCR genetics and structure, and the interactions between nonclassical or MHC-like molecules and TCR, where known, in evolutionary distant vertebrate species.
Overview of TCR recognition of MHCp
The first crystal structure of an MHC molecule was HLA-A2 (11); this structure was a seminal milestone in understanding the function of MHC proteins as antigen-presenting molecules. The two α helices positioned over a β-sheet platform provided an ideal structure by which peptides could be anchored and presented, forming a composite surface with the MHC molecule for recognition by a TCR. Later, as the first crystal structures of complexes between TCRs and MHCp were elucidated (12, 13), it was noted that the binding orientation situated the germline encoded CDR1 and CDR2 loops of the TCR over the α helices of the MHC molecule, whereas the junctionally-encoded CDR3 loops were generally positioned over the peptide. Subsequent TCR-MHCp structures, with only a few exceptions, all subscribed to this diagonal docking orientation (Fig. 2). Detailed reviews of this interaction have been previously published (1, 2).
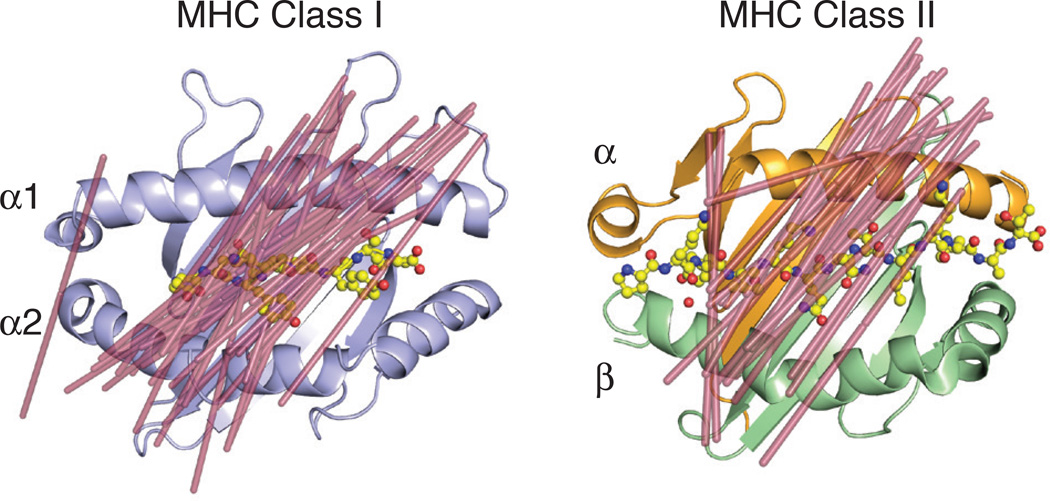
Fig. 2. MHC class I and class II restricted TCR docking orientations.
Shown are the docking orientations of αβ TCRs on their MHC class I (left, PDB ID: 2CKB) or class II (right, PDB ID: 1FYT) ligands. Lines were drawn (shown in raspberry) from the two external conserved cysteines in the Variable Ig domains (C22 of α chain and C23 of β chain) to demonstrate orientation of the TCR on the MHC surface. The following PDB IDs were used for the complex structures for the class I model: 1FO0, 1KJ2, 3RGV, 2CKB, 2OI9, 3PQY, 2OL3, 1AO7, 1BD2, 1LP9, 1OGA, 2BNQ, 3GSN, 3HG1, 3O4L, 3QDJ, 3QDM, 3UTS, 3VXM, 3VXR, 3VXU, 4G8G, 2NX5, 3MV7, 2AK4, 4JRY, 3DXA, 3KPS, 2YPL, 1MI5, 3FFC, 3SJV, 4MJI, 2ESV; and for the class II model: 3PL6, 4OZF, 4OZI, 4GG6, 4E41, 2IAN, 1FYT, 4H1L, 2WBJ, 1YMM, 1ZGL, 3O6F, 4P4K, 3C5Z, 3C60, 3C6L, 3RDT, 3MBE, 2PXY, 1U3H, 3QIB, 1D9K.
Identification of αβ T-cell lineages with restriction to MHC molecules outside of the classical class I and II proteins, among them invariant natural killer T (iNKT) cells and mucosal associated invariant T (MAIT) cells, raised the important question concerning the requirement for this diagonal docking orientation observed in αβ T cells restricted to classical MHC: do all T cells need to dock in the same way for T-cell signaling to occur? Or have these T cells coevolved with their respective ligands such that each lineage has a unique recognition strategy? Below we first review these MHC-like molecules, their interaction with their respective T-cell lineages, and what they can reveal about TCR-ligand coevolution. We later discuss T-cell lineages such as γδ T cells, which have examples of MHC-nonrestricted recognition, and how they fit into the general paradigm of T-cell recognition and activation.
CD1-restricted T cells specific for lipid antigens
Soon after identification of the CD1 loci as encoding several MHC-related β2m-associated molecules (14), it was noted that the putative antigen-binding α1 and α2 domains bore little sequence homology to those of classical MHC molecules (15). However, CD1 can be recognized by both the αβ and the γδ T-cell lineages and restrict T-cell responses to foreign microbial antigens, implying a similar antigen-presenting function as MHC (16–18). Subsequent studies surprisingly revealed that CD1 family members present lipids antigens to T cells (19), with the first CD1 crystal structures demonstrating how these molecules have ‘repurposed’ the α1/α2 domain of classical MHC molecules for the binding of lipids via deep hydrophobic tunnels, thus exposing polar head groups for T-cell contact (20–23). In terms of a host-pathogen arms race, escape from T-cell recognition of lipids would require more extensive pathogen evolution given the numerous enzymatic pathways involved in lipid synthesis, and thus more complicated than mutating than a single protein amino acid, which could otherwise permit escape from certain MHC-restricted αβ T-cell clones. All CD1 family members also present host-derived lipids to autoreactive T cells (24–27), seemingly at odds with the central immunological paradigm of self/non-self discrimination; however, it is becoming clear that autoreactivity is evolutionarily conserved and imparts CD1-specific cells with novel immunoregulatory functions (28).
The CD1 locus encodes several distinct genetic isoforms, all of which are nonpolymorphic within a particular species. The human CD1 locus encodes five separate isoforms: CD1a, CD1b, CD1c, CD1d, and CD1e. The first three are considered Group 1 CD1 molecules based on genomic location and homology, whereas CD1d is considered Group 2 CD1. CD1a–CD1d are cell-surface expressed glycoproteins that are directly implicated in lipid antigen presentation to T cells (19, 26, 29, 30), whereas CD1e remains localized to intracellular Golgi or vesicular compartments and regulates lipid loading into other CD1 family members (31, 32). As discussed in more detail below, the structures of human CD1a–CD1d allow each isoform to specialize in the presentation of distinct lipid classes, representing a different type of evolutionary diversification than the highly polymorphic classical MHC molecules (33, 34). However, mice and muroid rodents are unusual among mammals in only having 2 CD1 genes, CD1d1 and CD1d2, and functional antigen presentation to T cells has only been described for the former, which is simply known as CD1d (35, 36). It is widely regarded that studies of murine immunology authentically describe the workings of the human immune system. However, within the field of T-cell-CD1 recognition, there are examples of both remarkable conservation and surprising distinctions between species. In fact, there are CD1 family members and associated T-cell populations that are entirely species-specific. With the motivating goal of immunology being the understanding and treatment of human immunological disease, it is important to understand which aspects of CD1 biology and recognition deciphered from murine studies can be applied to human biology (37).
Overall architecture of human/mouse CD1a–e
Despite the global structural similarity between CD1 and MHC molecules, CD1 has several distinct features within its antigen-binding pocket that are specialized for its lipid antigen cargo. The α helices surrounding the antigen-binding groove are positioned farther from the β-platform that forms the base of the groove, creating a deeper pocket structure. Furthermore, the pocket architecture of CD1 molecules has evolved to specifically accommodate lipid acyl chains through their hydrophobic tunnels, differing from the classical MHC groove characterized by polar hydrogen-bond forming residues. This CD1 binding mode positions lipid polar head-groups, with tremendous potential chemical diversity, such they protrude outwards from the internal pockets and thus situated for T-cell recognition. Additionally, the various CD1 isoforms survey distinct intracellular vesicular compartments, maximizing the potential diversity of lipids subjected to immune surveillance (38).
Several recent reviews have detailed the structural details and lipid-binding repertoire of the ‘standard’ human and mouse CD1 family members (30, 33, 34, 39). In brief, the pocket sizes of CD1 molecules range in size from CD1a>CD1d>CD1c>CD1e>CD1b, with cavity sizes ranging from ~1350 Å (CD1a) to 2200 Å (CD1b). The pocket architecture differs substantially between these family members. CD1a possesses one well-defined pocket, called the A′ pocket based off alignment to the pockets of classical MHC molecules, and an exposed F′ pocket that is partially open to solvent, allowing lipids with larger head groups, such as lipopeptides, to protrude (22, 40). CD1b has the most extensive internal pocket structure, with a deeply buried T′ tunnel connecting the A′ and F′ pockets, forming a continuous channel suited for the presentation of extremely long lipid species, such as mycolic acids from mycobacterial species (21, 41). Furthermore, the width of the CD1b tunnels can accommodate lipid modifications such as methyl branches and cyclopropyl groups, which found among the structural unique mycobacterial cell wall lipids (42). CD1c is also suited for presentation of branched lipid species from mycobacteria and possesses an exit portal at the terminus of the A′ pocket, through which especially long lipids could protrude (23). The open F′ groove of CD1c permits presentation of lipopeptides, somewhat akin to CD1a. Mouse and human CD1d are structurally very similar, with two well-defined hydrophobic pockets that are suited for binding un-branched lipid acyl chains of up to 26 carbons (A′ pocket) and 18 carbons (F′ pocket) (20). Lastly, CD1e lacks clearly delineated pockets, but rather a side and solvent-exposed groove, which may relate to its ability as a lipid transfer chaperone (32, 43). Collectively, the human CD1 family binds and presents hugely diverse lipid species via the unique pocket architectures of the different genetic isoforms.
Evolutionary conservation of CD1 group members
CD1 genes have also been described in other mammalian species, in birds, and in reptiles (44–47). Although rodents only express Group 2 CD1 (CD1d) genes, and not CD1a–c, many other non-human mammals do have functional Group 1 CD1 molecules. Members of the Equus genus, which includes horses, zebras, and asses, have the largest known family of CD1 genes, with 13 genes total showing 60–83% identity to their human counterparts (48). Seven isoforms were classified as CD1a, two as CD1b, one as CD1c, one as CD1d, and two as CD1e (48) (Table 1). The largest differences between horse and human CD1 are found in the α1 and α2 helices, which are principally responsible for lipid binding and TCR contacts (48).
Table 1.
| Nonclassical class I molecules | T cel subsets | ||||
|---|---|---|---|---|---|
| Mammals | Placental mammals | Human | CD1a | αβ T cells | |
| CD1b | γδ T cells | ||||
| CD1c | iNKT cells | ||||
| CD1d | Type II NKT cells | ||||
| CD1e | MAIT cells | ||||
| MR1 | |||||
| Mouse | αβ T cells | ||||
| CD1d | γδ T cells | ||||
| T22/T10 | iNKT cells | ||||
| MR1 | Type II NKT cells | ||||
| MAIT cells | |||||
| Dog | CD1a (x2) | ||||
| CD1b | αβ T cells | ||||
| CD1c | γδ T cells | ||||
| CD1d | iNKT cells(80) | ||||
| CD1e | |||||
| MR1 | |||||
| Rabbit | CD1a (x2) | αβ T cells | |||
| CD1b | γδ T cells | ||||
| CD1d | iNKT cells? | ||||
| CD1e | |||||
| Artiodactyls | Sheep |
CD1a(49) | αβ T cells | ||
| Cow | CD1b (x3)(49) | γδ T cells | |||
| CD1d(50) | MAIT cells(125) | ||||
| CD1e(49) | iNKT cells?(81) | ||||
| MR1(137) | |||||
| Horse | CD1a (x7)(48) | ||||
| CD1b(48) | αβ T cells | ||||
| CD1c(48) | γδ T cells | ||||
| CD1d (x2)(48) | iNKT cells(81) | ||||
| CD1e (x2)(48) | |||||
| MR1 | |||||
| Marsupials | Opossum | CD1 in some species(53) | αβ T cells | ||
| MR1(120) | γδ T cells | ||||
| MAIT cells(120) | |||||
| Monotremes | Platypus | ? | VHδ γδ T cells(164) | ||
| TCRμ T cells(172, 173) | |||||
| Birds | Chicken | chCD1-1, chCD1-2(45, 46) | αβ T cells | ||
| γδ T cells | |||||
| YF1*7.1(139) | VHδ γδ T cells(170) | ||||
| Reptiles | Squamata |
CD1(47) | αβ T cells only(141) |
||
| Testudines, Crocodylia | αβ and γδ T cells(141) | ||||
| Amphibians | Frog | αβ T cells | |||
| XNC(84) | γδ T cells | ||||
| VHδ γδ T cells(171) | |||||
| XNC10-invariantT(83) | |||||
| Cartilaginous fish | Shark | Hefr-19 (class Ib)(88) | αβ T cells | ||
| Gici-11 (predicted class Ib)(88) | γδ T cells | ||||
| Sqac-UAA*NC1(89) | IgM/W γδ T cells(147) | ||||
| NAR-TCR γδ T cells(175) | |||||
Ruminants, including cows, also express multiple CD1 molecules, including CD1a, CD1e, and three CD1b isoforms with differences in their binding groove and cytoplasmic tails (Table 1). Although these species were originally thought to lack CD1d due to absence of a functional start codon (49), it was later found that cows do in fact express cell surface CD1d (50) (Table 1). Bovine CD1d is able to bind to glycosphingolipids with short fatty acid chain lengths, including C12-di-sulfatide, C16-αGalCer, and C18, but not longer C24 fatty acids (50, 51). The crystal structure of bovine CD1d in complex with C16-αGalCer confirmed that it has a flexible binding groove and plasticity in the A′ pocket due to changes in the conserved Trp40 residue (51). The A′ pocket was also considerably shorter than mouse and human CD1d, due to interaction between Trp166 and Thr100 inside the pocket, explaining the inability of bovine CD1d to bind fatty acids with longer chains (51). The crystal structure of another bovine CD1 isoform, CD1b3 also showed variations in the binding pocket compared to human CD1b. The T′ tunnel in this structure is closed due to the presence of valine instead of glycine at position 98, suggesting that like CD1d, CD1b3 might bind a skewed set of lipids (52). Additionally, there is a roof over the F′ pocket, which prevents presentation of alkyl chains toward the presumed TCR interface, as is seen in human CD1b (21, 52). It is unclear if the other CD1b isoforms may have more ‘normal’, human-like binding pockets. It is reasonable to assume that diverse microbial and self-lipids would be present in different species, leading to adaptations in the binding pockets of CD1 in both horses and ruminants.
Unlike most placental mammals (besides rodents), which have multiple CD1 genes, marsupials only possess one CD1 isoform, CD1. Marsupial CD1 is functionally expressed in some species including bandicoot (I. macrourus), but is likely a pseudogene in the opossum (M. domestica) (53) (Table 1). It is unclear why functional CD1 is not necessary in all marsupials or why these animals are lacking the multiple isoforms found in many other mammals. Perhaps the presence of another highly conserved nonclassical molecule, such as MR1, described in detail below, could functionally compensate for a lack of CD1 in these species. Alternatively, high expression of the unusual TCR, TCRµ, which is proposed to bind unprocessed/free antigen as discussed in detail below, could also alleviate the requirement of antigen presentation by MHC-like or nonclassical class I molecules in marsupials.
