Abstract
Genome-wide association studies (GWAS) have been a standard practice in identifying single nucleotide polymorphisms (SNPs) for disease susceptibility. We propose a new approach, termed integrative GWAS (iGWAS) that exploits the information of gene expressions to investigate the mechanisms of the association of SNPs with a disease phenotype, and to incorporate the family-based design for genetic association studies. Specifically, the relations among SNPs, gene expression, and disease are modeled within the mediation analysis framework, which allows us to disentangle the genetic effect on a disease phenotype into two parts: an effect mediated through a gene expression (mediation effect, ME) and an effect through other biological mechanisms or environment-mediated mechanisms (alternative effect, AE). We develop omnibus tests for the ME and AE that are robust to underlying true disease models. Numerical studies show that the iGWAS approach is able to facilitate discovering genetic association mechanisms, and outperforms the SNP-only method for testing genetic associations. We conduct a family-based iGWAS of childhood asthma that integrates genetic and genomic data. The iGWAS approach identifies six novel susceptibility genes (MANEA, MRPL53, LYCAT, ST8SIA4, NDFIP1, and PTCH1) using the omnibus test with false discovery rate less than 1%, whereas no gene using SNP-only analyses survives with the same cut-off. The iGWAS analyses further characterize that genetic effects of these genes are mostly mediated through their gene expressions. In summary, the iGWAS approach provides a new analytic framework to investigate the mechanism of genetic etiology, and identifies novel susceptibility genes of childhood asthma that were biologically meaningful.
Keywords: childhood asthma, integrative genomics, genome-wide association studies, mediation analysis, variance component tests
Introduction
Given the rapid increase of the available data on genetic variants, genome-wide association studies (GWAS) have been a common practice for investigating the associations of single nucleotide polymorphisms (SNPs) with complex diseases [Amos et al., 2008; Ferreira et al., 2008; Hung et al., 2008; Moffatt et al., 2007; Thorgeirsson et al., 2008; Wallace et al., 2008]. The success of these studies in numerous discoveries of new disease susceptibility loci further popularized the usage of GWAS, which, on the other hand, also incurred challenges and limitations. A well-acknowledged limitation is its agnostic style [Hunter and Chanock, 2010]: none of the biological knowledge was encoded in standard GWAS analyses. Multimarker analyses have been advocated to integrate the biological information into statistical learning and to decrease the number of tests. Successful examples are multiple SNP analyses or SNP-set analyses [Kwee et al., 2008; Liu et al., 2008; Wu et al., 2010] that have been shown to have better performance than the standard single SNP analyses in reanalyzing breast cancer and Alzheimer’s disease GWAS datasets [Cruchaga et al., 2014;Wu et al., 2010].
To move beyond GWAS for disease risk, gene expression can also be construed as a molecular phenotypic trait. Such a type of genetic association studies focusing on identifying SNPs that are associated with gene expression or namely expression quantitative trait loci (eQTL) are so-called eQTL studies. As both GWAS and eQTL studies become popular in genetic research, considerable interest emerges in integrating the two. Studies that substantiate the notion have been conducted in many diseases, including asthma [Cusanovich et al., 2012; Moffatt et al., 2007], osteoporosis [Hsu et al., 2010], type 2 diabetes [Zhong et al., 2010], skin cancer [Zhang et al., 2012], glioblastoma, and Crohn’s disease [Xiong et al., 2012]. These studies consider SNP-disease and SNP-expression associations separately. For example, the most commonly used two-stage approach is to identify the top GWAS SNPs that are also eQTL SNPs. This type of analyses was supported by the evidence that trait-associated SNPs are more likely to be functional and thus eQTL [Nicolae et al., 2010]. The role of gene expression in the biological process from SNPs to disease phenotype remains unclear. Even though eQTL studies may be helpful in reranking or filtering the top SNPs to enrich true positives, whether the association of those eQTL SNPs with gene expression can be translated into a mechanistic contribution to the disease risk is rarely addressed.
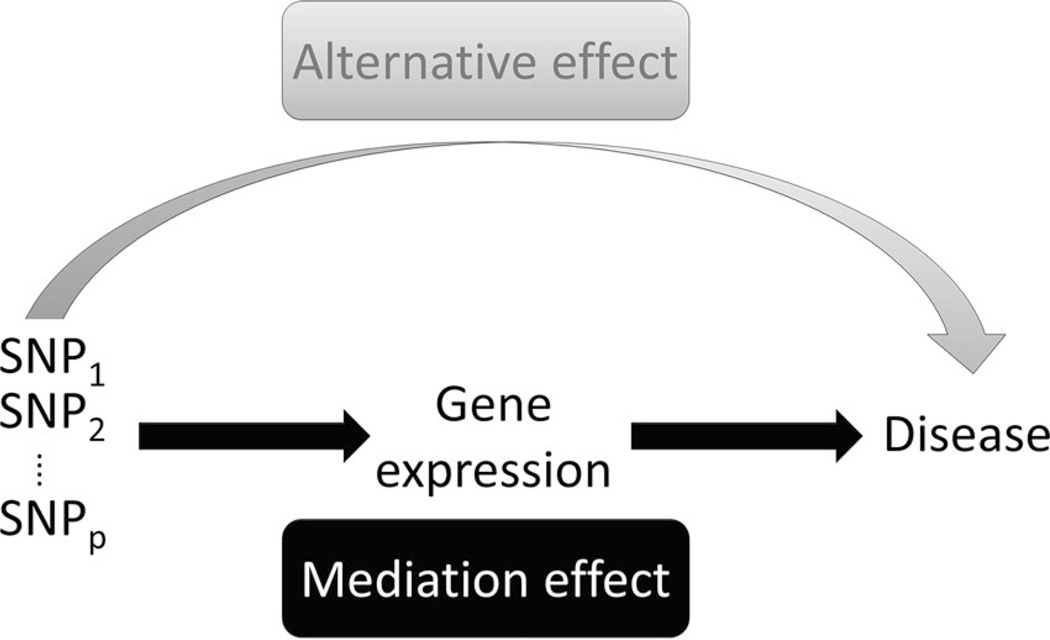
This article ismotivated by a family-based GWAS of childhood asthma, in which the association between SNPs at the ORMDL3 gene and the risk of childhood asthma was discovered in an MRCA (Medical Research Council Asthma) dataset, a family-based case-control study, and validated in many other datasets [Moffatt et al., 2007]. In the MRCA study, mRNA expression data were also collected, and it was reported that SNPs at the ORMDL3 gene were highly associated with its expression value in an eQTL study [Dixon et al., 2007;Moffatt et al., 2007]. The association of the GWAS SNPs and expression of the ORMDL3 gene has also been validated in a molecular study [Berlivet et al., 2012]. Based on these work, we propose an integrative approach to conduct GWAS, termed iGWAS (integrative GWAS) where we jointly analyze SNPs and gene expression on disease risk as a biological process, illustrated by a mediation model (Fig. 1). Moreover, we are interested in studying whether the effect of genes on asthma risk is mediated through gene expression, or though alternative biological mechanisms.