Non-mammalian species including birds were also found to express CD1. Chickens have two CD1 molecules, one, chCD1–2, with a shallow binding groove that likely only binds single chain lipids (54), and another, chCD1-1, with dual A′ and F′ pockets for binding to two- or three-chain lipids of varying lengths (55) (Table 1). Birds are likely capable of presenting a variety of lipids via these two CD1s. chCD1-1, with its deep dual pockets, is more similar to human CD1 molecules than CD1–2, and specifically resembles CD1b and CD1d (55). While chCD1–2 resembles CD1a, but with a smaller A′ pocket and narrower groove (54).
Neither chCD1-1 nor chCD1–2 are classified as true group 1 or group 2 CD1 molecules, but based on the overall similarity between chCD1-1 and human CD1 molecules, it was hypothesized that chCD1–2, with its shallower binding pocket, may be reminiscent of a primordial CD1 molecule. Recent work on CD1 in reptiles, however, suggests that many lizard CD1 molecules also contain dual binding pockets and most resemble mammalian CD1d and chicken chCD1-1 (47) (Table 1). The short binding pocket of chCD1–2 may therefore be a chicken specific adaptation, rather than an example of a primordial CD1 molecule, although understanding the structure of CD1 molecules from more evolutionarily distinct species could help to clarify this relationship. Several species of reptile also contain additional CD1 family members with little homology to either chicken or mammalian CD1, suggesting that like chCD1–2, some of their genes have diversified since speciation for binding to a variety of lipid structures (47). As suggested for horses and ruminants, these binding pocket adaptations likely allow for the binding of a diverse array of lipids in non-mammalian species. It is not entirely clear, however, which T-cell subsets might be responsible for CD1-lipid specific responses in these animals or if they are analogous to the CD1-reactive T cells in mice and humans discussed below.
CD1d recognition by human and mouse invariant natural killer T cells
Both mice and humans possess a T cell population characterized by a highly restricted TCR repertoire with particular enrichment of a DN coreceptor phenotype and expression of natural killer (NK) cell markers, underlying their classification as NKT cells (56–58). These invariant NKT (iNKT) cells (also called type I NKT cells, a distinction that is described below) utilize an invariant α chain devoid of junctional diversity, specifically a Vα14-Jα18 in mice and Vα24-Jα18 in humans, and limited diversity of Vβ chains. These TCRs specify iNKT cell recognition of CD1d (25, 59), and are potently stimulated by the glycosphingolipid α-galactosylceramide (α-GalCer) (29, 35), which was first described as a sea sponge-derived anti-tumor natural product and then subsequently identified as both a lipid of human intestinal commensal bacteria and mammalian cell membranes themselves (60–62). Recognition of αGalCer and other agonist lipids, which include certain self-lipids (63, 64), is likely involved in thymic selection and induces rapid and potent IL-4 and IFN-γ secretion in human and mouse iNKT cells alike (65, 66).
In addition to similarity in function and antigen specificity, mouse and human NKT cells also share a surprising CD1d cross-species reactivity. Certain CD1d–autoreactive mouse iNKT TCR-expressing hybridomas are stimulated equally well by mouse and human CD1d–expressing APCs, a feature not shared by hybridomas expressing other non-Vα14/Vβ8 TCRs (29). Human iNKT cells can also recognize mouse CD1d molecules; in both cases, the particular Vβ chain usage and CDR3β residues may shape this feature. Furthermore, the affinities of a human iNKT TCR for human and mouse CD1d-αGalCer complexes are within a similar range, which are effectively equalized by substitutions in the CDR3β loop (67).
The structures of unliganded human iNKT TCRs provided the first clues for the basis of iNKT TCR cross-reactivity (67). Though only 2/6 of iNKT TCR CDR1α residues are identical between humans and mice, they form an adjoining patch of shared residues with the highly similar CDR3α loop, in which 10/12 residues are identical. Furthermore, the 4/6 conserved residues of the CDR2β loop are structurally juxtaposed, forming a contiguous surface of identical residues shared between the human and mouse iNKT TCRs, and collectively suggesting a conserved importance of these loops for ligand recognition. The iNKT TCR αGalCer-CD1d complex crystal structure validates this model perfectly (68). The CDR1α loop is positioned directly over the α-GalCer galactose headgroup, forming several hydrogen bonds with this sugar moiety. The invariant CDR3α loop dominates recognition of the CD1d α-helices, centered over the F′ pocket, yet also directly contacts the α-GalCer headgroup. Surprisingly, the lipid antigen itself is solely contacted by germline-encoded residues, distinguishing this structure dramatically from those of classical αβ TCR-pMHC molecules (Fig. 3A,C). Of note, the residues contacting α-GalCer are either identical, biochemically similar, or simply backbone-mediated, in a comparison of the human and mouse iNKT TCR sequences, highlighting the evolutionary conservation of this interaction. In fact, a remarkable number of human and mouse iNKT TCRs in complex with various lipids exhibit a nearly identical docking mode, a feature that has been reviewed extensively (69, 70). Of note, the CD1d residues contacted by the iNKT TCRs are also highly conserved between mouse and human (Fig. 4A). The conservation of the docking mode and contributing amino acids is especially surprising given the diversity of lipids thus far identified within crystallized structures. To accommodate different sugar anomers, modifications, and linkage numbers, as well as glycerophospholipids, the iNKT TCR reorients the polar sugar head group to accommodate these diverse chemical features. This is particularly striking in the case of the iNKT TCR-isoglobotrihexosylceramide (iGB3)-CD1d complex, in which the outward protruding tri-sugar moiety is flatted 90° versus the unliganded CD1d–iGB3 complex, resolving the prior uncertainty of how this lipid could potentially act as an agonist ligand (71–73).
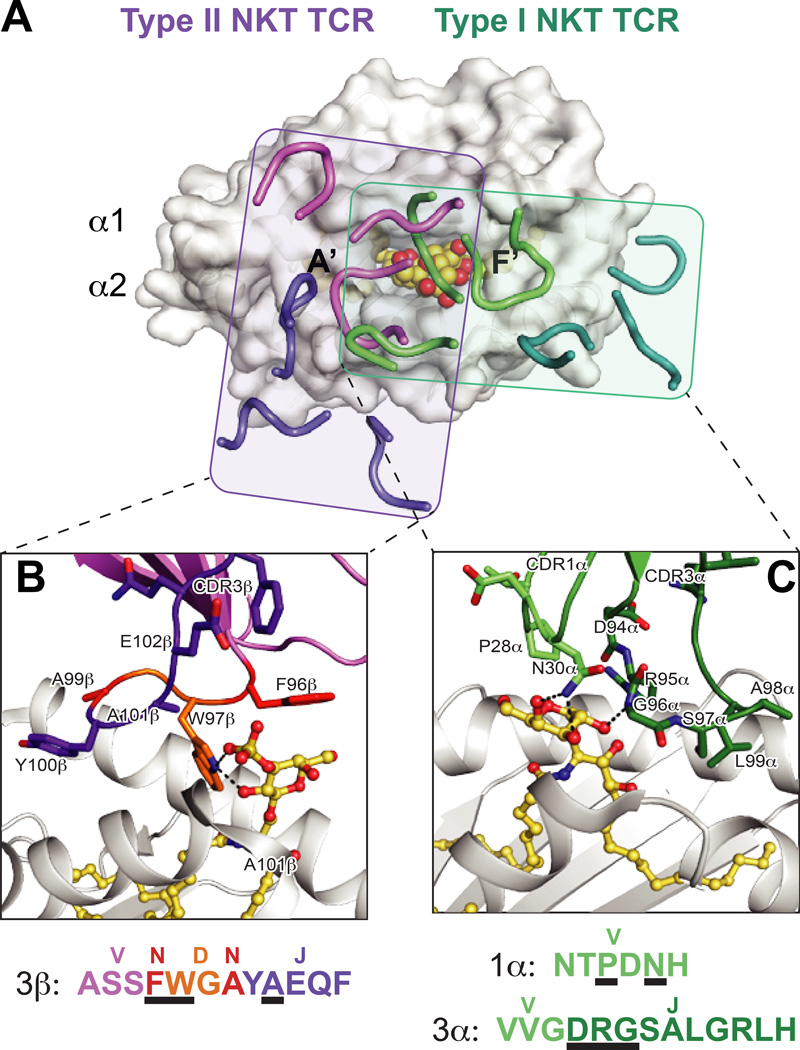
Fig. 3. Differential docking modes and lipid antigen contacts of Type I and Type II NKT TCRs.
(A) Footprints of the Type I (iNKT) and Type II NKT TCRs on CD1d. The complex structures of the murine iNKT and Type II NKT TCR structures are aligned by CD1d. Shown is the surface of murine CD1d-αGalCer (PDB ID: 3HE7) with the CDR loops of the murine Vβ7 iNKT TCR (PDB ID: 3HE7) in green (α chain) and teal (β chain), and type II NKT TCR (PDB ID: 4EI5) in purple (α chain) and pink (β chain). The rough borders of the TCR/CD1d interfaces are shown as shaded boxes colored teal (Type I) and purple (Type II). (B) Detail of the mouse Type II NKT TCR-CD1d–sulfatide interface. Residues that contact the sulfatide antigen are shown and colored according to origin of encoding nucleotides (pink = V, purple = J, orange = D, red = non-templated). CD1d is shown in grey cartoon, sulfatide is in yellow ball-and-sticks. Hydrogen bonds are shown in black dashed lines. Lower panel details the amino acids of the CDR3β loop, colored according to origin as above, with black underlines denoting with residues contact sulfatide. The major sulfatide contacts are through either non-templated or D-encoded TCR residues. (C) Detail of the mouse Vβ7 iNKT TCR –CD1d-αGalCer interface. Residues that contact the αGalCer antigen are shown and colored according to origin of encoding nucleotides (light green = V, dark green = J). . CD1d is shown in grey cartoon, αGalCer is in yellow ball-and-sticks. Hydrogen bonds are shown in black dashed lines. Lower panel details the amino acids of the CDR1α and CDR3α loops, colored according to origin as above, with black underlines denoting with residues contact αGalCer. All contacts with the lipid antigen and germline encoded.
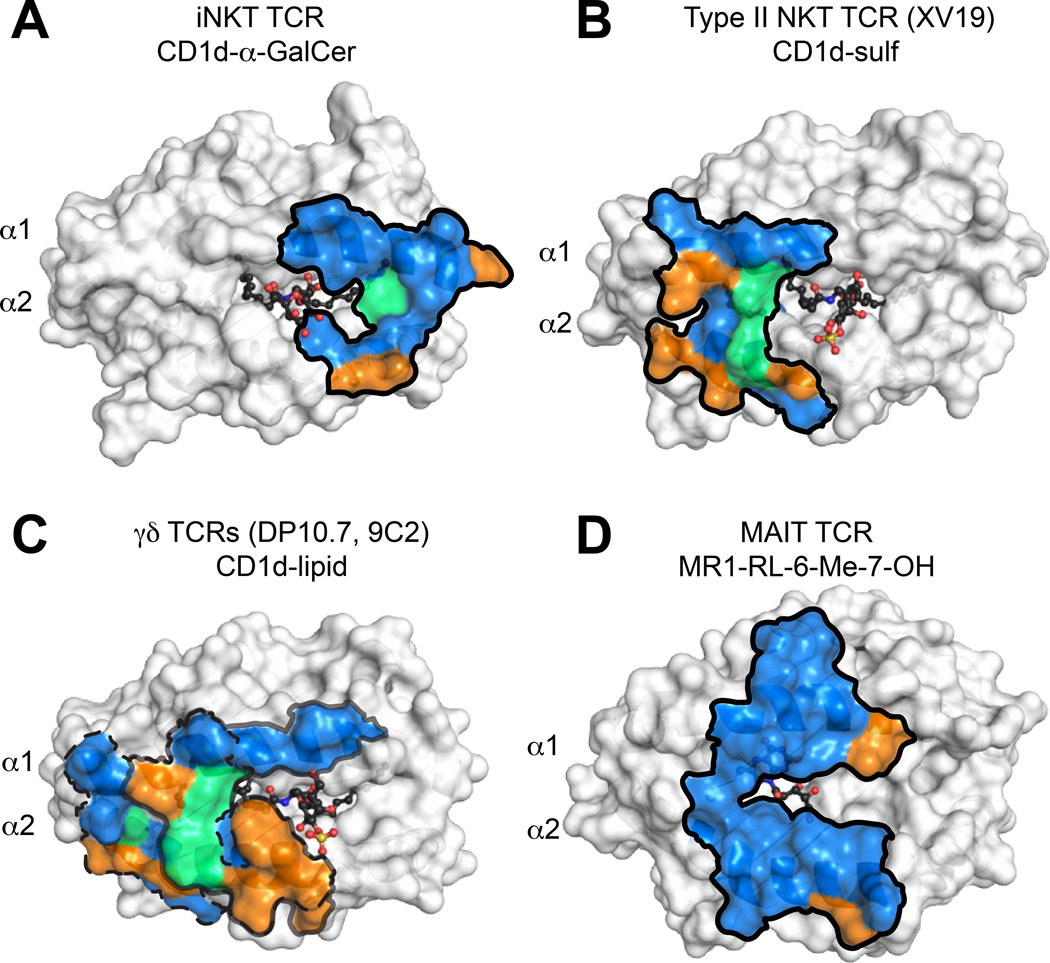
Fig. 4. Evolutionary conservation of TCR footprints on antigen presenting molecules.
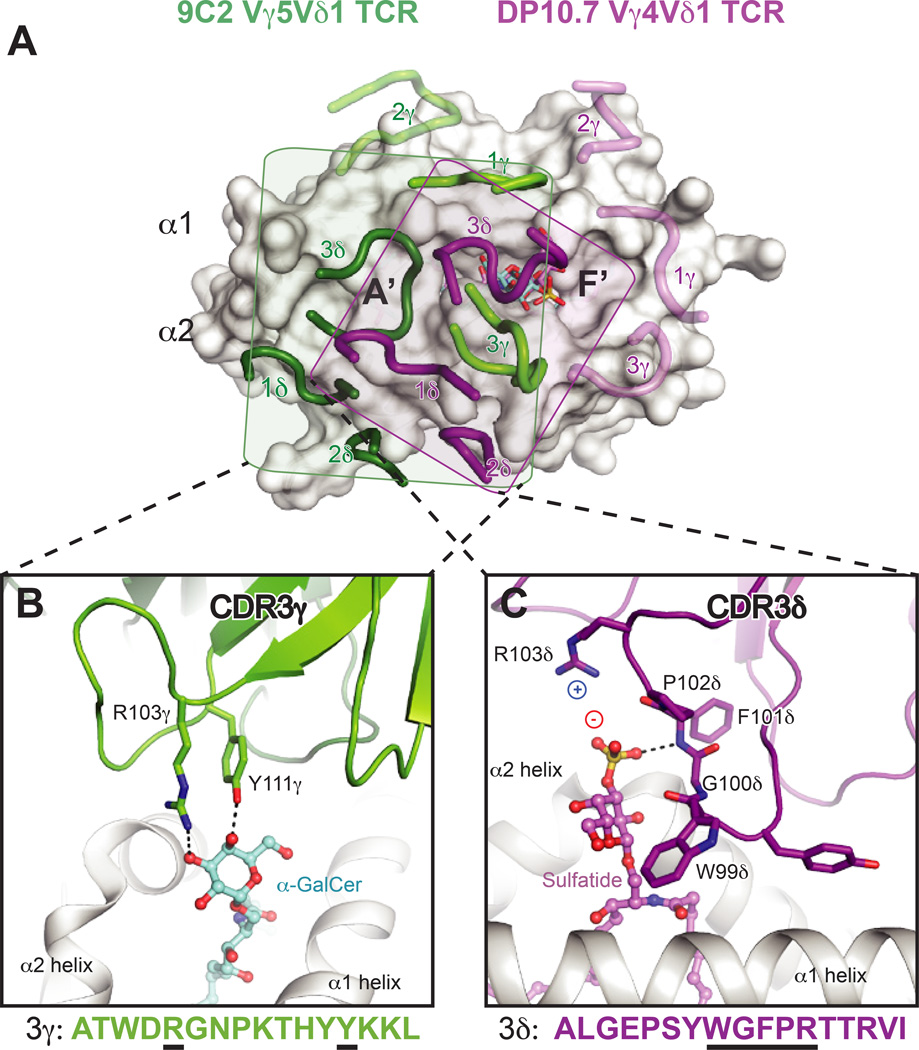
Shown are the surfaces of CD1d or MR1 in white, with residues that contact the TCR colored blue, green or orange. Blue = residues identical in mice and humans, green = residues with similar biochemical properties in mice and humans, orange = no conservation. Dark outlines define total contact borders. (A) Human CD1d-αGalCer with residues contacted by the human iNKT TCR colored as above. (B) Mouse CD1d–sulfatide with residues contacted by the mouse XV19 TCR colored as above. (C) Footprints of the human DP10.7 and 9C2 γ TCR on human CD1d–lipid surface (sulfatide shown). The border of the DP10.7 TCR footprint on CD1d is shown in grey; border of 9C2 TCR footprint on CD1d is shown in dashed line. (D) Human MR1-RL-6-Me-7-OH with residues contacted by the human MAIT TCR colored as above.