Figure 1.
Directed acyclic diagram (DAG) of the mediation model. The gray path indicates the alternative effect, and the black path indicates the mediation effect.
The iGWAS approach is developed within the framework of causal mediation modeling [MacKinnon, 2008; Robins and Greenland, 1992] using counterfactuals [Rubin, 1978]. The model can be illustrated as a directed acyclic graph (DAG) [Robins, 2003], which provides intuitive interpretation of how SNPs and gene expression coordinate to influence on the development of diseases (Fig. 1). Rather than focusing on agnostic associations, the iGWAS approach considers the coordinated biological process from genetics to gene transcription and then to disease outcome. In particular, we decompose the etiological mechanism for the total genetic effect (TE) into the genetic effect on disease risk mediated through gene expression (mediation effect, ME) and the genetic effect through other biological pathways or environmental risk factors (alternative effect, AE). We developed previously a testing procedure for the TE of SNPs and gene expression in population-based case-control studies [Huang et al., 2014]. However, this method focuses only on the TE.
In this paper, we are primarily interested in examining ME and AE. The characterization of the ME and AE is critical in understanding the etiological mechanisms of genetic effects, and will in turn assist in generating new hypotheses that are more biologically plausible. Inclusion of gene expression data can also better explain the heterogeneity of the human genome that genetic data alone are not able to capture and thus increase statistical power of identifying disease susceptibility genes. Furthermore, the existing method for TE was developed under population-based studies that study subjects are independent, and failed to accommodate the family design of the MRCA data. To bridge these gaps, we develop in this paper a general analytic framework, iGWAS, that integrates genetic and genomic data to examine ME, AE, as well as TE and incorporates the family design.
This paper makes both methodological and scientific contributions. Methodologically, we propose a new analytic framework, iGWAS, that facilitates to study the ME (eQTL genetic effect on a phenotype mediated through gene expression) and the AE (genetic effect through other biological pathways or environment-mediated mechanisms), and incorporates the family design. Moreover, we develop robust tests for ME and AE of SNPs using both SNP and gene expression data. Scientifically, our proposed iGWAS approach identifies novel susceptibility genes of childhood asthma and characterizes their mechanisms through gene expression.
Methods
Integrative Genome-Wide Association Studies (iGWAS)
Causal Mediation Model and Null Hypotheses
We utilize the causal mediation model to investigate the etiologic mechanism of genetic effect. We jointly model the effect of a set of SNPs associated with expression of a gene (i.e., eQTL SNPs) and the corresponding gene expression on occurrence of a disease using a logistic regression model, adjusting for covariates. For subject i, the probability of having a dichotomous outcome (e.g., Y = 1/0 for case/control) is associated with q covariates (Xi, with the first covariate to be 1, i.e., the intercept), p SNP genotypes (Si, e.g., cis-eQTL SNPs of a gene), one mRNA expression of a gene (Gi), and p cross-product interactions between the SNPs and gene expression as:
| (1) |
where βX = (βX1, …, βXq)T, βS = (βS1, …, βSp)T, βG and βC = (βC1, …, βCp)T are regression coefficients for the covariates, SNP genotypes, gene expression, and interactions of the SNP genotypes and gene expression, respectively. We next consider another regression model for the gene expression (Gi), which depends on the q covariates (Xi) and p SNP genotypes (Si):
| (2) |
where αX = (αX1, …, αXq)T and αS = (αS1, …, αSp)T are the regression coefficients for the covariates and SNP genotypes, respectively; and εGi follow a normal distribution with mean 0 and variance . We here focus on eQTLs by restricting analyses to SNPs that are associated with the gene expression, i.e., αS ≠ 0.
Assuming confounding effects are properly controlled after accounting for the covariates, the TE of comparing two genotypes of the SNPs s1 vs. s0 is defined as: TE = logit{P (Y = 1|S = s1, X)} − logit{P (Y = 1|S = s0, X)}. Under the rare disease assumption for a binary disease phenotype, the TE of the SNPs on the log odds ratio (OR) of disease risk comparing two genotype values s1 vs. s0 can be expressed using the regression coefficients in models (1) and (2) as [Huang et al., 2014]:
We can also express the ME (indirect effect) of the SNP set (s1 vs. s0)mediated by the gene expression G, and AE (direct effect) of the SNP set through other biological pathways, on the log OR of disease risk as [Huang et al., 2014]:
| (3) |
| (4) |
Note that the TE can be decomposed into ME and AE. Although direct and indirect effects have been defined and used in the causal mediation literature [Robins and Greenland, 2012], we rename them here as AE and ME to better reflect their biological interpretation. The indirect effect or ME of an SNP set S in our setting is the effect of eQTL SNPs on disease risk mediated through gene expression (the black path in Fig. 1), whereas the direct effect or AE is the effect on disease risk independent of expression of the gene, but perhaps through other genes or other mechanisms (the gray path in Fig. 1). We have shown that identification of the TE requires a weaker assumption than those required for the ME and AE [Huang et al., 2014].
As we focus on the eQTL SNPs, the SNPs with nonzero association with the gene expression, or equivalently, αS ≠ 0, it can be easily shown that
| (5) |
| (6) |
provided that there does not exist a perfect cancellation for effects with βS ≠ 0, βG ≠ 0, and βC ≠ 0. It follows that the equivalence for TE under the null would be:
| (7) |
Note different genetic models (dominant, recessive, or additive) follow the same null hypotheses (5)–(7): no effects for subjects carrying two different genotypes. Regardless of the genetic models, the above equivalence applies. However the coefficients may have different interpretations under different models. For example, under the additive models, βs is the log OR comparing between subjects carrying two minor alleles and those with one allele, as well as between one and zero minor allele; and βs under the dominant models is the log OR between the presence and absence of a minor allele. For implementation, different genetic models can be easily incorporated by altering the genetic coding in S, e.g., 0, 1, 2 for zero, one, and two minor alleles, respectively, under additive model.
Testing Procedure for the AE Under Family Design
As the number of SNPs (p) in a gene can be large and some may be highly correlated due to linkage disequilibrium, the conventional test such as the likelihood ratio test that uses large degrees of freedom has limited power. We resort to an empirical Bayes approach [Lin, 1997] by assuming the regression coefficients of individual SNP effects, βSj (j = 1, …, p), follow an arbitrary distribution with mean 0 and variance τS, and the SNP-by-expression interaction coefficients βCj follow another arbitrary zero-mean distribution with variance τC. The resulting model (1) becomes a logistic mixed model [Breslow and Clayton, 1993]. The null hypothesis of no AE (5) hence is equivalent to a joint test of the two variance components, τS and τC, in the induced logistic mixed model:
For family data, one can obtain the scores for τS and τC under the induced logistic mixed model: UτS = (Y − μ̂0, AE)T R*−1SSTR*−1(Y − μ̂0, AE) and UτC = (Y − μ̂0, AE)T R*−1CCT R*−1(Y − μ̂0, AE), where Y = (Y1, …, Yn)T, S = (S1, …, Sn)T, C = (C1, …, Cn)T = (G1S1, …, GnSn)T; the outcome risk under the null (element-wise calculation), and β̂X0 and β̂G0 are the estimators of βX and βG, respectively, under the null model: , an ni by ni covariance matrix represents the within-family correlation with ni being the number of subjects within family i and m being the number of families, and . With scores for the parameters of interest, τS and τC, we propose a test statistic as a weighted sum of the two scores, w1UτS + w2UτC:
where the weights (w1 and w2) are chosen to be the inverse of their respective standard deviation of UτS and UτC to make them comparable on the same scale. The unknown true covariance R* can be replaced by a working covariance R, which leads to a sum of L 2 norms of two estimating equations for βs and βc:
Examples of possible working covariance structures include a sample covariance matrix, a diagonal matrix assuming working independence or a kinship matrix.
The test statistic is derived according to the outcome model (1) where the disease risk is determined by SNPs, gene expression, and their cross-product interactions (i.e., the full model). We may specify a parsimonious model with only main effects by assuming that the disease risk depends on SNPs and gene expression without interactions (βC = 0). Following the same development as described above, we obtain a similar test statistic under the main effect model:
Testing Procedure for the ME Under Family Design
The null hypothesis of no ME (6) in the induced logistic mixed model is equivalent to:
We construct the test statistics for the ME for the above null hypothesis under model (1) as a weighted sum of L2 norms of estimating equations for βG and βC:
and under the main effect model, one can obtain the test statistic:
where (element-wise), and β̂X0 and β̂S0 are the estimators of βX and βS, respectively, under the null model:
| (8) |
However, the maximum likelihood estimators of (βx, βs) may not be stable in the presence of a large number of SNPs and high correlations due to linkage disequilibrium. Thus, we estimate (βx, βs) by fitting the null model (8) using ridge regression via the penalized log-likelihood: , where li is the estimating equation for the null model (8) constructed as log-likelihood of Yi assuming working independence for subjects Yij (j = 1, …, ni) within unit i, and λ is a tuning parameter. The tuning parameter λ can be selected using cross-validation or generalized cross-validation (GCV) [O’Sullivan, 1994]. Using the asymptotic distribution of and and the GCV function presented in the supplementary material, one can construct a perturbation procedure (discussed in the following) to calculate P-values of and .
Testing Procedure for the TE Under Family Design
The null hypothesis of no total SNP effect (7) is equivalent to a joint test of the two variance components, τS and τC, and the scalar regression coefficient for gene expression effect, βG in the induced logistic mixed model:
With scores for the three parameters of interest, τS, τC, and βG, we propose a test statistic as a weighted sum of L 2 norms of three estimating equations for βS, βG, and βC:
| (9) |
Again, test statistics can be derived under other parsimonious models, e.g., the main effect model by assuming that the disease risk depends on SNPs and gene expression without interactions (βC = 0), or the SNP-only model by assuming that the disease risk depends only on SNPs (βG = 0, βC = 0). Following the same development, we obtain similar test statistics for the two simpler models, denoted as and :
where μ̂0 = (μ̂01, …, μ̂0n)T, , and β̂X0 is the maximum likelihood estimator of βX under the null model: .