The immutable and evolutionarily conserved docking angles of iNKT TCR-CD1d complexes, largely without any contribution of non-templated residues in CDR3 loops as are key for conventional αβ TCR-pMHC recognition, parallels the ‘innate-like’ attributes of these cells. Such an assembly of iNKT cells with a shared antigen specificity could respond en masse to agonist lipid ligands without prior need for clonal expansion, influencing a nascent immune response with their copious cytokine production. With regards to infection, certain pathogen-derived α-linked glycolipids can stimulate NKT cells (74–76), and again biochemical and structural studies have validated high affinity TCR-lipid-CD1d interactions and typical iNKT TCR docking modes (77, 78). The ability of iNKT TCRs to recognize certain glycolipids from gram-negative bacterial lacking the potent innate-immune stimulatory lipopolysaccharide suggests they may have evolved as a bridge between the innate and adaptive immune systems, perhaps in a similar role as the Toll-like receptors (TLRs) upon various innate immune system cells. Yet unlike the bona fide innate immune receptors, iNKT TCRs are inherently autoreactive, blurring the lines for their role as a potential innate-like pathogen sensor. Reductionist studies in the murine system have painted a landscape of distinct iNKT cell functions, yet a unified model of their specific roles in human health is currently still being unraveled (79).
iNKT cell populations in diverse vertebrate species
Despite the conservation of CD1, and especially CD1d, in many species, the role of T-cell-specific responses to these molecules outside of mice and humans is not entirely clear. iNKT-like cells using similar Vα and Jα segments to human and mouse iNKT cells have also been identified in canines, based on binding to CD1d/αGalCer (80), and a similar TCR α chain to TRAV10V/Vα24 has been described in horses, pigs, cows, sheep, and rabbits (81) (Table 1). However, only horses and pigs were found to contain sequences homologous to the canonical CDR3 regions of human and mouse iNKT cells (81). These species all express CD1d, so it is possible that they still have functional CD1-restricted iNKT cells but with TCR sequence motifs that differ from mouse and human iNKT cells. Originally, the lack of conserved NKT rearrangements in bovine species, together with the presumed non-functional CD1d was taken to mean that these animals likely did not have this invariant population (81). It is now known that cows do express surface CD1d (50), although with a slightly smaller binding pocket than human and mouse CD1d, as discussed above (50, 51). It is therefore possible that a bovine NKT cell population has also been overlooked, especially since the altered binding pocket of cow CD1d might present a different subset of lipids and thus bind to a different invariant CDR3 repertoire in these animals.
Vβ genes from species that do not express CD1 molecules, when paired with human invariant Vα chains are able to bind to mammalian CD1d (82). This conserved binding raises the possibility that there is a precedent for binding to such monomorphic molecules in other species, and it is likely that other species have similar systems with invariant populations that recognize, if not CD1, than other functionally similar molecules, as has been shown for XNC-specific invariant T-cell responses in amphibian species, discussed in more detail below.
Invariant T cells in non-mammalian species
Recent findings that T cells expressing an invariant Vα6-Jα1.43 rearrangement are highly expressed in Xenopus tadpoles and are specific for a nonclassical MHC class I molecule, XNC10 (83), have extended the study of invariant T cells to non-mammalian species. XNC10 is a member of the Xenopus nonclassical (XNC) family of nonclassical class I molecules, which are highly conserved among Xenopus species (84–86) (Table 1). Residues in some XNC molecules that are thought to be important for peptide binding are not well conserved between XNC classes or with human classical MHC sequences (86). Therefore it is possible that these molecules bind non-peptide ligands or that each class of XNC proteins bind to different antigens and may function to activate different types of cells. XNC-specific invariant T cells were shown to play an important role in tadpole immune responses to viruses, and thus likely contribute significantly to early immune responses in the frog (83). Development of XNC10-iT invariant T cells is dependent on expression of XNC10, which may be expressed in the thymus before metamorphosis, and before significant MHC class Ia expression in the tadpole thymus (83, 86, 87). There is currently no structure of Xenopus Vα6-Jα1.43 TCRs bound to XNC10, but it will be interesting to see how similar this interaction is to structurally characterized invariant T-cell/nonclassical MHC complexes from mammals.
Divergent non-classical or MHC-like class I molecules have also been suggested in other non-mammalian species, such as sharks (88, 89) (Table 1), although it is not clear if there is a similar invariant T-cell population in these animals that may recognize such molecules, or if as discussed below, some population of less invariant cells, such as γδ T cells or other type II NKT-like cells may be specific for these or other nonclassical class I molecules.
CD1d recognition by human and mouse type II, non-invariant NKT cells
In mammals a second class of CD1d–autoreactive cells was identified roughly contemporaneously with invariant NKT cells. Unlike iNKT cells, this second population does not utilize an invariant TCR α chain and highly restricted β chain, nor respond to the prototypical iNKT cell antigen αGalCer (27, 36), but rather surveys a likely non-overlapping repertoire of self-lipids of potent stimulatory capacity (90, 91). Another important distinction is that the non-Vα14 CD1d–reactive cells do not exhibit the same cross-species reactivity characteristic of iNKT cells, implying a different TCR-CD1d recognition mechanism (29). This population of ‘non-invariant’ αβ TCR+, CD1d–restricted but not αGalCer reactive cells, has been coined type II NKT cells to clarify their distinct specificities and functions from those of ‘type I’ iNKT cells (92). Though type II NKT cells are defined on their non-reactivity to αGalCer, the breadth of their self-lipid surveillance is unclear. A defined population of murine CD1d–restricted cells specific for the self-glycosphingolipid sulfatide has been identified, and though the type II NKT TCR repertoire exhibits certain Vα gene usage enrichments, they are a heterogeneous population likely capable of diverse lipid antigen discrimination (91, 93–95).
Based on the identification of type II NKT cells specific for the self-lipid sulfatide and a responsive murine hybridoma clonal TCR (XV19 or Hy19.3) (27, 94, 96), two groups solved crystal structures of type II NKT TCR complexes (97, 98). Though the lipid antigens are slightly different (lysosulfatide versus sulfatide, the former of which contains a single sphingosine-derived acyl chain), the overall structures are nearly identical. In contrast to the α-chain biased docking of iNKT TCRs, the type II NKT TCRs equally utilize the α and β chains, situated over the A′ pocket of CD1d at a roughly perpendicular docking angle, and therefore overall more similar to classical αβ TCR-pMHC complexes (2, 99) (Figs 2, 3A). Furthermore, unlike the iNKT TCR-lipid-CD1d structures, junctional residues of the CDR3β loop contact the sulfatide/lysosulfatide head group, typical of more adaptive-like modalities (Figure 3B). However, given that only the head group of sulfatide/lysosulfatide is exposed from the CD1d binding groove and thus affording a small available surface for TCR recognition, only the β chain makes contact with the lipid antigen, an important distinction from classical TCR-pMHC structures in which larger exposed peptide surface is generally contacted by both the α and β chain CDR3 loops.
Though only two type II NKT TCR complex structures have been determined, it is unlikely that human type II TCRs would share such a conserved docking mode within and across species, as do type I NKT TCRs. Most obviously, Type II NKT cells utilize multiple α and β chains, and thus potential CD1d–lipid contacting CDR loop residues are more diverse. Additionally, several of the murine CD1d residues contacted by the XV19 TCR are non-synonymous in human CD1d (Figure 4B), and in fact mutating murine residues to the human counterpart markedly reduced reactivity (98). This contrasts with the iNKT TCR system, in which many of the key CD1d energetic hotpots are identical between species (100) (Fig. 4A). Lastly, the lipid repertoire of human and mouse Type II NKT TCRs may be quite different. Though sulfatide was identified as a ligand for human αβ TCRs, the particular CD1 restriction was not identified, and sulfatide can in fact be presented by the CD1a–c molecules (24, 26). When human peripheral blood T cells were stained with CD1d–sulfatide tetramers directly ex vivo without any prior stimulation that could potentially introduce repertoire bias, few αβ TCR+ cells were identified, but rather γδ lineage cells expressing a Vδ1+ TCR were found (101). It is unclear why human and mouse T cells would segregate recognition of certain lipids to distinct T-cell lineages. The basis of human γδ TCR-CD1d recognition will be discussed below.
CD1d recognition by human and mouse γδ T cells
γδ T cells are an enigmatic T-cell population abundant in epithelial tissue sites, but notably less prevalent in the thymus, spleen, and lymph nodes, which have served as a major basis of immunological studies. They comprise less than 5% of CD3+ thymocytes in adult humans and mice, but this in fact because a major developmental window of γδ T cells occurs early in fetal ontogeny, with a subsequent decline in thymic γδ T-cell output post-birth, both in mice and humans (102–104). It is clear that γδ T cells are not focused towards pMHC recognition despite early efforts to identify such a phenomenon (105–107). A diverse array of γδ T-cell ligands has been uncovered instead, leading to the suggestion that the γδ TCR may be suited for more ‘antibody-like’ recognition (108, 109). However, many MHC-like and nonclassical molecules have also been identified as γδ TCR ligands, several of which present antigen akin to the MHC and MHC-like molecules recognized by αβ T cells. These include CD1d as mentioned above, which is also one of the few γδ TCR ligands identified in both humans and mice (101, 110, 111).
Two recent crystal structures of human Vδ1+ TCRs in complex with CD1d–lipids (αGalCer and sulfatide) have illuminated how γδ TCRs may recognize antigen in the context of a presenting molecule, akin to modes of αβ TCR recognition. These structures have been comprehensively reviewed elsewhere (112–114). In both structures, the δ chain is either preferentially or exclusively used to contact the CD1d–lipid surface, which coincides with the enrichment of Vδ1 TCRs among CD1d–specific γδ T cells and absence of an apparent Vγ chain preference. The CDR1δ loops are important for recognition of the CD1d α2-helix, with variable contribution of the CDR2δ loop. Furthermore, residues of the CDR3 loops are important for lipid head group recognition, though either the γ or δ chain appear capable of imparting the particular lipid specificity (Fig. 5). Though the Vδ1-encoded residues are important for contacting the CD1d α-helices in both structures, their interactions with CD1d are quite distinct. The majority of described CD1d–specific γδ T cells utilize the Vδ1 chain, and thus use a ‘semi-invariant TCR’ (101). Other semi-invariant innate-like T-cell populations, like mucosal associated invariant T (MAIT) cells recognizing the MHC-related-1 (MR1) protein, discussed in detail later, adopt remarkably conserved binding modes when comparing the aggregate structural database. These perplexing features and dissimilarity of the two γδ TCR-CD1d structures indicate that this may not be an evolutionarily conserved germline-specified interaction mode. In both γδ TCR-CD1d–lipid structures, many of the important CD1d contacts are completely different in mouse CD1d, and thus murine CD1d–specific γδ TCRs may dock differently despite some homology of the mouse Vδ6 and human Vδ1 chains (Fig. 4C). Thus a cross-species conserved ligand surface (either CD1d or MR1) likely underlies the nearly immutable docking of iNKT and MAIT TCRs within and across species (Fig. 4A ,D), and the lack thereof for γδ TCR-CD1d interfaces may reflect that these are not ‘hard-wired’ interactions. The relation of human Vδ1+ T cell CD1d recognition to biological function remains to be determined, though overall these TCRs are autoreactive and can be identified as a previously activated state when isolated from human blood and intestinal tissue, similar to the autoreactive type I and type II NKT cell subsets described previously.
Fig. 5. Differential interaction modes of human γδ TCRs with CD1d–lipid complexes.
(A) Footprints of the DP10.7 (PDB ID: 4mng) and 9C2 TCRs (PDB: 4lhu) upon CD1d–lipid complexes. The two complex structures were aligned via CD1d to show the different orientation of the CDR loops upon the CD1d surface. CD1d is shown as a white surface. The 9C2 TCR CDR loops are shown in light green (γ) or dark green (δ), and the corresponding lipid recognized by this TCR, αGalCer, is shown in aquamarine sticks. The DP10.7 TCR CDR loops are shown in pink (γ) and purple (δ), and the corresponding lipid contacted, sulfatide, is shown in pink. The rough borders of each of the TCR footprints in depicted by a shaded box for the 9C2 (box in green) and DP10.7 TCR (purple). For both TCRs, loops that do not contact CD1d–lipid are shown as transparent. (B) Detail of the 9C2 TCR-αGalCer interface. The CDR3γ loop is shown in green, and residues that contact αGalCer are labeled and shown as sticks. Below, the CDR3γ amino acid sequence is depicted, with residues that contact αGalCer underlined in black. (C) Detail of the DP10.7 TCR sulfatide interface. The CDR3δ loop is shown in purple, and residues that contact αGalCer are labeled and shown as sticks. Below, the CDR3δ amino acid sequence is depicted, with residues that contact αGalCer underlined in black. For (B) and (C), hydrogen bonds are shown as dash lined, and salt-bridge interactions are highlighted by showing the charge of the involved residues/moieties.
T-cell recognition of group 1 CD1 molecules
T cells responding to the group 1 CD1d molecules CD1a–c were described several years before the group 2 (CD1d)-restricted NKT cells (16, 18), yet their functions and mechanisms of antigen recognition remain more elusive, largely to their exclusivity to humans and certain other non-muroid mammals. The development of CD1-humanized mice has demonstrated a protective role for CD1-restricted cells in mycobacterial responses, which behave more similarly to adaptive-like pMHC-specific αβ T cells in terms of response kinetics (115). Similarly to CD1d, group 1 CD1 family members appear to present both self and non-self-lipid antigens to diverse human T-cell subsets of both the αβ and γδ lineages (30, 116).
Structural details of group 1 recognition have been described only very recently. Crystal structures of a CD1a–specific TCR clone bound to self-lipids (lysophosphatidylcholine or endogenous mammalian cell lipids, likely a single fatty acid) revealed an orthogonal docking over the A′ roof of the TCR (117) (Fig. 6A). Surprisingly, the TCR did not contact the lipid antigen itself, but other experiments indicated this TCR bound most potently to “permissive” lipids that did not obstruct the A′ roof interactions (Fig. 6B). It is unclear whether this docking mode is characteristic among CD1a–specific TCRs, especially among a population of CD1a–restricted lipopeptide (dideoxymycobactin)-specific T cells, in which the very bulky peptide head group is required for recognition, contrasting with its likely ability to block binding of the autoreactive CD1a–specific clone described above (40, 118) (Fig. 6C). Additionally, mutagenesis studies indicate likely diverse binding modes of CD1c–restricted αβ TCRs (119). These diverse TCRs preferentially recognize CD1c bound to the mycobacterial phosphomycoketide lipids, and suggest docking of the TCR directly over the central lipid portal of CD1c, in contrast to the A′ pocket-focused CD1a–specific and type II NKT TCRs. However, the footprints were not uniform, indicating that these TCRs do not exhibit a uniform lock-and-key docking mode like iNKT TCRS.