Perturbation Procedure and Omnibus Test Under Family Design
In this section, we develop perturbation procedure for AE, ME, and TE. For illustration, we focus on the TE, and stress that procedures for AE and ME can be constructed following the same development. Under the null model that there is no effect of a gene on disease, i.e., under the null hypothesis (7), tests under the full model , main effect model , and SNP-only model are all valid tests. If there indeed exists an effect of the gene, the test assuming the correct model is expected to perform optimally with the highest power, while a test under the mis-specified model is likely to lose power. However, we do not know which model is the true underlying model, so it is desirable to develop an omnibus test that maximizes testing power by searching all three candidate models. Specifically, we calculate the P-values under each of the three models, and construct an omnibus test statistic using the minimum of the three P-values. We then calculate the P-value of the omnibus test statistic using a perturbation procedure [Cai et al., 2000; Huang et al., 2014; Parzen et al., 1992].
With the asymptotic distribution derived for in the supplement material (III. Asymptotics of Q statistics), we construct a perturbation procedure to perform hypothesis tests. Let , where , Xi, Si, Gi, Ci are q by ni, p by ni, 1 by ni, p by ni matrices, respectively; Yi = (Yi1, …, Yini)T, are the corresponding estimates under the null, β̂X0 is the estimator of βX under the null model for the correlated family data, and Zi are independent standard normal random variables. By generating independent Z = (Z1, …, Zm) repeatedly, the perturbed realization of in (9) under the null can be obtained, {, b = 1, …, B} where expression of A can be found in supplement material (III), and B is the number of perturbations. The P-value of QSGC can then be approximated using the tail probability of the perturbed realizations by comparing and the observed . Similar perturbation procedures can be constructed for ME with the asymptotic distribution of QME (see supplement material) or AE. The advantage of our proposed method is that we account for the within-family correlation by introducing a working correlation matrix R. Furthermore, our method does not require a correct specification of R. Through the perturbation procedure, our method protects the type I error under any structure of R, and gains power if the structure is close to the truth. The family design can also introduce ascertainment bias: families are not randomly selected from population, but instead ascertained according to disease status. Previous studies have shown that the bias in type I error rate is negligible using variance component tests based on estimating equations [Schifano et al., 2012]. We also account for ascertainment in our simulation studies.
Using the perturbation procedure, we can also calculate the P-value of the omnibus test statistic. Specifically, denote by pS, pSG, and pSGC, the P values of the test statistics , and , respectively; we define the minimum P-value of pS, pSG, and pSGC as the test statistic for the omnibus test. With the perturbed realization of the distribution for the three statistics , and , the underlying distribution of this minimum P-value can then be approximated [Huang et al., 2014]. By comparing the observed minimum P-value with the approximated distribution, the omnibus P-value can be calculated as the tail probability of the distribution. One can also use the perturbation procedure to calculate the P-value of the omnibus test for ME (or AE) constructed using the minimum of the P-values of and (or and ). R codes that implement the proposed testing procedures for the TE, ME, and AE are available upon request.
Simulation and Asthma Data Analysis
We detailed the simulation and data analysis in the supplement material (I. Numerical Simulation, and II. Asthma iGWAS Data Analysis). Briefly, we randomly simulated SNPs and gene expression from chromosome 10 for 100 cases and 100 controls, mimicking the family design of the MRCA data and accounting for ascertainment. We simulated 500,000 datasets to estimate size of the test and 2,000 datasets to estimate statistical power. We conducted an integrative GWAS using the MRCA asthma dataset [Moffatt et al., 1986]. The MRCA asthma dataset is a GWAS for childhood asthma with a family design where both genome-wide SNP and gene expression data are available. The MRCA data were collected from families of the British descent and consists of 378 subjects: 266 cases and 112 controls. Control subjects are either siblings or parents of the cases. Genome-wide SNP genotypes were obtained using the Illumina 300K SNP array and gene expressions were measured using the Affymetrix HU133A 2.0 expression array. Our analyses focused on the SNPs that are associated with the nearby gene within 1 Mb on the same chromosome (cis-eQTL) with false discovery rate (FDR) [Benjamini and Hochberg, 1995] less than 1%, which has been published using similar datasets [Liang et al., 2008]. We then grouped these eQTL SNPs with their corresponding gene expression as an SNPs-expression set. The iGWAS analyses were applied to a total of 11,198 such sets. P-values were calculated with 5,000 resampling perturbation and approximated with the method based on the mixture of normal distributions [Cai et al., 2012].
Results
Detail simulation results were provided in the supplement material (IV. Numerical Results for AE, ME, and TE; supplementary Table S1, and Figs. S1 and S2). In summary, numerical simulation showed that under the null, the iGWAS for ME, AE, and TE protect the small type I error (e.g., 5 × 10−5); and under the alternative, the omnibus tests are robust to different underlying models and approach the optimal power from tests with correct model specification.