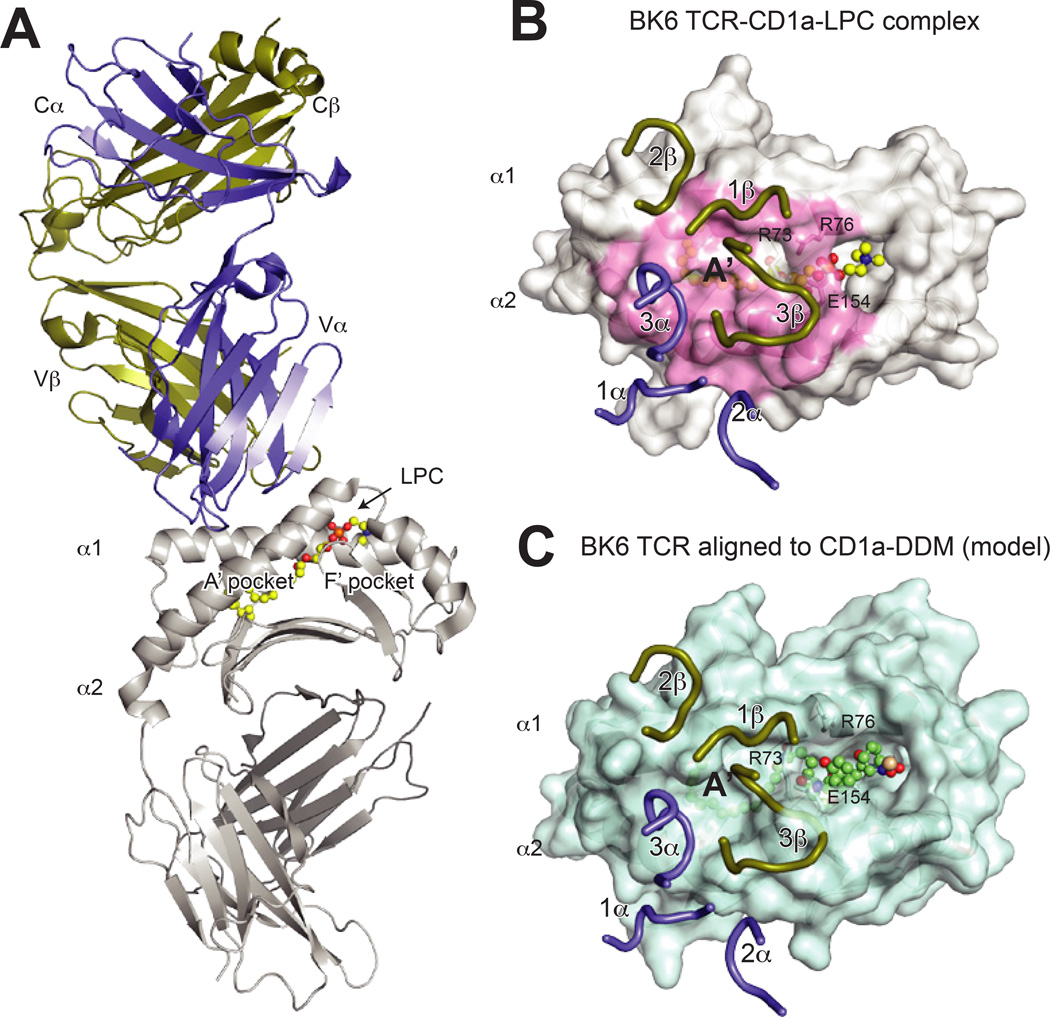
Fig. 6. Structure of the CD1a–LPC-BKT αβ and implications for lipopeptide recognition.
(A) Overall structure of the BK TCR-CD1a–lysophophatidylcholine (LPC) complex (PDB 4×6c). The TCR is shown in purple (α chain) and gold (β chain), CD1a is shown in grey and LPC is shown in yellow ball and sticks. (B) Surface view of the BK6 TCR CDR loops upon the CD1a–LPC surface. The TCR chains are colored as above, and the CD1a surface contacted by the TCR is colored in pink. Notably, the lipid head group is not contacted by the CDR loops, which instead are positioned over the A′ roof of CD1a. The residues R73, R76 and E154, which form this roof via a salt-bridge network, are indicated. (C) Alignment of the BK6-CD1a complex (aligned via CD1a) to the CD1a- didehydroxymycobactin (DDM) complex (PDB ID: 1xz0). Shown is the CD1a–DDM surface with BK6 TCR CDR loops positioned as for the CD1a–LPC structure. The larger head-group of DDM disrupts the CD1a A′ roof formed by R73, R76 and E154, and also repositions additional α helical residues, which would clash with TCR CDR loops. As a result, the contact interface as in the CD1a–LPC complex would be altered, implying that CD1a–specific TCRs specific for larger lipid species would need to undergo CDR loop conformational changes or adopt different binding modes.
Recognition of vitamin-metabolites by MR1-restricted T cells
MR1 recognition by MAIT cells
MR1 is an MHC-like protein that is widely conserved across most mammalian species with exceptionally high amino acid conservation and very low intra-species variation (120, 121). In humans, MR1 is genetically encoded near the CD1 locus on Chromosome 1, whereas in mouse, the syntenic region has been split (122, 123) between Chromosome 3, where CD1d is encoded and Chromosome 1, where MR1 is encoded (124). MR1 was shown to be the ligand for a conserved, semi-invariant population of T cells called MAIT cells (125, 126) due to their expression of a semi-invariant TCR and their location in mucosal tissues. MAIT cells have an effector-memory phenotype and typically express CD161 and CD26 (127, 128), are double negative or express the CD8αα co-receptor, but are most known for the expression of a semi-invariant TCR frequently composed of a Vα7.2 (TRAV1–2)/Jα33 α-chain paired with either Vβ13 (TRBV6) or Vβ2 (TRBV20) β-chains (125). More recently, using a MR1 tetramer-based approach or antigen-specific stimulation and cell sorting, the MAIT TCR repertoire was shown to include additional Jα and Vβ regions (129, 130).
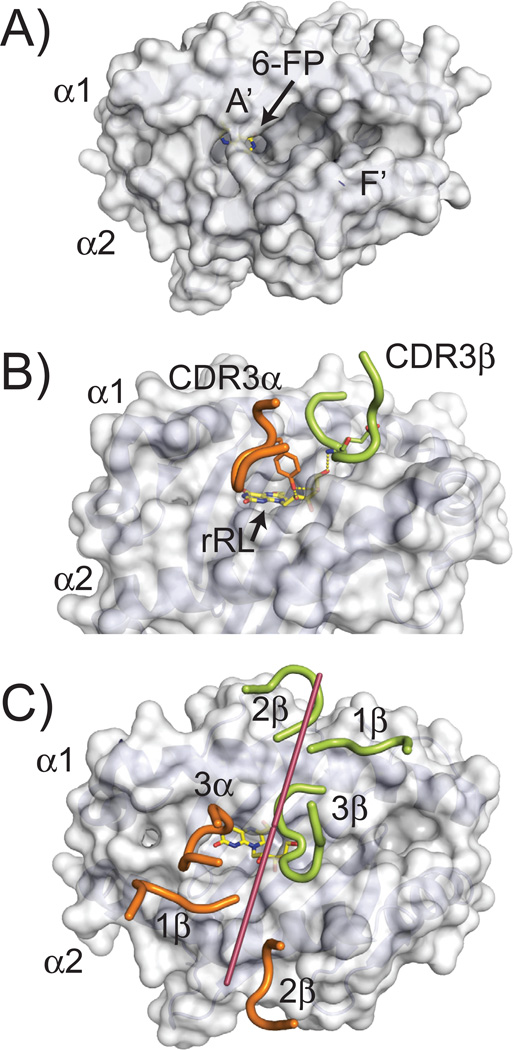
The initial structure of MR1 provided insight into the class of ligands that are likely presented by this class I-like protein. To date, two crystal structures of MR1 [human and cow (131, 132)] have been defined, and both have highly similar structures with identical cavity architectures that have Cα backbone structures most closely related to the classical class I molecules. Despite this similarity, MR1’s putative antigen-presenting site is much smaller, composed of two main pockets, termed A′ and F′, due to their similarity in location to the A′ and F′ cavities in CD1 molecules (Fig. 7A). The A′ cavity is lined with aromatic and basic amino acid residues, restricting its size and providing an overall positive charge to the pocket. Identified within the A′ of the original human structure, was a derivative of folic acid, 6-FP, which was attached via a covalent Schiff base to a lysine at position 43 within the cavity (131). Electron density consistent with 6-FP, or a related derivative, was also observed in the bovine MR1 structure (132). 6-FP, upon exogenous addition to MR1 expressing cells, has been shown to upregulate MR1 expression on the cell surface, however it is non-stimulatory to MAIT cells (131).
Fig. 7. MR1 ligand presentation and recognition by MAIT TCRs.
(A) Surface representation of human MR1 (PDB ID: 4GUP) showing the cavity structure of its antigen binding groove. Two main cavities are apparent, called A′ and F′. The 6-FP ligand identified in the crystal structure, is barely visible, colored in yellow (carbon atoms) and blue (nitrogen atoms). (B) Positioning of the CDR3α and CDR3β loops of a MAIT TCR in the complex structure with MR1 presenting the rRL ligand (PDB ID: 4LCC). The residues in these loops that contact the rRL ligand are shown, Y95 for the CDR3α loop and E99 for CDR3β. Hydrogen bonds established between these residues and the ribityl chain of the rRL ligand are shown as dashed yellow lines. (C) Orientation of the MAIT TCR CDR loops while docked onto the MR1 structure. Similar representation as to Figure 2; line was drawn (shown in raspberry) from the two external conserved cysteines in the Variable Ig domain of the MAIT TCR (C22 of α chain and C23 of β chain) to demonstrate orientation of the TCR on the MR1 surface. CDRα loops are colored in pink; CDRβ loops in green. The rRL ligand is shown as sticks in yellow (carbon atoms) and blue (nitrogen atoms).
A series of riboflavin intermediates have been shown to stimulate MAIT cells when presented by MR1 (131, 133). One of these intermediates, 6,7-dimethyl-8-D-ribityllumazine (DMRL), is the direct precursor to riboflavin in the biosynthetic pathway, whereas others are variants formed upon complexation of DMRL’s precursor, 5-amino-ribityl uracil (5-A-RU), with metabolic adducts (rRL-6-CH2OH (reduced 6-hydroxymethyl-8-D-ribityllumazine), 6-methyl-7-hydroxy-8-D-ribityllumazine (RL-6-Me-7-OH), 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil (5-OE-RU), and 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU)). 5-OP-RU is an unstable transition-state of rRL-6-CH2OH and 5-OE-RU a variant of 5-OP-RU; these transitory molecules are proposed to be captured by MR1 through Schiff-base attachment with the K43 residue of MR1 (shown to complex with 6-FP) and presented to MAIT cells in a stimulatory fashion (133). The stimulatory small molecules all possess a ribityl chain that extends out of the MR1 binding cavity in such a way to be engaged by the MAIT TCR. The CDR3 α and β loops are directly positioned over the opening to the A′ cavity and several complex structures with MAIT TCRs have shown engagement of the ribityl chains from the above-mentioned stimulatory compounds by residues within the CDR3 α and β loops (Fig. 7B). To date, riboflavin derivatives have been the only MR1-presented, molecular family that have been defined as stimulatory for MAIT cells, although MAIT cells expressing different MAIT TCRs demonstrate selective reactivity to a range of pathogens (130). This is highly suggestive that other ligand families exist and that the diversity characterized in the MAIT TCR repertoire is used in this ligand discrimination.
The docking architecture of all MAIT TCR/MR1 complexes to date has been almost completely conserved (133–135), including a xeno-reactive complex between bovine MR1 and a human MAIT TCR (132). The MAIT TCR engages MR1 in a diagonal docking orientation reminiscent of conventional αβ TCR/MHCp interactions, where the β chain is positioned over the α1 helix of MR1 and the α chain over the α2 helix. Both CDR3 loops are centrally positioned over the horizontal mid-line of MR1, close to the opening of the A′ and F′ cavities (Fig. 7C). The xeno-reactive complex between bovine MR1 and a human MAIT TCR may be the most informative in regards to how ‘innate-like’ this docking actually is, as there was no TCR contact with antigen in this structure. Instead, several residues that differ between human and bovine MR1 likely enhanced the affinity of the interaction, both producing cross-reactivity in functional assays (125) but also providing an enhanced interaction suitable for crystallization. Because this complex [and one using ‘humanized MR1’ (136)] was essentially identical to that of human MR1 and human MAIT TCRs, it is likely this conserved footprint represents a preferred docking orientation that positions the CDR3 α and β loops for contact with antigens extending from the opening to the A′ (and presumably F′) cavities. The striking cross-species docking similarity of MAIT TCR-MR1 complexes is highlighted the very high evolutionary conservation of MR1 residues within the interface (Fig. 4D).
MR1 molecules and MAIT cells in nonhuman/mouse species
As mentioned above, MAIT cells and MR1 molecules have been well documented in ruminants, including sheep and cows in addition to mice and humans (125, 137) (Table 1). The MR1 sequence is highly conserved between cow and human MR1, with 85% similarities in AA sequence, which likely explains both the crossreactivity of an α-human MR1 antibody with bovine MR1 (137), and the xenoreactive MAIT cell activation by MR1 (138). In contrast to CD1, where the α1 and α2 helices showed the greatest diversity between mammalian species (48), α1 and α2 helices and binding pocket are the most well conserved in MR1 (120). It is reasonable to assume that metabolites targeted for presentation by MR1 may be more conserved than lipid structures (of both self and microbial origin) targeted by CD1. Cows also express an invariant Vα19Vα33 TCRα chain similar to human MAIT TCR chains (125), which differ from human sequences by only 2 or 3 AA residues throughout CDR1, CDR2 and CDR3 (137), and presumably interact with MR1 molecules in cows. Unlike invariant T cells in frogs, however, bovine MAIT cells are found at relatively low levels in neonatal animals, only increasing after animals are approximately 3 weeks old (137), suggesting that invariant cells may be less important in ruminants immediately after birth.
Marsupials also express MR1 molecules (120) (Table 1), and similar to α1 and α2 domain conservation between human and mouse MR1 (121, 124) and human and cow MR1 (137), marsupials and eutherian mammalian MR1 are also highly conserved, with 74–78% sequence identity to mouse and human homologs (120). Many of the differences that do exist between species are predicted to lie in peripheral regions of the α-helices, and to have side chains pointing away from the groove, rather than towards it (120). Although it has yet to be directly demonstrated that marsupial T cells actually recognize MR1/antigen, TCR segments with similar contact residues to the TRAJ33 segment used by human MAIT TCRs have also been found in marsupials (120), suggesting that T cells expressing similar TCRα gene segments may also interact with MR1 in non-placental mammalian species. This conservation, together with the presence of MR1 across multiple mammalian species also implies that the MAIT cell lineage is broadly conserved throughout mammalian evolution.
Although an orthologue of MR1 has not definitively been found in non-mammalian species (120), a similar molecule, YF1*7.1 has been identified in chickens (Table 1). This molecule has a typical architecture of an MHC class I molecule, but with a smaller binding groove that is hydrophobic, suggesting that it likely presents non-peptide ligands (139). The closest relative of chicken YF1*7.1 may be mammalian MR1, although the structures are somewhat different in that YF1*7.1 has a Loop1-β2M salt bridge likely present in classical MHC class I molecules from birds, amphibians and reptiles (139). This connection and the Loop1-Loop2 salt bridge found in in mammalian MHC class I are both predicted to be missing from MHC-like class I molecules including MR1 (139). Therefore, if YF1*7.1 is the chicken equivalent of MR1, it is much less conserved than MR1 across mammalian species. It is possible that other species have co-opted other MHC-like class I molecules to present similar metabolite-based antigens for their own subsets of invariant T cells, similar to what has been seen in frogs with the XNC family.
Innate-like T cells of unknown ligand specificity: γδ T cells in non-human and non-mouse models
All jawed vertebrates have both TCR α, β, γ and δ gene segments as well as conventional polymorphic, MHC class I and II molecules. αβ TCRs likely bind to classical MHC molecules in similar ways across species (82), however there is larger variation in the structure, sequence and repertoire of γδ TCRs and nonclassical molecules; little is known about these T-cell subsets in other species. Although γδ T-cell populations are notoriously different between species, with very little overlap between the mouse and human systems, and even a surprising diversity within primate species (140), T cells expressing a γδ TCR are found in all jawed vertebrates [except some reptiles (141)] (Table 1) and are thus likely playing an important role in the immune system of all animals with a traditional adaptive immune system. In light of recent structural insights into the binding of TCRs from non-canonical T cells, including γδ T cells, to MHC-like class I molecules in humans, discussed above, we examine what is known about these cells in non-human and non-mouse models. Since human iNKT and γδ TCRs can recognize CD1 molecules (101, 112, 113), it is worth speculating that a subset of non-human/mouse γδ TCRs might also be specific for the non-classical MHC and MHC-like molecules found in these species.
Frequency of γδ T cells in the periphery
Humans, mice, and dogs are all considered ‘γδ low’ animals, with low numbers of γδ T cells in peripheral blood, whereas ‘γδ high’ animals including sheep, cows, rabbits and chickens have a high proportion of peripheral γδ T cells (142, 143). Adult sheep blood contains 20–30% γδ T cells, while in young lambs it can be even higher, up to 60% (143), and γδ T cells in chickens make up 10% of thymocytes, 15% of peripheral lymphocytes and 25% of spleen cells (142). Despite this great discrepancy in γδ T-cell number in peripheral blood of different species, both γδ low and γδ high animals have high numbers of skin and mucosal tissue-resident γδ T cells. It is currently unclear as to whether there are distinct functions for blood, skin, and mucosal resident γδ T cells, although their distribution in these tissues strongly suggest there is some form of specialized surveillance for these populations. Interestingly, in sheep γδ T cells are found at much higher levels in non-wooly skin compared to wool-covered skin (143), suggesting that perhaps γδ T cells are more necessary in exposed skin that is likely to encounter a higher number of pathogens. In support of this model, sheep breeds resistant to infection with the parasite, Haemonchus contortus had higher numbers of skin γδ T cells at 28 days after infection than non-resistant breeds (144).
Ruminants could require high numbers of γδ cells to act quickly to handle the high burden of bacterial, viral, and fungal pathogens these animals are exposed to in their environment. Newborn ruminants are far more precocious than many other mammals immediately after birth, and therefore interact with their environment, and any nearby pathogens, from a very early stage. Due to a difference in placental structure, young ruminants lack the same level of passive maternal immune protection as humans and other mammals, so they rely more on their early immune systems, such as γδ T cells, for early protection of neonates. Ruminants and artiodactyls are not, however the only γδ high species, because as mentioned, rabbits, and chickens (142, 145) also have high γδ T cells as do beluga whales (Delphinapterus leucas) (146). Based on staining of γ and δ TCR transcripts in shark thymus, it is likely that γδ T cells, in general, make up a large proportion of total T cells in the shark periphery, but it is not clear if they are truly a γδ high species (147). Perhaps these other γδ high species, similar to ruminants, are also exposed to high bacterial and viral burdens early in life and would thus benefit from high γδ T numbers for early defense. This is a particularly attractive model in thinking about marine animals, since they would be challenged with water-borne bacteria and viruses virtually from birth. It is unclear, however, whether the large numbers of γδ T cells found in these animals recognize antigen in the context of MHC-like molecules such as CD1d in these species.
Genomic organization, number, and diversity of γδ TCR gene segments
From comparisons across a wide range of divergent species it is evident there is little conservation in γδ TCR gene segment organization and arrangement. In some species, the gene fragments are located in a single array, such as in humans where there exist 6 functional Vγ (TRGV) genes (of 14 total). The J and C gene segments in humans exist as 2 J-C gene clusters (148). In contrast, in the mouse genome, the 6 functional TRGV genes are arranged in 3 cassettes, with cassette 1 (TRGC1) containing 4 TRGV genes and the other two cassettes with 1 TRGV gene each. There also exists a fourth non-functional cluster (TRGC3) (149, 150). Sheep contain two completely distinct γ loci, with 13 TRGV genes between the two loci in 6 total cassettes (151), as well as two distinct Vδ gene families. Other ruminants including goats, cattle and river buffalo also contain two γ loci (151). Rabbits (Oryctolagus cuniculus) have 10 total TRGV genes, two TRGJ genes, and one TRGC gene with a loci structure more similar to humans than mice and dogs (152). In sharks, the genomic organization of the T-cell receptor locus, including the γδ genes, is organized in a normal, translocon configuration, similar to most mammalian loci (153), in contrast to the immunoglobulin locus in cartilaginous fish, which is found in a cluster organization. It is unclear whether these different gene organization patterns are the product of selection, although it is tempting to speculate that different patterns of gene segment organization could be selected to ensure a preferential rearrangement between certain gene segments, generating populations of T cells with biased TCR repertoires.
Thymic selection of γδ T cells
Development of MHC restricted T-cell populations is assumed to occur in the thymus, where antigen-presenting molecules are expressed, presumably in concert with relevant self-selecting ligands, for education of the developing T-cell populations. The presence of particular γδ T-cell populations dependent on thymic selection may imply a role of an antigen-presenting molecule in ligand recognition. In sheep, for example, γδ development is dependent on a functional thymus, with Vγ genes Vγ1 and Vγ2.1 absent from animals without a thymus, and larger rearrangement differences noted in Vδ genes. Particularly the Vδ repertoire of thymectomized sheep is quite different from normal animals and adult animals without a thymus express more fetal Vδ genes (Vδ1.12, Vδ1.13, Vδ3 and Vδ4) (154). In addition to a skewing of γδ T-cell numbers and gene usage, when thymectomy was performed prenatally, the few remaining γδ T cells functioned abnormally later in life (143). Chicken γδ T cells are highly expressed in the thymus of embryonic animals (142), and swine γδ T cells also develop in the thymus and peripheral cells are short lived after thymectomy, suggesting that γδ T cells in pigs and chickens are also thymus dependent and may therefore be selected on a thymic antigen-presenting molecule (155, 156).
The question of thymic development is also relevant to early, invariant αβ T cells. Interestingly, as discussed above, the frog has an almost completely invariant early T-cell repertoire. In the frog model, these invariant T cells with germline encoded rearrangements may be necessary early, perhaps for early/quick responses before the adaptive immune system is fully developed. One model suggested that the limited diversity would prevent autoimmune recognition in the absence of strong negative selection, since classical MHC class Ia molecules are not expressed early in these animals (86). However, invariant sequences in mammals are generally thought to encode TCRs that are specific to self-molecules, so it is hard to reconcile how these invariant repertoires necessarily protect from autoimmunity based on specificity alone. Mammalian TCRs from invariant T cells and γδ T cells with high self-reactivity may emerge from the thymus ‘pre-activated’ for similarly quick responses, but it is unclear how these cells exist without leading to autoimmune pathologies. Understanding the thymic development pathways of these unusual TCRs in non-human/mouse mammals and non-mammalian systems will help clarify the purpose of these cells in early immune responses.
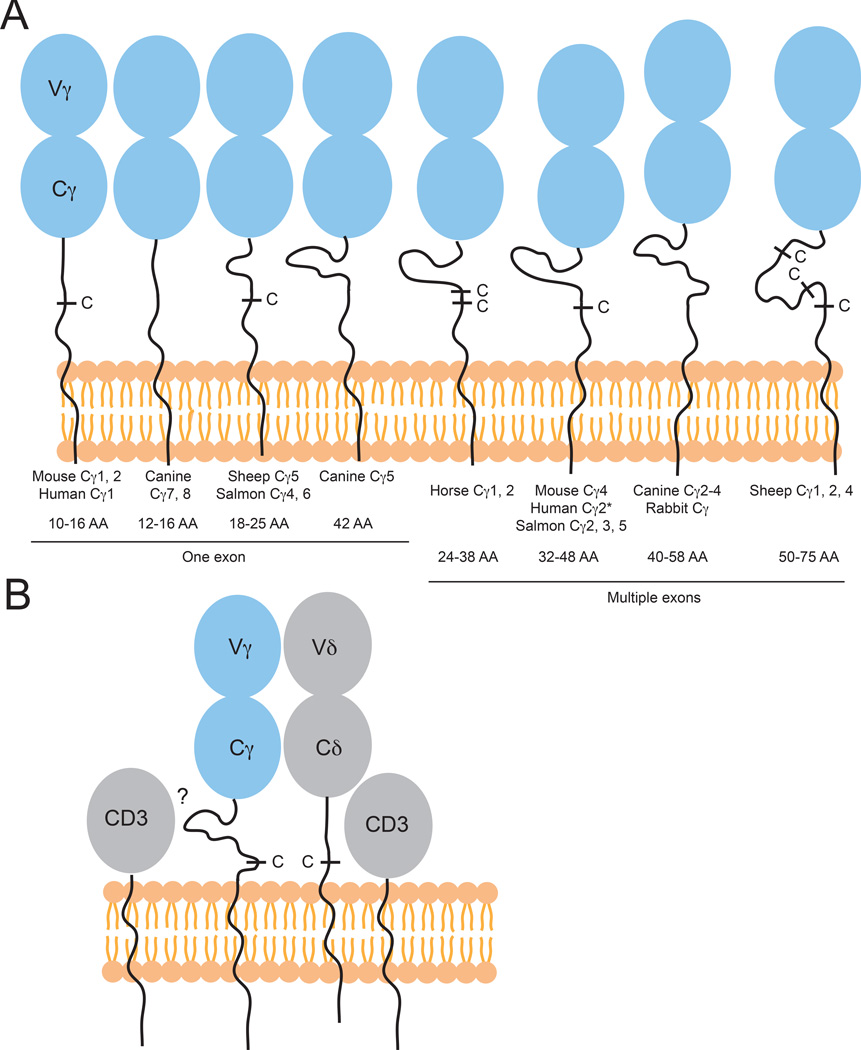
Structure of γδ TCRs: CDR3 lengths
The CDR3 loops in αβ TCRs recognizing MHCp in almost all cases have been shown to play an important role in peptide scanning. Additionally, in all of the subsequent structures of ‘non-conventional’ αβ and γδ TCRs in complex with their respective ligands, at least one of the junctionally encoded CDR3 loops have been shown to contact ligand. Furthermore, the CDR3 loops usually play central roles in antigen recognition by antibodies. Thus CDR3 loop diversity and length are often related to antigen recognition capacity. Due to the ability to use multiple Dδ segments during gene rearrangement, human Vδ CDR3 loops are long and highly diverse (157, 158) with between 8–21 amino acids (AA) on average, a length more similar to IgH genes, which average 2–25 AA. CDR3γ lengths, encoded by Vγ-Jγ gene rearrangements, are generally more similar to CDR3α/β lengths, which do not exceed 12 AA (Fig. 8). It has been suggested that, due to these long, IgVH-like CDR3δ sequences, γδ T may recognize antigen differently than αβ T cells and that they may see a wider variety of antigens than traditional MHC-presented peptides (159–161). These long CDR3δs are also found in many other species: sheep Vδ rearrangement can lead to long and diverse CDR3s, with 1–18 extra residues/rearrangement, but with shorter Vγ junctions (154), and horse (Equus caballus) CDR3δ rearrangements are also unusually long, between 1–19 AA, and almost always contain at least one glycine residue (162). Long CDR3δs are also frequently found in non-mammalian species, for example skate CDR3δs have a range of 2–13 AA (163) and nurse sharks also have longer CDR3s in TCRδ than any of the other chains (147) (Fig. 8). Use of multiple D segments in rearrangement has also been shown in other species including platypus, leading to CDR3s of 10–20 AA in length (164). Interestingly, these platypus Vδs also often encode an extra cysteine residue near the end of the V domain, which together with additional cysteines in CDR3, may provide stability to these long CDR3 loops, as is also seen in platypus IgV regions (164, 165) (Fig. 8).
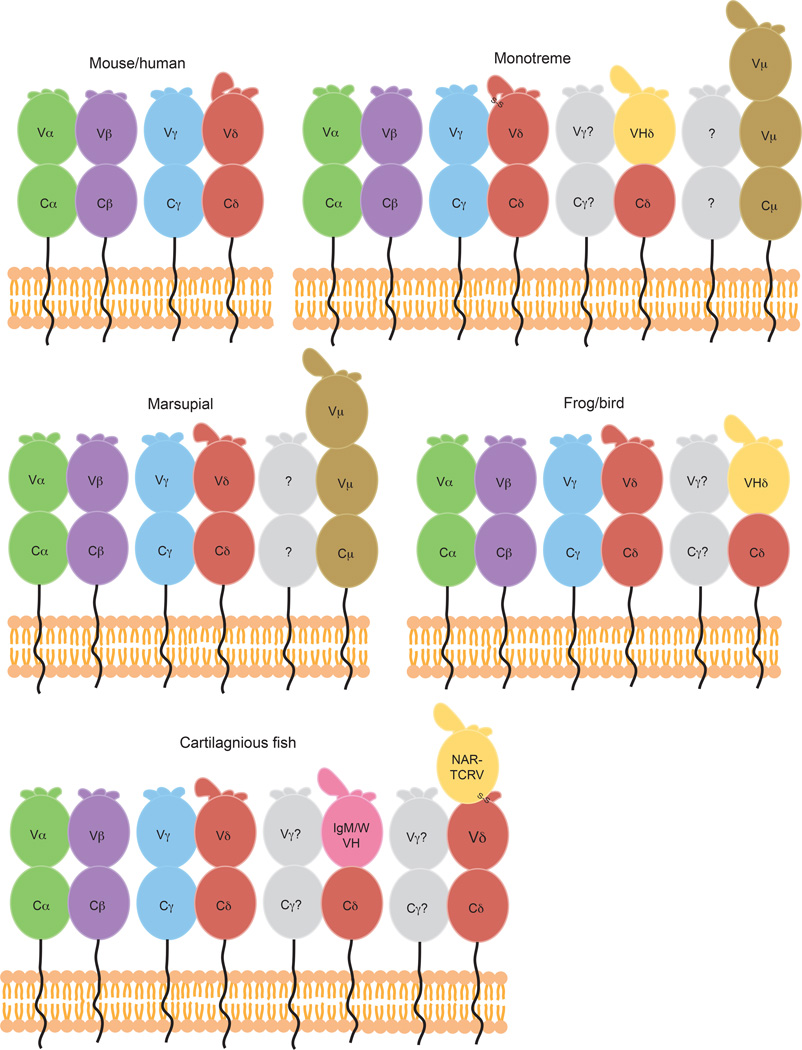
Fig. 8. Comparison of TCR chains present in diverse species.
Cartoon diagrams of the variable TCR chains present in a variety of species including mice and humans, monotremes, marsupials, frogs and birds, and cartilaginous fish. Traditional TCRα (green), β (purple), γ (blue), and δ (red) chains are found in all species. TCRδ chains with VH-like TCRVδ domains are shown in yellow, and those expressing true IgVH domains via transrearrangements in pink. Potential binding partners for unusual TCRδ chains are colored grey. Predicted structure of three-domain TCR chains, NAR- TCR and TCRµ (brown) are shown in B, C, and E, with the NAR-TCRV domain depicted yellow due to its similarity with IgNARV domains. CDR1, 2, and 3 loops depicted at the top of each TCRV domain, with larger loops representing longer CDR3s found in some Vδ domains. Predicted disulfide linkages due to extra cysteine residues in monotreme TCRδ CDR3 (B) and between terminal and supporting shark NARTCRV domains (E) are labeled.