Family-Based iGWAS of Childhood Asthma
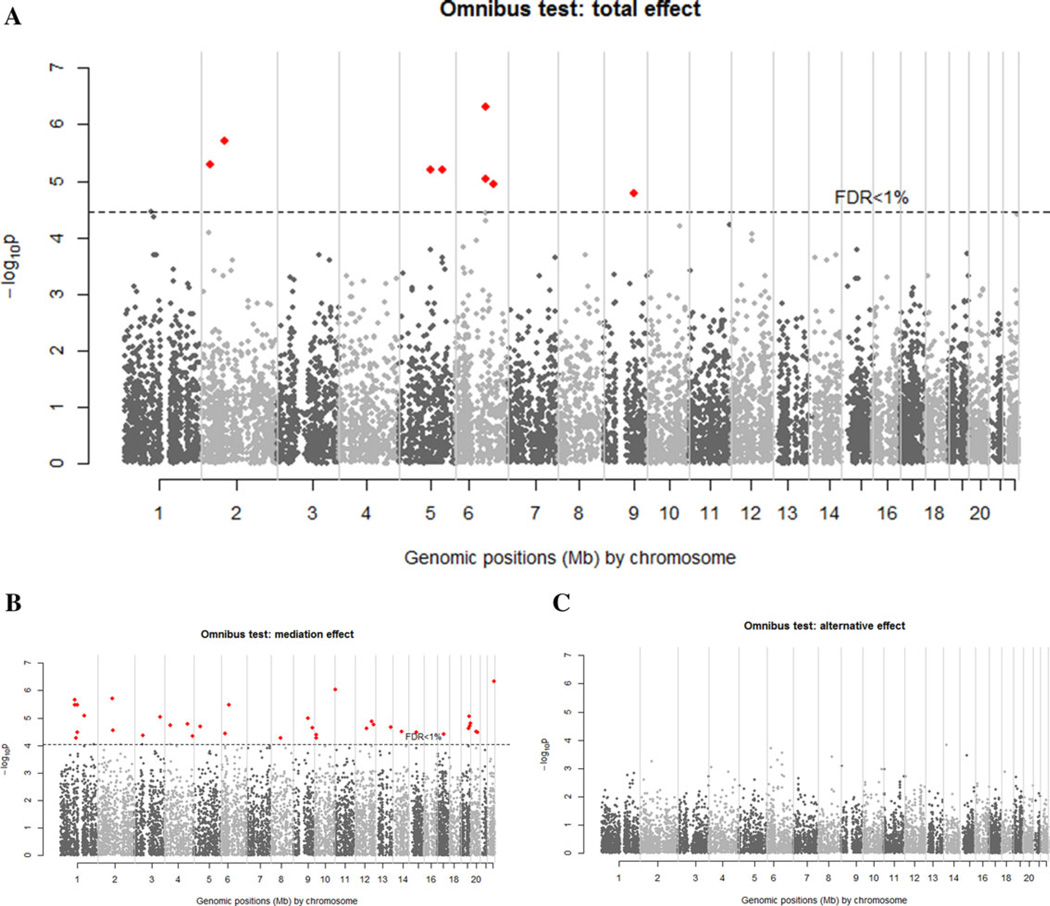
We grouped eQTL SNPs and their corresponding gene expression probes into 11,198 sets and then performed the proposed gene-centric iGWAS analyses on these sets one at a time. We first focused on examining 11,198 TEs of SNPs and gene expression. For the SNP-only analyses, the set contained only eQTLs without gene expression. We used 1% of FDR as the cut-off for genome-wide statistical significance. For the SNP-only analyses, there were no gene or transcript with FDR < 1% (supplementary Fig. S3A). There were 14 transcripts, 2 transcripts, and 8 transcripts with FDR < 1% in genome wide analyses using the main effect model (QSG), full model with interaction (QSGC), and omnibus test (Qomb), respectively (supplementary Figs. S3B and S3C, and Fig. 2A). The genome-wide results were consistent with our findings from numerical simulations: the omnibus test (Qomb) and main effect model (QSG) that incorporated the information of gene expression were more powerful tests compared to the conventional SNP-only approach (QS).
Figure 2.
Manhattan plots of genome-wide scan of the asthma data using SNPs and gene expression data with the gene-centric iGWAS approach. Analysis is restricted to eQTLs. (A) Omnibus P-values for total effects; (B) omnibus P-values for mediation effects; (C) omnibus P-value for alternative effects. Red dots indicate the transcripts found from the omnibus test with FDR < 1%.
In addition to increasing power of identifying susceptibility genes, a more important advantage of the iGWAS approach is that it facilitates to unravel the mechanism of genetic effects. Specifically, we investigated the mechanistic contribution of SNPs and gene expression to asthma risk by studying the ME of eQTL SNPs on asthma risk through gene expression and the AE of eQTL SNPs on asthma risk through other mechanisms. To study the ME and AE, we investigated the main effect model and full model. The P-values of omnibus tests for MEs and AEs in scanning the genome are presented in Figures 2B and C, and those of main effect models and full models are presented in supplementary Figures S4 and S5. There are 36 genes with significant MEs at FDR < 1%, but no gene with AE survives the same cut-off. The results indicated that the effect of most asthma susceptibility genes may be mediated through their own gene expressions. The 36 genes with significant MEs (FDR < 1%) and the top 10 genes with the most significant AEs are presented in supplementary Tables S2 and S3.
iGWAS Results for Candidate Genes
The nominal P-values of the eight transcripts with FDR < 1% in omnibus tests for TEs are presented in Table 1. The eight transcripts corresponded to six unique genes and an RNA transcript not within a gene (229319_at). The MANEA gene showed up twice in the list because the eQTL SNPs-expression set was grouped for each expression probe, and genes with multiple probes may be grouped into different sets. For the TEs of the eight transcripts, the model with main effects of SNPs and gene expression (QSG) performed the best, except for MRPL53 and MANEA (219003_s_at) where the full model (QSGC) performed even better. The omnibus P-values were very close to the smallest P-values among the three candidate models (QS, QSG, and QSGC). The SNP-only effect of PTCH1 was not significant (P = 0.32), but its SNP-expression joint effect was very significant (P = 8.5 × 10−6). Even with the conservative Bonferroni adjustment for the 11,198 sets (P < 4.5 × 10−6), MANEA (219003_s_at) (P = 4.8 × 10−7) and MRPL53 (P = 1.9 × 10−6) were still statistically significant.
Table 1.
P-values of alternative effect, mediation effect, and total effect for genes with most significant total effect (FDR < 1%) using the asthma data. Ch, chromosome; QS, test with only SNP-set, the SNP-only model; QSG, test with both SNP set and gene expression (without interaction), the main effect model; QSGC, test with SNP set, gene expression, and their cross-product interactions, the full model; Qomb, omnibus test for QS (if applicable), QSG, and QSGC
| Alternative effect | Mediation effect | Total effect | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe | Gene | Ch | Number of eQTL |
|
|
|
|
|
|
|
|
|
|
||||||||||
| 219003_s_at | MANEA | 6 | 159 | 2.9 × 10−4 | 2.6 × 10−4 | 2.8 × 10−4 | 0.0079 | 0.0076 | 0.0082 | 4.8 × 10−7 | 2.1 × 10−6 | 2.9 × 10−7 | 2.8 × 10−7 | ||||||||||
| 225523_at | MRPL53 | 2 | 175 | 5.7 × 10−4 | 5.8 × 10−4 | 6.2 × 10−4 | 0.055 | 0.053 | 0.060 | 1.9 × 10−6 | 1.0 × 10−5 | 5.2 × 10−6 | 1.0 × 10−6 | ||||||||||
| 226996_at | LYCAT | 2 | 193 | 0.19 | 0.18 | 0.18 | 4.7 × 10−4 | 4.6 × 10−4 | 5.0 × 10−4 | 5.2 × 10−6 | 0.0065 | 4.0 × 10−6 | 6.0 × 10−5 | ||||||||||
| 230261_at | ST8SIA4 | 5 | 185 | 0.012 | 0.012 | 0.013 | 1.9 × 10−4 | 2.0 × 10−4 | 2.0 × 10−4 | 6.3 × 10−6 | 0.0020 | 4.1 × 10−6 | 1.4 × 10−4 | ||||||||||
| 222422_s_at | NDFIP1 | 5 | 48 | 0.11 | 0.11 | 0.11 | 0.0011 | 0.0010 | 0.0012 | 6.4 × 10−6 | 0.0031 | 4.0 × 10−6 | 4.1 × 10−5 | ||||||||||
| 1554193_s_at | MANEA | 6 | 67 | 0.0017 | 0.0018 | 0.0020 | 0.012 | 0.011 | 0.012 | 9.0 × 10−6 | 6.0 × 10−5 | 4.8 × 10−6 | 1.4 × 10−5 | ||||||||||
| 229319_at | — | 6 | 148 | 0.29 | 0.29 | 0.28 | 2.9 × 10−4 | 3.0 × 10−4 | 3.0 × 10−4 | 1.2 × 10−5 | 0.018 | 7.5 × 10−6 | 6.70 × 10−5 | ||||||||||
| 209815_at | PTCH1 | 9 | 13 | 0.92 | 0.91 | 0.93 | 1.0 × 10−5 | 1.8 × 10−5 | 1.3 × 10−5 | 1.6 × 10−5 | 0.32 | 8.5 × 10−6 | 2.2 × 10−4 | ||||||||||
As shown in Table 1, all except MRPL53 had very significant ME, no matter under the main effect model or full model, and the omnibus P-values were very close to smallest P-values. The AE was significant in MANEA (P = 2.9 × 10−4 and 0.0017), MRPL53 (P = 5.7 × 10−4), and ST8SIA4 (P = 0.012), which indicated that eQTL SNPs of these genes may act through other mechanisms in addition to mRNA expression of the genes, to affect the asthma risk.
Nominal P-values for genes containing SNPs reported in previous asthma GWAS [Ferreira et al., 2011; Moffatt et al., 2007; Ober et al., 2008; Torgerson et al., 2011] are presented in Table 2. The iGWAS approach confirmed the significant TEs from ORMDL3 (P = 0.0081), CHI3L1 (P = 0.0055), and IL6R (P = 0.040). The effects of CHI3L1 and IL6R were mostly through gene expression (P = 0.0013 and 0.018, respectively), while the effect of ORMDL3 gene seemed to be mainly through alternative biological mechanisms (P = 0.023). Of note, the strategy used in previous GWAS to identify these genes was single SNP analysis, while our reanalysis utilized the proposed gene-centric approach.
Table 2.
Nominal P-values of alternative effect, mediation effect, and total effect for genes reported in previous GWAS of asthma risk
| Alternative effect | Mediation effect | Total effect | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe | Gene | Ch | Number of eQTL |
|
|
|
|
|
|
|
|
|
|
||||||||||
| 223259_at | ORMDL3 | 17 | 209 | 0.027 | 0.028 | 0.027 | 0.037 | 0.037 | 0.040 | 0.094 | 0.065 | 0.11 | 0.068 | ||||||||||
| 235136_at | ORMDL3 | 17 | 93 | 0.023 | 0.023 | 0.022 | 0.31 | 0.30 | 0.35 | 0.053 | 0.034 | 0.12 | 0.058 | ||||||||||
| 240701_at | ORMDL3 | 17 | 36 | 0.048 | 0.050 | 0.047 | 0.11 | 0.11 | 0.12 | 0.0081 | 0.015 | 0.0052 | 0.0080 | ||||||||||
| 209395_at | CHI3L1 | 1 | 14 | 0.20 | 0.19 | 0.19 | 0.0013 | 0.0011 | 0.0019 | 0.0055 | 0.48 | 0.0030 | 0.018 | ||||||||||
| 209396_s_at | CHI3L1 | 1 | 14 | 0.24 | 0.23 | 0.25 | 0.0081 | 0.0074 | 0.0090 | 0.025 | 0.47 | 0.015 | 0.053 | ||||||||||
| 226333_at | IL6R | 1 | 23 | 0.67 | 0.70 | 0.66 | 0.018 | 0.017 | 0.021 | 0.040 | 0.76 | 0.021 | 0.072 | ||||||||||
| 205945_at | IL6R | 1 | 29 | 0.89 | 0.88 | 0.90 | 0.71 | 0.65 | 0.87 | 0.94 | 0.88 | 0.92 | 0.94 | ||||||||||
| 206618_at | IL18R1 | 2 | 66 | 0.66 | 0.68 | 0.64 | 0.98 | 0.95 | 1.0 | 0.81 | 0.68 | 0.91 | 0.85 | ||||||||||
| 209349_at | RAD50 | 5 | 12 | 0.84 | 0.82 | 0.85 | 0.83 | 0.74 | 0.99 | 0.92 | 0.84 | 0.95 | 0.95 | ||||||||||
Abbreviation and notations are the same as Table 1.