Conventional TCR α and β chains are thought to have a restricted range of CDR3 lengths to allow them to scan the peptide antigen, while still accommodating binding to MHC framework regions. These long CDR3s identified in γδ T-cell lineage suggests that TCRδ chains are not constrained by such requirements and have often been used in arguments against γδ T-cell recognition of antigen in the context of MHC-like folds. However, structures of human γδ TCRs in complex with CD1d molecules prove that γδ TCRs with long CDR3δs are capable of binding to a traditional MHC class I-like structure (112, 113), although the binding footprints of these TCRs onto CD1d were considerably different than traditional αβ TCR/MHC docking orientations, perhaps in part because of these long CDR loops (114). Furthermore, recognition of the MHC-like molecule T22 is almost entirely dictated by the CDR3δ loop of reactive γδ T cells in the mouse, albeit in a highly ‘unconventional’ docking orientation (166). It is important to determine how these long CDR3 loops of γδ TCRs contribute to antigen binding, and what the role of having stabilized CDR3 loops, as suggested in the platypus, might be when compared to more flexible long loops. Does the stabilization of these loops make for a better binding platform for traditional, MHC-like molecules in these animals, despite their long CDR3 sequences? Or might these T cells simply recognize free antigen, as has been suggested for some human and mouse γδ cells (109, 167, 168) and is likely for certain δ chains with V regions that strongly resemble IgVH domains?
Structure of γδ TCRs: VH-like TCR Vδs
While the enhanced length and diversity of the CDR3δ loops may argue for an antibody-like recognition of a repertoire of structurally diverse antigens, early crystal structures of the Vδ domain of γδ TCRs showed that the CDR1δ and CDR3δ loop structures looked very CDR3α-like (160, 169). CDR3 loops in both Vα and Vδ are folded back towards CDR1 and 2, rather than pointing away from the center of the domain, as is seen in VH CDR3s. In contrast, the structure of human Vδ framework regions and the CDR2δ loop are more similar to IgVH than Vα. Specifically, in some cases the c′′ loop of Vδ forms hydrogen bonds with c′, as is seen in VH and VL domains (159, 169). Other species, in addition to expressing “conventional” γδ TCRs, express γδ TCRs that contain V domains with even more IgVH-like qualities.
Frogs (Xenopus tropicalis) and birds, including the zebra finch (Taeniopygia guttata) and chicken (Gallus gallus) express TCRδV domains that, based on nucleotide alignments and phylogeny, appear to be virtually indistinguishable from IgVH domains (170, 171); they are therefore referred to as ‘VHδ‘ domains (Fig. 8, Table 1). In both frogs and zebra finch, these VHδs are located within the normal α/δ locus, but in chickens and turkeys they are found in a separate locus (170). Whether or not they are linked to the α locus, these VHδs only appear to recombine upstream of Cδ, and not IgC domains. While no true VHδ-like domains have yet been found in placental mammals or in marsupials, VHδ-like domains have been described in the duckbill platypus, a mammalian monotreme (164)(Table 1); these VHδs have long CDR3s and utilize multiple D segments (Fig. 8). Therefore these alternate Vδ domains are widely distributed across vertebrate classes yet appear to be relatively rare in the mammalian lineage.
VHδs are true TCR V domains that share high levels of similarity to VH, rather than a rearrangement with the actually IgH locus. However, some species of sharks do have true trans-rearrangements with IgVH domains. Apart from being simply VH-like, bona fide IgV domains from IgM and IgW can rearrange to TCRδ and α chains in the nurse shark (147) (Fig. 8, Table 1), although the functional significance of these rearrangements is still being assessed. It has also been suggested that shark T cells may undergo antibody-like somatic hypermutation (SHM) (153), further adding to the idea of antibody-like binding in these unusual TCRs. Perhaps these antibody-like TCRδ chains are potentially more important for non-mammalian species, like birds, sharks and frogs, because of a bias away from MHC class I-like restricted T cells during evolution. Characterization of the MHC-like molecules within these species, and the T cells that respond to them, will be an important contribution to resolving this conundrum.
TCRµ and NAR-TCR
The platypus VHδ domains described above are the first example of a true VH-like TCR V domain in mammals; however other ‘unusual’ TCR domains have also been characterized in nonplacental mammals and non-mammalian species. Nonplacental mammals (monotremes and marsupials) express a fifth class of TCR chain, TCRµ, in addition to traditional α, β, γ, and δ TCR chains (172, 173) (Fig. 8, Table 1). TCRµ is made up of three, rather than two, Ig domains, including one Cµ and two Vµ domains. The terminal Vµ undergoes VDJ recombination in marsupials, whereas the second Vµ is invariant (172). In platypus, recombination occurs in both Vµ domains, although the rearrangement of the second Vµ does not involve D segments so junctionally encoded diversity is limited within this domain (173). In both species, Vµ domains are more closely related to VH than to TCR V domains (173, 174).
The first description of a three-domain TCR chain, NAR-TCR, was in shark (175) (Fig. 8, Table 1), but NAR-TCR and TCRµ likely evolved separately as a result of convergent evolution in both sharks and mammals, since the extra V domain of NAR-TCR is unrelated to TCRµ or traditional IgH chains, and instead is related to IgNAR V domains (172, 175). IgNAR is a heavy chain-only antibody isotype present in cartilaginous fish able to bind antigen via induced-fit solely through its single, terminal NAR V domain (176, 177). Unlike the IgW/M trans-rearrangements described above, NAR-TCR V gene segments are found within the δ locus, upstream of some TCRδ V and C domains (175). Both the NAR-TCR V, and the supporting Vδ, are diverse and undergo double rearrangement during V-D-J recombination. NAR-TCR transcripts makeup roughly half of the TCRδ amplification products from shark spleen, thymus and PBL, suggesting they are found at high levels in these tissues and are thus likely playing an important role in the shark immune response (175).
Although there are no crystal structures of either NAR-TCR or TCRµ, the extra V domain is predicted to be unpaired, and free to bind antigen in an antibody-like fashion, with the second Vδ or Vµ domains playing mostly a structural and supportive role rather than functioning in an antigen binding capacity (173, 175) (Fig. 8). Both NAR-TCR V and the supporting shark Vδ domains contain an extra cysteine residue, presumably for stabilizing the interface between the two domains (175)(Fig. 8). Interestingly, the frog VHδ domains described above also possess an extra, unpaired cysteine, in a similar location but without a third domain present it is unclear what the role of it might be in antigen binding (171). No three-domain TCRγ chains have been identified in sharks or marsupials, strengthening the idea that both NAR-TCRV and the terminal Vµ may bind antigen as a single-domain (174, 175). Supporting V and C domains likely pair with a normal TCRγ chain, but exactly which Vγ chains are capable of interaction with these unusual TCRs is unclear (Fig. 8). Understanding the γ chain pairing and structure of these three-domain TCRδ chains will be instrumental in understanding the role of the extra domain in antigen binding.
It has frequently been suggested that some γδ TCRs may be able to recognize antigen in an Ig-like manner, directly, and outside of the context of presenting molecules (167). While there is evidence for this in several models, for example binding of γδ TCRs to PE (168), and the Vγ9Vδ2 T-cell population that respond to phosphoantigens (178) clearly this is not the case for all human γδ TCRs, since some have been shown to bind to MHC-like antigen-presenting molecules (112, 113, 166). However the presence of these unusual TCR chains including VHδ in frogs, birds and nonplacental mammals, NAR-TCRδ and trans-rearranged Ig/TCRs in sharks, and Vµ in nonplacental mammals may support the model that these TCRs bind free antigen in an antibody-like manner, although this remains to be shown definitively (170). Rather than binding free-floating antigen in a strictly antibody-dependent fashion, however, these unusual TCRs may see unprocessed or presented antigen in a cell-surface dependent manner, for example, by binding budding virus from infected cells or antigen bound to parasites (175). VH-like TCRs have not been described in placental mammals, and so it is interesting to speculate about the role of Ig-like TCRs and the mode of antigen binding by γδ TCRs in these animals and how it might relate to the γδ T-cell compartment in humans and mice.
Structure of γδ TCRs: γδ TCR C domains
TCR binding and signaling involves more than simply docking to MHC, MHC-like, or nonclassical class I MHC, and/or free antigen, as it also involves TCR constant (C) domains, CD3 molecules and downstream cytoplasmic signals. TCR C genes contain exons for not only the Ig fold, but also the cytoplasmic tail, transmembrane region and connecting piece/hinge between the cell surface and IgC fold, all of which contribute to interactions with CD3 molecules and downstream signaling molecules. These regions are yet another important evolutionary adaptation relevant to the concept of TCR function and are thus reviewed briefly here. In general, the connecting piece is short in both human and mouse TCR C domains; however, the connecting piece of the human Cγ TRG-C2 is longer than the others and can contain either two or three exons of 16 AA each, depending on the population (179) (Figure 9). Mice also possess one Cγ with a slightly longer connecting piece, TRG-C4 (149), but while these long connecting pieces are relatively rare in mice and humans, they are found frequently in other species (Figure 9).
Fig. 9. High variability of Cγ connecting pieces both within and across species.
(A) Differences in length and features of the connecting piece/hinge of TCRγ chains in multiple species. Vγ and Cγ domains are shown in blue, with plasma membrane in orange. Examples are roughly organized from shortest to longest connecting pieces, with ranges of amino acid lengths and exon usage displayed beneath each gene name. Presence of cysteine resides marked with C. (*) Indicates allelic difference in human Cγ2 of either 2 or 3 exons. (B) Cartoon showing hypothetical regions of interaction between the TCRγ connecting piece and CD3.
Dogs have six functional Cγ genes, with variations in the length of exon 1 as well as differences is the number of exons making up the connecting piece (either one or two) (180). The rabbit connecting piece is comprised of two 20 AA exons (152), and horses have two Cγ genes of different lengths, TRGC1 and 2. Of these, TRGC1 is longer with 18 more AA in the connecting piece than TRGC2 (162). Atlantic salmon (Salmo salar) have six Cγ domains, more than most other animals, and each has a unique connecting piece and may play different roles in downstream responses (181). Sheep have 5 different Cγ genes, each also with a unique connecting piece ranging from 25–75 AA (154, 182), and cows can use unusually long connecting pieces (27–55 AA) as well (183).
The long Cγ connecting pieces in ruminants can include two additional cysteines, not normally present in Cγ domains, C140 and C156 (184), which may either form tertiary structures or interesting bonds with other cell surface receptors (Fig. 9A). Artiodactyl connecting pieces often also contain a TTE(K)PP motif, which is some cases is repeated two or even three times as in several pig and cow genes. Neither human nor mouse Cγ genes have this motif (184). It is currently unknown how these long connecting pieces influence the way TCR chains interact with each other as well as other cell-surface molecules. However, due to their location, they could potentially influence anything from pairing with the δ chain and CD3 binding, perpetuation of TCR signals, to tissue homing (Fig. 9B). In contrast to this extreme variability of Cγ, the connecting region within the Cδ genes are relatively more conserved in these species (182). This provides an interesting dichotomy, where this conservation of the Cδ region contrasts with the potential high diversity of the Vδ domain due to multiple Dδ segment usage during V-D-J rearrangement. Perhaps this could indicate a bias of Vδ towards antigen binding while the partner γ chain contributes more to V region pairing and stabilization, while Cγ domain variation may modulate interactions with signaling molecules via the diverse connecting pieces. In this way Cγ domains could function to modulate T-cell signaling and potentially alter cellular effector functions.
In addition to the diversity of connecting pieces, there are several other variations in γδ C domains between species. Rabbit constant regions are also shorter at 109 amino acids, with a short 12 amino acid FG loop (152), which is midway between the very short, 7 AA, loop in the structure of human γδ TCR, and the 19 AA-long loop found in human Cβ chains (185). In αβ T cells, this loop is important for development, thymic selection, and T-cell function (186, 187), and has recently been shown to be important for sustained αβ TCR/MHC bonds during TCR/MHCp interactions and T-cell signaling (188). It has also been suggested to be important for CD3ɛ binding, suggesting that invariant CD3 chains may bind differently to TCRs with short FG loops (189). Nurse sharks are missing one of the two canonical Ig-domain stabilizing cysteines in the TCR Cα domain; this intra-domain di-sulfide is broadly conserved in most Ig domains and aids in stabilizing the core Ig domain through connection of the two beta sheets. While absent in the nurse shark TCR Cα, it is present in the TCR Cγ, δ, and β chains (147). It has been noted that mammalian TCR Cα domains, while containing both conserved cysteines, are less compact and more flexible than other Ig folds (12), and that this flexibility may be important for signal transduction. While it is tempting to speculate that the loss of this cysteine in the nurse shark may endow this domain with a different type of structural flexibility, it is unclear how this would extend to alternative functioning of the nurse shark TCR Cα. The variety of differences in the γδ C regions of different species may suggest different binding or activation requirements in different animals, but without a better understanding of both the structure of these unusual TCRs, and their true antigens it will be difficult to fully realize the meaning of these variations.
In all species Vδ chains are highly diverse and contain longer CDR3 chains than TCRα, β or γ. Non-mammalian species and non-placental mammals including sharks, frogs, birds, opossums, and platypus can express unusual versions of δ chains, VHδ, NAR-TCR, or TCRµ, all of which may be likely to bind free antigen in an antibody-like manner. In contrast, γ chains often have more ‘normal’ CDR3 lengths and domain structures, but show a greater diversity in Cγ chains between and within species, with several non-human and non-murine species having a variety of diverse connecting pieces, which could presumably affect pairing with the TCRδ chain or CD3 molecules and downstream signaling or effector functions.
Summary of innate-like T-cell recognition: implications for evolutionary considerations
T cells, collectively, share similarities in structure of their of their TCR, the components that contribute to signal transduction and many of their effector functions, yet the antigen pool from which they survey their environment is now recognized as being incredibly diverse. We have reviewed above those T-cell populations where antigen recognition has been well studied, including recognition of peptides presented in the context of classical MHC, lipid in the context of CD1 molecules and small molecule metabolites presented by MR1 molecules. Within these categories there are examples of varying degrees of conservation of recognition, such as with the general diagonal docking orientation with conventional αβ TCRs recognizing pMHC, or the strict docking of iNKT and MAIT cells to their respective ligands CD1d and MR1. We also see, or suspect, divergent recognition schemes, such as with recognition of CD1a and CD1c, both of which appear to be recognized by T cells with diverse TCRs, likely using different footprints to probe the recognition surface, and Vδ1 γδ TCR recognition of CD1d. The conservation of CD1 and MR1 molecules in divergent species, and the presence of non-classical/MHC-like proteins in less-well characterized species suggest there are many T-cell recognition systems present throughout vertebrate evolution and we are likely only viewing the tip of the iceberg with our studies of human and mouse. Outside of the T-cell populations that are restricted to recognition of antigen within the context of an antigen-presenting molecule are those that may recognize antigen directly. Evidence for this exists for some γδ T cells in both human and mice directly recognizing antigen in an antibody-like fashion (104) whereas the structural adaptations that have occurred in TCRs of more divergent species, such as VHδ, NAR-TCR, or TCRµ suggest these domains may be specialized to recognize free antigen, with no prerequisite of antigen processing or presentation.
Thus, the coevolution of the TCR and antigen is challenging to dissect; did TCRs originally evolve as membrane-associated antibody-like structures, with the diversity inherent in their receptors focused on direct recognition of diverse antigens? Or did the TCR structure originally evolve as a sensor of self-molecules, with antigen presentation of variable ligands (peptides, lipids, metabolites, etc.), and recognition of free antigen coming later? Other fascinating questions revolve around the almost uniform presence of both αβ and γδ across all vertebrate species [even analogous populations in the jawless fishes (190)], with the γδ lineage more prone to alterations in TCR structure and antigen recognition. Which of these lineages came first, αβ or γδ, and how did they become specialized in this way? Important steps towards answering these questions include better characterization of these lineages in divergent species and a clearer understanding of the antigens these T-cell populations respond to (both MHC and non-MHC related). Furthermore, exploring how T cells signal in these different populations and across species will also add to our understanding of T-cell evolution; is there a single conserved way that T cells signal, such as a clear formation of an immunological synapse? How would this function in T cells that recognize free antigen? Do they signal with a more B cell-like strategy? These and related questions are central to understanding not only the evolution of T cells, but also their place in the evolution of the adaptive immune system.
Acknowledgements
This work was supported by the National Institutes of Health grants to E. J. Adams: R01_AI073922 and R01_AI115471.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Garcia KC, Adams EJ. How the T cell receptor sees antigen--a structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 3.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 7.Van Laethem F, et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell. 2013;154:1326–1341. doi: 10.1016/j.cell.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Tikhonova AN, et al. alphabeta T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity. 2012;36:79–91. doi: 10.1016/j.immuni.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins EJ, Riddle DS. TCR-MHC docking orientation: natural selection, or thymic selection? Immunol Res. 2008;41:267–294. doi: 10.1007/s12026-008-8040-2. [DOI] [PubMed] [Google Scholar]
- 11.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 12.Garcia KC, et al. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 13.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 14.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 15.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 16.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 1989;341:447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 17.Balk SP, et al. Oligoclonal expansion and CD1 recognition by human intestinal intraepithelial lymphocytes. Science. 1991;253:1411–1415. doi: 10.1126/science.1716785. [DOI] [PubMed] [Google Scholar]
- 18.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4-8- T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 19.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 21.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 22.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 23.Scharf L, et al. The 2.5 a structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shamshiev A, Donda A, Carena I, Mori L, Kappos L, De Libero G. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29:1667–1675. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 25.Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 26.Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nat Rev Immunol. 2001;1:177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 29.Brossay L, et al. CD1d–mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams EJ. Lipid presentation by human CD1 molecules and the diverse T cell populations that respond to them. Current opinion in immunology. 2014;26:1–6. doi: 10.1016/j.coi.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 32.Facciotti F, et al. Fine tuning by human CD1e of lipid-specific immune responses. Proc Natl Acad Sci U S A. 2011;108:14228–14233. doi: 10.1073/pnas.1108809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Libero G, Mori L. Novel insights into lipid antigen presentation. Trends Immunol. 2012;33:103–111. doi: 10.1016/j.it.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol. 2013;31:529–561. doi: 10.1146/annurev-immunol-032712-095912. [DOI] [PubMed] [Google Scholar]
- 35.Kawano T, et al. CD1d–restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 36.Burdin N, et al. Selective ability of mouse CD1 to present glycolipids: alpha-galactosylceramide specifically stimulates V alpha 14+ NK T lymphocytes. Journal of immunology. 1998;161:3271–3281. [PubMed] [Google Scholar]
- 37.Berzins SP, Ritchie DS. Natural killer T cells: drivers or passengers in preventing human disease? Nat Rev Immunol. 2014;14:640–646. doi: 10.1038/nri3725. [DOI] [PubMed] [Google Scholar]
- 38.Salio M, Silk JD, Cerundolo V. Recent advances in processing and presentation of CD1 bound lipid antigens. Current opinion in immunology. 2010;22:81–88. doi: 10.1016/j.coi.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Ly D, Moody DB. The CD1 size problem: lipid antigens, ligands, and scaffolds. Cellular and molecular life sciences : CMLS. 2014;71:3069–3079. doi: 10.1007/s00018-014-1603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zajonc DM, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22:209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Batuwangala T, et al. The crystal structure of human CD1b with a bound bacterial glycolipid. Journal of immunology. 2004;172:2382–2388. doi: 10.4049/jimmunol.172.4.2382. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Alles LF, et al. Structural reorganization of the antigen-binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proc Natl Acad Sci U S A. 2011;108:17755–17760. doi: 10.1073/pnas.1110118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Alles LF, et al. Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc Natl Acad Sci U S A. 2011;108:13230–13235. doi: 10.1073/pnas.1105627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dascher CC. Evolutionary biology of CD1. Current topics in microbiology and immunology. 2006;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- 45.Salomonsen J, et al. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proceedings of the National Academy of Sciences of the United States of America. 2005 doi: 10.1073/pnas.0409213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller MM, et al. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proceedings of the National Academy of Sciences of the United States of America. 2005 doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, et al. Analysis of the reptile CD1 genes: evolutionary implications. Immunogenetics. 2015 doi: 10.1007/s00251-015-0837-2. [DOI] [PubMed] [Google Scholar]
- 48.Dossa RG, Alperin DC, Hines MT, Hines SA. The equine CD1 gene family is the largest and most diverse yet identified. Immunogenetics. 2014;66:33–42. doi: 10.1007/s00251-013-0741-6. [DOI] [PubMed] [Google Scholar]
- 49.Van Rhijn I, et al. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. Journal of immunology (Baltimore, Md : 1950) 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen TK, et al. The bovine CD1D gene has an unusual gene structure and is expressed but cannot present α-galactosylceramide with a C26 fatty acid. International immunology. 2012;25:91–98. doi: 10.1093/intimm/dxs092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Guillaume J, Pauwels N, Van Calenbergh S, Van Rhijn I, Zajonc DM. Crystal structures of bovine CD1d reveal altered αGalCer presentation and a restricted A’ pocket unable to bind long-chain glycolipids. PloS one. 2012:7. doi: 10.1371/journal.pone.0047989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Girardi E, et al. Crystal structure of bovine CD1b3 with endogenously bound ligands. Journal of immunology (Baltimore, Md : 1950) 2010;185:376–386. doi: 10.4049/jimmunol.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker ML, Miller RD. Evolution of mammalian CD1: marsupial CD1 is not orthologous to the eutherian isoforms and is a pseudogene in the opossum Monodelphis domestica. Immunology. 2007 doi: 10.1111/j.1365-2567.2007.02545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zajonc DM, Striegl H, Dascher CC, Wilson IA. The crystal structure of avian CD1 reveals a smaller, more primordial antigen-binding pocket compared to mammalian CD1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17925–17930. doi: 10.1073/pnas.0809814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dvir H, Wang J, Ly N, Dascher CC, Zajonc DM. Structural basis for lipid-antigen recognition in avian immunity. Journal of immunology (Baltimore, Md : 1950) 2010;184:2504–2511. doi: 10.4049/jimmunol.0903509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dellabona P, et al. In vivo persistence of expanded clones specific for bacterial antigens within the human T cell receptor alpha/beta CD4-8- subset. J Exp Med. 1993;177:1763–1771. doi: 10.1084/jem.177.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180:1763–1771. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Valpha24+ CD4-CD8- T cells. J Exp Med. 1997;186:109–120. doi: 10.1084/jem.186.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morita M, et al. Structure-activity relationship of alpha-galactosylceramides against B16-bearing mice. Journal of medicinal chemistry. 1995;38:2176–2187. doi: 10.1021/jm00012a018. [DOI] [PubMed] [Google Scholar]
- 61.Wieland Brown LC, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS biology. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kain L, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian alpha-linked glycosylceramides. Immunity. 2014;41:543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson BL, Teyton L, Bendelac A, Savage PB. Stimulation of natural killer T cells by glycolipids. Molecules. 2013;18:15662–15688. doi: 10.3390/molecules181215662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gapin L, Godfrey DI, Rossjohn J. Natural Killer T cell obsession with self-antigens. Current opinion in immunology. 2013;25:168–173. doi: 10.1016/j.coi.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 66.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kjer-Nielsen L, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Borg NA, et al. CD1d–lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 69.Girardi E, Zajonc DM. Molecular basis of lipid antigen presentation by CD1d and recognition by natural killer T cells. Immunological reviews. 2012;250:167–179. doi: 10.1111/j.1600-065X.2012.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams EJ, Lopez-Sagaseta J. The immutable recognition of CD1d. Immunity. 2011;34:281–283. doi: 10.1016/j.immuni.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 71.Pellicci DG, et al. Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12:827–833. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zajonc DM, Savage PB, Bendelac A, Wilson IA, Teyton L. Crystal structures of mouse CD1d–iGb3 complex and its cognate Valpha14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. Journal of molecular biology. 2008;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu ED, Girardi E, Wang J, Zajonc DM. Cutting edge: structural basis for the recognition of beta-linked glycolipid antigens by invariant NKT cells. Journal of immunology. 2011;187:2079–2083. doi: 10.4049/jimmunol.1101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 75.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 76.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 77.Wang J, et al. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci U S A. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, et al. The V{alpha}14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med. 2010 doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berzins SP, Smyth MJ, Baxter AG. Presumed guilty: natural killer T cell defects and human disease. Nat Rev Immunol. 2011;11:131–142. doi: 10.1038/nri2904. [DOI] [PubMed] [Google Scholar]
- 80.Nobutaka Y, Kenichi M, Tohishiro T, Annabelle T, Yasuyuki I. Identification of canine natural CD3-positive T cells expressing an invariant T-cell receptor alpha chain. Veterinary Immunology and Immunopathology. 2009 doi: 10.1016/j.vetimm.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 81.Looringh van Beeck FA, et al. Functional CD1d and/or NKT cell invariant chain transcript in horse, pig, African elephant and guinea pig, but not in ruminants. Molecular immunology. 2009;46:1424–1431. doi: 10.1016/j.molimm.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scott-Browne JP, Crawford F, Young MH, Kappler JW, Marrack P, Gapin L. Evolutionarily conserved features contribute to αβ T cell receptor specificity. Immunity. 2011;35:526–535. doi: 10.1016/j.immuni.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edholm E-SS, et al. Nonclassical MHC class I-dependent invariant T cells are evolutionarily conserved and prominent from early development in amphibians. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14342–14347. doi: 10.1073/pnas.1309840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flajnik MF, Kasahara M, Shum BP, Salter-Cid L, Taylor E, Du Pasquier L. A novel type of class I gene organization in vertebrates: a large family of non-MHC-linked class I genes is expressed at the RNA level in the amphibian Xenopus. EMBO J. 1993;12:4385–4396. doi: 10.1002/j.1460-2075.1993.tb06123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goyos A, Sowa J, Ohta Y, Robert J. Remarkable conservation of distinct nonclassical MHC class I lineages in divergent amphibian species. Journal of immunology (Baltimore, Md : 1950) 2010 doi: 10.4049/jimmunol.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edholm ES, Goyos A, Taran J, De Jesus Andino F, Ohta Y, Robert J. Unusual evolutionary conservation and further species-specific adaptations of a large family of nonclassical MHC class Ib genes across different degrees of genome ploidy in the amphibian subfamily Xenopodinae. Immunogenetics. 2014;66:411–426. doi: 10.1007/s00251-014-0774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flajnik MF, Kaufman JF, Hsu E, Manes M, Parisot R, Du Pasquier L. Major histocompatibility complex-encoded class I molecules are absent in immunologically competent Xenopus before metamorphosis. Journal of immunology. 1986;137:3891–3899. [PubMed] [Google Scholar]
- 88.Bartl S, Baish MA, Flajnik MF, Ohta Y. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. Journal of immunology (Baltimore, Md : 1950) 2003;159:6097–6104. [PubMed] [Google Scholar]
- 89.Wang C, Perera TV, Ford HL, Dascher CC. Characterization of a divergent non-classical MHC class I gene in sharks. Immunogenetics. 2003;55:57–61. doi: 10.1007/s00251-003-0542-4. [DOI] [PubMed] [Google Scholar]
- 90.Chiu YH, et al. Distinct subsets of CD1d–restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med. 1999;189:103–110. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park SH, Weiss A, Benlagha K, Kyin T, Teyton L, Bendelac A. The mouse CD1d–restricted repertoire is dominated by a few autoreactive T cell receptor families. J Exp Med. 2001;193:893–904. doi: 10.1084/jem.193.8.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 93.Brossay L, Tangri S, Bix M, Cardell S, Locksley R, Kronenberg M. Mouse CD1-autoreactive T cells have diverse patterns of reactivity to CD1+ targets. Journal of immunology. 1993;160:3681–3688. [PubMed] [Google Scholar]
- 94.Jahng A, Maricic I, Aguilera C, Cardell S, Halder RC, Kumar V. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d–reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Behar SM, Podrebarac TA, Roy CJ, Wang CR, Brenner MB. Diverse TCRs recognize murine CD1. Journal of immunology. 1999;162:161–167. [PubMed] [Google Scholar]
- 96.Blomqvist M, et al. Multiple tissue-specific isoforms of sulfatide activate CD1d–restricted type II NKT cells. Eur J Immunol. 2009;39:1726–1735. doi: 10.1002/eji.200839001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patel O, et al. Recognition of CD1d–sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol. 2012;13:857–863. doi: 10.1038/ni.2372. [DOI] [PubMed] [Google Scholar]
- 98.Girardi E, et al. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat Immunol. 2012;13:851–856. doi: 10.1038/ni.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang JH, Reinherz EL. The structural basis of alphabeta T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunological reviews. 2012;250:102–119. doi: 10.1111/j.1600-065X.2012.01161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wun KS, et al. A minimal binding footprint on CD1d–glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bai L, et al. The majority of CD1d–sulfatide-specific T cells in human blood use a semiinvariant Vdelta1 TCR. Eur J Immunol. 2012;42:2505–2510. doi: 10.1002/eji.201242531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McVay LD, Carding SR. Generation of human gammadelta T-cell repertoires. Crit Rev Immunol. 1999;19:431–460. [PubMed] [Google Scholar]
- 103.Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 104.Chien YH, Meyer C, Bonneville M. gammadelta T Cells: First Line of Defense and Beyond. Annu Rev Immunol. 2014;32:121–155. doi: 10.1146/annurev-immunol-032713-120216. [DOI] [PubMed] [Google Scholar]
- 105.Matis LA, Cron R, Bluestone JA. Major histocompatibility complex-linked specificity of gamma delta receptor-bearing T lymphocytes. Nature. 1987;330:262–264. doi: 10.1038/330262a0. [DOI] [PubMed] [Google Scholar]