Single-Locus Analyses
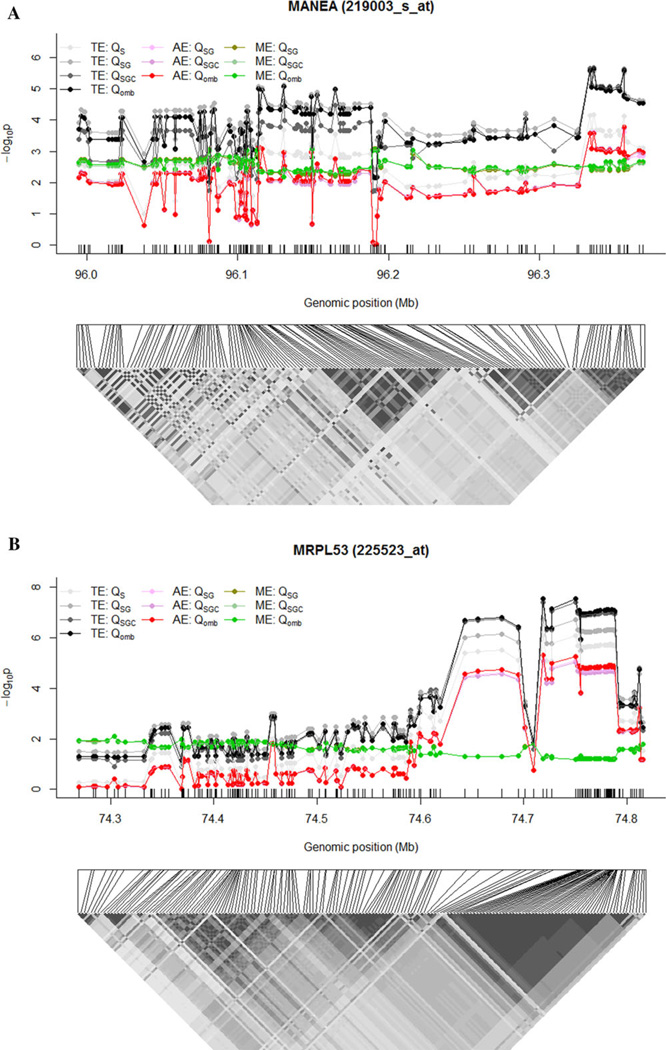
We also conducted single-locus analyses for the top eight transcripts as well as the candidate genes identified from previous GWAS. In the single-locus analyses, we analyzed each single SNP and the gene expression associated with the SNP, studying AEs, MEs, and TEs. Again, AEs and MEs were studied under the main effect model and full model, whereas TEs were investigated under the SNP-only model, main effect model, and full model. We present the P-values from different tests for AEs, MEs, and TEs of MANEA (219003_s_at) and MRPL53, the two most significant genes in Fig. 3. Results of the remaining six transcripts are shown in supplementary Figures S6–S11. The ME or AE was prominent in certain linkage disequilibrium blocks, in particular, for MRPL53, MANEA, and ST8SIA4. The median (first quartile-third quartile) of the proportion (%) of mediation for all eQTL SNPs was 11.9% (8.8–13.6%) for MANEA (219003_s_at), 3.9% (1.0–4.4%) for MANEA (1554193_s_at), 8.7% (3.8–13.7%) for MRPL53, 50.6% (31.6–53.7%) for LYCAT, 17.7% (13.7–20.7%) for ST8SIA4, 54.1% (33.6–54.5%) for NDFIP1, 95.0% (78.8–100.0%) for 229319_at, and 48.4% (25.9–100.0%) for PTCH1. The high proportions of mediation observed in NDFIP1, PTCH1, LYCAT, and 229319_at were consistent with their highly significant MEs and nonsignificant AEs (Table 1, supplementary Figs. S6, S8, S10, and S11). Detailed results from single-locus analyses are presented in supplementary Tables S4–S11, and the patterns support the findings of gene-centric analyses presented in Table 1. The results of single-locus analyses for the genes identified from the previous GWAS (ORMDL3, CHI3L1, IL6R, IL18R1, and RAD50) are presented in supplementary Tables S12–S20. The findings from single-locus analyses are consistent with the results using the iGWAS approach.
Figure 3.
Plots of P-values of AE, ME, and TE in single-locus analyses for MANEA (219003_s_at) (A) and MRPL53 (B). Lower panel represents the linkage disequilibrium structure for SNPs within each gene, measured as r2 ranging from 0 (white) to 1 (black). QS, tests assuming an SNP-only model; QSG, tests assuming a model with main effects of SNPs and gene expression; QSGC, tests assuming a model with main effects and their interactions; Qomb, omnibus tests.
Discussion
We proposed in this paper an integrative approach, iGWAS, that is able to analyze multiplatform genomic data under the family-based design. The model can be presented as a causal diagram (Fig. 1), which was set up based on the central dogma of molecular biology that DNA can be transcribed to mRNA expression and mRNA can then be translated to be protein to affect the phenotypic trait such as disease risk. The mediation diagram provides an intuitive illustration of our hypothesis. The iGWAS approach is integrative in different aspects. The model not only integrates different types of genomic data, i.e., SNP and gene expression data, but also incorporates different types of genetic/genomic association studies to delineate clinical outcome rather than perform a GWAS, an expression microarray study, and an eQTL study separately. Moreover, the iGWAS approach integrates biological knowledge into the computational model, as illustrated in the causal mediation diagram.