- 106.Bluestone JA, Cron RQ, Cotterman M, Houlden BA, Matis LA. Structure and specificity of T cell receptor gamma/delta on major histocompatibility complex antigen-specific CD3+, CD4-, CD8- T lymphocytes. J Exp Med. 1988;168:1899–1916. doi: 10.1084/jem.168.5.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matis LA, Fry AM, Cron RQ, Cotterman MM, Dick RF, Bluestone JA. Structure and specificity of a class II MHC alloreactive gamma delta T cell receptor heterodimer. Science. 1989;245:746–749. doi: 10.1126/science.2528206. [DOI] [PubMed] [Google Scholar]
- 108.Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Born WK, Kemal Aydintug M, O’Brien RL. Diversity of gammadelta T-cell antigens. Cell Mol Immunol. 2013;10:13–20. doi: 10.1038/cmi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dieude M, et al. Cardiolipin binds to CD1d and stimulates CD1d–restricted gammadelta T cells in the normal murine repertoire. J Immunol. 2011;186:4771–4781. doi: 10.4049/jimmunol.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mangan BA, et al. Cutting edge: CD1d restriction and Th1/Th2/Th17 cytokine secretion by human Vdelta3 T cells. J Immunol. 2013;191:30–34. doi: 10.4049/jimmunol.1300121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luoma AM, et al. Crystal structure of Vdelta1 T cell receptor in complex with CD1d–sulfatide shows MHC-like recognition of a self-lipid by human gammadelta T cells. Immunity. 2013;39:1032–1042. doi: 10.1016/j.immuni.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Uldrich AP, et al. CD1d–lipid antigen recognition by the gammadelta TCR. Nat Immunol. 2013;14:1137–1145. doi: 10.1038/ni.2713. [DOI] [PubMed] [Google Scholar]
- 114.Luoma AM, Castro CD, Adams EJ. gammadelta T cell surveillance via CD1 molecules. Trends Immunol. 2014;35:613–621. doi: 10.1016/j.it.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Felio K, et al. CD1-restricted adaptive immune responses to Mycobacteria in human group 1 CD1 transgenic mice. J Exp Med. 2009;206:2497–2509. doi: 10.1084/jem.20090898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Lalla C, et al. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur J Immunol. 2011;41:602–610. doi: 10.1002/eji.201041211. [DOI] [PubMed] [Google Scholar]
- 117.Birkinshaw RW, et al. alphabeta T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol. 2015;16:258–266. doi: 10.1038/ni.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kasmar AG, et al. Cutting Edge: CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. Journal of immunology. 2013;191:4499–4503. doi: 10.4049/jimmunol.1301660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roy S, et al. Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alphabeta T cells. Proc Natl Acad Sci U S A. 2014;111:E4648–E4657. doi: 10.1073/pnas.1408549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsukamoto K, Deakin JE, Graves JA, Hashimoto K. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics. 2013;65:115–124. doi: 10.1007/s00251-012-0666-5. [DOI] [PubMed] [Google Scholar]
- 121.Yamaguchi H, Hirai M, Kurosawa Y, Hashimoto K. A highly conserved major histocompatibility complex class I-related gene in mammals. Biochemical and biophysical research communications. 1997;238:697–702. doi: 10.1006/bbrc.1997.7379. [DOI] [PubMed] [Google Scholar]
- 122.Albertson DG, Fishpool R, Sherrington P, Nacheva E, Milstein C. Sensitive and high resolution in situ hybridization to human chromosomes using biotin labelled probes: assignment of the human thymocyte CD1 antigen genes to chromosome 1. EMBO J. 1988;7:2801–2805. doi: 10.1002/j.1460-2075.1988.tb03135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moseley WS, Seldin MF. Definition of mouse chromosome 1 and 3 gene linkage groups that are conserved on human chromosome 1: evidence that a conserved linkage group spans the centromere of human chromosome 1. Genomics. 1989;5:899–905. doi: 10.1016/0888-7543(89)90132-8. [DOI] [PubMed] [Google Scholar]
- 124.Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. Journal of immunology (Baltimore, Md : 1950) 1998;161:4066–4077. [PubMed] [Google Scholar]
- 125.Tilloy F, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 127.Martin E, et al. Stepwise development of MAIT cells in mouse and human. PLoS biology. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sharma PK, et al. High expression of CD26 accurately identifies human bacteria-reactive MR1-restricted MAIT cells. Immunology. 2015 doi: 10.1111/imm.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Reantragoon R, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. 2013;210:2305–2320. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gold MC, et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. 2014;211:1601–1610. doi: 10.1084/jem.20140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 132.Lopez-Sagaseta J, et al. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E1771–E1778. doi: 10.1073/pnas.1222678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Corbett AJ, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 134.Eckle SB, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med. 2014;211:1585–1600. doi: 10.1084/jem.20140484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Patel O, et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nature communications. 2013;4:2142. doi: 10.1038/ncomms3142. [DOI] [PubMed] [Google Scholar]
- 136.Lopez-Sagaseta J, Dulberger CL, McFedries A, Cushman M, Saghatelian A, Adams EJ. MAIT recognition of a stimulatory bacterial antigen bound to MR1. Journal of immunology. 2013;191:5268–5277. doi: 10.4049/jimmunol.1301958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Goldfinch N, Reinink P, Connelley T, Koets A, Morrison I, Van Rhijn I. Conservation of mucosal associated invariant T (MAIT) cells and the MR1 restriction element in ruminants, and abundance of MAIT cells in spleen. Vet Res. 2010;41:62. doi: 10.1051/vetres/2010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang S, et al. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8290, 8295. doi: 10.1073/pnas.0903196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hee CS, et al. Structure of a Classical MHC Class I Molecule That Binds “Non-Classical” Ligands. PLoS biology. 2010 doi: 10.1371/journal.pbio.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kazen AR, Adams EJ. Evolution of the V, D, and J gene segments used in the primate gammadelta T-cell receptor reveals a dichotomy of conservation and diversity. Proc Natl Acad Sci U S A. 2011;108:E332–E340. doi: 10.1073/pnas.1105105108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Olivieri DN, von Haeften B, Sánchez-Espinel C, Faro J, Gambón-Deza F. Genomic V exons from whole genome shotgun data in reptiles. Immunogenetics. 2014;66:479–492. doi: 10.1007/s00251-014-0784-3. [DOI] [PubMed] [Google Scholar]
- 142.Sowder JT, Chen CL, Ager LL, Chan MM, Cooper MD. A large subpopulation of avian T cells express a homologue of the mammalian T gamma/delta receptor. J Exp Med. 1988;167:315–322. doi: 10.1084/jem.167.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hein WR, Dudler L, Morris B. Differential peripheral expansion and in vivo antigen reactivity of alpha/beta and gamma/delta T cells emigrating from the early fetal lamb thymus. Eur J Immunol. 1990;20:1805–1813. doi: 10.1002/eji.1830200827. [DOI] [PubMed] [Google Scholar]
- 144.Gonzalez JF, et al. Fecundity in adult Haemonchus contortus parasites is correlated with abomasal tissue eosinophils and gammadelta T cells in resistant Canaria Hair Breed sheep. Veterinary parasitology. 2011;178:286–292. doi: 10.1016/j.vetpar.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 145.Sawasdikosol S, et al. Selection of rabbit CD4- CD8- T cell receptor-gamma/delta cells by in vitro transformation with human T lymphotropic virus-I. J Exp Med. 1993;178:1337–1345. doi: 10.1084/jem.178.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.De Guise S, Bernier J, Martineau D, Béland P, Fournier M. Phenotyping of beluga whale blood lymphocytes using monoclonal antibodies. Developmental and comparative immunology. 1996;21:425–433. doi: 10.1016/s0145-305x(97)00021-9. [DOI] [PubMed] [Google Scholar]
- 147.Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. Journal of immunology (Baltimore, Md : 1950) 2010;184:6950–6960. doi: 10.4049/jimmunol.0902774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lefranc MP, et al. Molecular mapping of the human T cell receptor gamma (TRG) genes and linkage of the variable and constant regions. Eur J Immunol. 1989;19:989–994. doi: 10.1002/eji.1830190606. [DOI] [PubMed] [Google Scholar]
- 149.Vernooij BT, Lenstra JA, Wang K, Hood L. Organization of the murine T-cell receptor gamma locus. Genomics. 1993;17:566–574. doi: 10.1006/geno.1993.1373. [DOI] [PubMed] [Google Scholar]
- 150.Raulet DH. The Structure, Function, and Molecular Genetics of the gamma/delta T Cell Receptor. Annual review of immunology. 1989 doi: 10.1146/annurev.iy.07.040189.001135. [DOI] [PubMed] [Google Scholar]
- 151.Massari S, Lipsi MR, Vonghia G, Antonacci R, Ciccarese S. T-cell receptor TCRG1 and TCRG2 clusters map separately in two different regions of sheep chromosome 4. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 1998;6:419–420. doi: 10.1023/a:1009245830804. [DOI] [PubMed] [Google Scholar]
- 152.Massari S, Ciccarese S, Antonacci R. Structural and comparative analysis of the T cell receptor gamma (TRG) locus in Oryctolagus cuniculus. Immunogenetics. 2012;64:773–779. doi: 10.1007/s00251-012-0634-0. [DOI] [PubMed] [Google Scholar]
- 153.Chen H, et al. Characterization of arrangement and expression of the T cell receptor gamma locus in the sandbar shark. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8591–8596. doi: 10.1073/pnas.0811283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hein WR, Dudler L. Divergent evolution of T cell repertoires: extensive diversity and developmentally regulated expression of the sheep gamma delta T cell receptor. The EMBO journal. 1993;12:715–724. doi: 10.1002/j.1460-2075.1993.tb05705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Binns RM, Pabst R, Licence ST. Subpopulations of T lymphocytes emigrating in venous blood draining pig thymus labelled in vivo with fluorochrome. Immunology. 1988;63:261–267. [PMC free article] [PubMed] [Google Scholar]
- 156.Binns RM, Pallares V, Symons DB, Sibbons P. Effect of thymectomy on lymphocyte subpopulations in the pig Demonstration of a thymus-dependent ‘null’ cell. International archives of allergy and applied immunology. 1977;55:96–101. doi: 10.1159/000231915. [DOI] [PubMed] [Google Scholar]
- 157.Allison JP, Havran WL. The Immunobiology of T Cells with Invariant gammadelta Antigen Receptors. Annual review of immunology. 1991 doi: 10.1146/annurev.iy.09.040191.003335. [DOI] [PubMed] [Google Scholar]
- 158.Lafaille JJ, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S. Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell. 1989;59:859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- 159.Hayday AC. [gamma][delta] cells: a right time and a right place for a conserved third way of protection. Annu Rev Immunol. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 160.Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Arevalo JH, Hassig CA, Stura EA, Sims MJ, Taussig MJ, Wilson IA. Structural analysis of antibody specificity. Detailed comparison of five Fab’-steroid complexes. Journal of molecular biology. 1994;241:663–690. doi: 10.1006/jmbi.1994.1543. [DOI] [PubMed] [Google Scholar]
- 162.Schrenzel MD, Ferrick DA. Horse (Equus caballus) T-cell receptor alpha, gamma, and delta chain genes: nucleotide sequences and tissue-specific gene expression. Immunogenetics. 1995 doi: 10.1007/BF00178585. [DOI] [PubMed] [Google Scholar]
- 163.Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW. alpha, beta, gamma, and delta T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 164.Parra ZE, Lillie M, Miller RD. A model for the evolution of the mammalian t-cell receptor alpha/delta and mu loci based on evidence from the duckbill Platypus. Molecular biology and evolution. 2012;29:3205–3214. doi: 10.1093/molbev/mss128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Johansson J, Aveskogh M, Munday B, Hellman L. Heavy chain V region diversity in the duck-billed platypus (Ornithorhynchus anatinus): long and highly variable complementarity-determining region 3 compensates for limited germline diversity. Journal of immunology (Baltimore, Md : 1950) 2002;168:5155–5162. doi: 10.4049/jimmunol.168.10.5155. [DOI] [PubMed] [Google Scholar]
- 166.Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. doi: 10.1126/science.1106885. [DOI] [PubMed] [Google Scholar]
- 167.Chien YH, Jores R, Crowley MP. Recognition by gamma/delta T cells. Annual review of immunology. 1995;14:511–532. doi: 10.1146/annurev.immunol.14.1.511. [DOI] [PubMed] [Google Scholar]
- 168.Zeng X, et al. γδ T cells recognize a microbial encoded B cell antigen to initiate a rapid antigen-specific interleukin-17 response. Immunity. 2012;37:524–534. doi: 10.1016/j.immuni.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li H, Lebedeva MI, Llera AS, Fields BA, Brenner MB, Mariuzza RA. Structure of the Vdelta domain of a human gammadelta T-cell antigen receptor. Nature. 1998;391:502–506. doi: 10.1038/35172. [DOI] [PubMed] [Google Scholar]
- 170.Parra ZE, Mitchell K, Dalloul RA, Miller RD. A second TCRδ locus in Galliformes uses antibody-like V domains: insight into the evolution of TCRδ and TCRµ genes in tetrapods. Journal of immunology (Baltimore, Md : 1950) 2012;188:3912–3919. doi: 10.4049/jimmunol.1103521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Parra ZE, Ohta Y, Criscitiello MF, Flajnik MF, Miller RD. The dynamic TCRδ: TCRδ chains in the amphibian Xenopus tropicalis utilize antibody-like V genes. Eur J Immunol. 2010;40:2319–2329. doi: 10.1002/eji.201040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Parra ZE, Baker ML, Schwarz RS, Deakin JE, Lindblad-Toh K, Miller RD. A unique T cell receptor discovered in marsupials. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9776–9781. doi: 10.1073/pnas.0609106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang X, Parra ZE, Miller RD. Platypus TCRµ provides insight into the origins and evolution of a uniquely mammalian TCR locus. Journal of immunology (Baltimore, Md : 1950) 2011;187:5246–5254. doi: 10.4049/jimmunol.1101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Parra ZE, et al. Comparative genomic analysis and evolution of the T cell receptor loci in the opossum Monodelphis domestica. BMC genomics. 2008;9:111. doi: 10.1186/1471-2164-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Criscitiello MF, Saltis M, Flajnik MF. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5036–5041. doi: 10.1073/pnas.0507074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 177.Stanfield RL, Dooley H, Verdino P, Flajnik MF, Wilson IA. Maturation of shark single-domain (IgNAR) antibodies: evidence for induced-fit binding. Journal of molecular biology. 2007;367:358–372. doi: 10.1016/j.jmb.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 178.Gu S, Nawrocka W, Adams EJ. Sensing of Pyrophosphate Metabolites by Vgamma9Vdelta2 T Cells. Front Immunol. 2014;5:688. doi: 10.3389/fimmu.2014.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Buresi C, Ghanem N, Huck S, Lefranc G, Lefranc MP. Exon duplication and triplication in the human T-cell receptor gamma constant region genes and RFLP in French, Lebanese, Tunisian, and black African populations. Immunogenetics. 1988;29:161–172. doi: 10.1007/BF00373641. [DOI] [PubMed] [Google Scholar]
- 180.Massari S, et al. The deduced structure of the T cell receptor gamma locus in Canis lupus familiaris. Molecular immunology. 2009;46:2728–2736. doi: 10.1016/j.molimm.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 181.Yazawa R, et al. Functional adaptive diversity of the Atlantic salmon T-cell receptor gamma locus. Molecular immunology. 2008;45:2150–2157. doi: 10.1016/j.molimm.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 182.Hein WR, Dudler L, Marcuz A, Grossberger D. Molecular cloning of sheep T cell receptor gamma and delta chain constant regions: unusual primary structure of gamma chain hinge segments. Eur J Immunol. 1990;20:1795–1804. doi: 10.1002/eji.1830200826. [DOI] [PubMed] [Google Scholar]
- 183.Takeuchi N, Ishiguro N, Shinagawa M. Molecular cloning and sequence analysis of bovine T-cell receptor gamma and delta chain genes. Immunogenetics. 1991;35:89–96. doi: 10.1007/BF00189517. [DOI] [PubMed] [Google Scholar]
- 184.Ciccarese S, Lanave C, Saccone C. Evolution of T-cell receptor gamma and delta constant region and other T-cell-related proteins in the human-rodent-artiodactyl triplet. Genetics. 1997;145:409–419. doi: 10.1093/genetics/145.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Allison TJ, Winter CC, Fournié J-J, Bonneville M, Garboczi DN. Structure of a human γδ T-cell antigen receptor. Nature. 2001 doi: 10.1038/35081115. [DOI] [PubMed] [Google Scholar]
- 186.Sasada T, Touma M, Chang H-C, Clayton LK, Wang J-h, Reinherz EL. Involvement of the TCR Cβ FG Loop in Thymic Selection and T Cell Function. J Exp Med. 2002 doi: 10.1084/jem.20020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Touma M, Chang H-CC, Sasada T, Handley M, Clayton LK, Reinherz EL. The TCR C beta FG loop regulates alpha beta T cell development. Journal of immunology (Baltimore, Md : 1950) 2006;176:6812–6823. doi: 10.4049/jimmunol.176.11.6812. [DOI] [PubMed] [Google Scholar]
- 188.Das DK, et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proceedings of the National Academy of Sciences. 2015 doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang J, et al. Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody. The EMBO journal. 1998;17:10–26. doi: 10.1093/emboj/17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Das S, Li J, Hirano M, Sutoh Y, Herrin BR, Cooper MD. Evolution of two prototypic T cell lineages. Cell Immunol. 2015 doi: 10.1016/j.cellimm.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]