The iGWAS approach has several advantages. First, with the enriched genomic information compared to single one alone, a better statistical power has been illustrated in both numerical studies and a family-based GWAS of childhood asthma using the family-based MRCA data. The iGWAS outperforms the SNP-only approach and identifies novel genes for asthma susceptibility that have not been reported. Second, the iGWAS approach makes an analytic investigation into the biological mechanism of genetic effects possible. The ME represents a cis-regulating effect that SNPs regulate expression of a nearby gene; and possible mechanisms behind the AE include a trans-regulating effect that SNPs affect expression of a distant causal gene, or a structural related effect that SNPs alter the biochemical structure of their gene product instead of their expression level. Third, although the iGWAS was developed under the family design, it can accommodate a wide range of study designs such as nonfamily-based case-control studies, cohort studies, or longitudinal studies with repeat measurement. Finally, the iGWAS approach may have translational utilities. For example, for genes with significant MEs, the expression levels may serve as diagnostic biomarkers to screen and prevent the disease. Moreover, potential therapeutic agents such as small RNAs that can repress expressions of the specific genes may be developed to counteract the MEs.
Because our method is developed under the framework of mediation modeling based on causal inference, we need to make untestable no-unmeasured confounding assumptions to draw conclusions beyond association. If we are interested in studying the TE, the only assumption is that there is no unmeasured confounding for the effect of eQTL SNPs on the outcome after adjusting for the covariates, which is the same as that made in the conventional SNP-only GWAS. However, if we are interested in the ME and AE, stronger assumptions are required [Huang et al., 2014].
The iGWAS approach confirmed the genes that have been reported to be associated with asthma risk such as ORMDL3 (P = 0.0081), CHI3L1 (P = 0.0055), and IL6R (P = 0.040) [Ferreira et al., 2011;Moffatt et al., 2007]. In addition to confirming their overall genetic effects, we also found that the genetic effects of CHI3L1 and IL6R were mediated by gene expression (Table 2) and that genetic loci of ORMDL3 may have alternative mechanisms independent of acting through its gene expression (P = 0.023). Our approach also identified six novel genes (MANEA, MRPL53, LYCAT, ST8SIA4, NDFIP1, and PTCH1). NDFIP1 has been reported to be a candidate susceptible gene in asthma GWAS but with weak effect, OR = 1.11 [Ferreira et al., 2011]. As we found that most of the genetic effect of NDFIP1 was through gene expression, the heterogeneity due to not accounting for gene expression may explain the modest effect in the previous study. NDFIP1 mediates peripheral immune tolerance by inducing cell cycle exit in CD4 T lymphocytes [Altin et al., 2014]. PTCH1, patched homolog 1, has been found to be associated with lung function in the general population and lung function abnormalities in white and African Americans with asthma [Li et al., 2011]. ST8SIA4, sialyltransferase, is predominantly expressed in immune cells [Kolker et al., 2012]. It plays an important role in substrate recognition that modulates cell adhesion and signaling [Zapater and Colley, 2012]. MANEA, α-endomannosidase, has been reported to be associated with psychiatric disorders including substance dependence and anxiety disorders [Farrer et al., 2009; Jensen et al., 2014; Yu et al., 2008]. Studies in nonpsychiatric disorders, however, are very limited. Consistent with our findings, a 3′UTR SNP, rs113503, has been shown to be related to mRNA expression of MANEA [Jensen et al., 2014]. LYCAT, lysocardiolipin acyltransferase 1, is expressed in developing lung of mouse [Wang et al., 2010], but its association with asthma or other immune-related disorders has not been reported. MRPL53, mitochondrial ribosomal protein L53, is involved in a ribonucleotide complex [Wessels et al., 2013]. The molecular mechanism of these candidate genes in relation to the development of asthma warrants further examination.
Supplementary Material
Footnotes
Supporting Information is available in the online issue at wileyonlinelibrary.com.
References
- Altin JA, Daley SR, Howitt J, Rickards HJ, Batkin AK, Horikawa K, Prasad SJ, Nelms KA, Kumar S, Wu LC, et al. Ndfip1 mediates peripheral tolerance to self and exogenous antigen by inducing cell cycle exit in responding CD4+ T cells. Proc Natl Acad Sci USA. 2014;111(6):2067–2074. doi: 10.1073/pnas.1322739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- Berlivet S, Moussette S, Ouimet M, Verlaan DJ, Koka V, Al Tuwaijri A, Kwan T, Sinnett D, Pastinen T, Naumova AK. Interaction between genetic and epigenetic variation defines gene expression patterns at the asthma-associated locus 17q12-q21 in lymphoblastoid cell lines. Hum Genet. 2012;131(7):1161–1171. doi: 10.1007/s00439-012-1142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow N, Clayton D. Approximate inference in generalized linear mixed models. J Am Stat Assoc. 1993;88:9–25. [Google Scholar]
- Cai T, Wei L, Wilcox M. Semiparametric regression analysis for clustered failure time data. Biometrika. 2000;87:867–878. [Google Scholar]
- Cai T, Lin X, Carroll RJ. Identifying genetic marker sets associated with phenotypes via an efficient adaptive score test. Biostatistics. 2012;13(4):776–790. doi: 10.1093/biostatistics/kxs015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, Harari O, Norton J, Budde J, Bertelsen S, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505(7484):550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Billstrand C, Zhou X, Chavarria C, DeLeon S, Michelini K, Pai AA, Ober C, Gilad Y. The combination of a genome-wide association study of lymphocyte count and analysis of gene expression data reveals novel asthma candidate genes. Hum Mol Genet. 2012;21(9):2111–2123. doi: 10.1093/hmg/dds021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39(10):1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Kranzler HR, Yu Y, Weiss RD, Brady KT, Anton R, Cubells JF, Gelernter J. Association of variants in MANEA with cocaine-related behaviors. Arch Gen Psychiatry. 2009;66(3):267–274. doi: 10.1001/archgenpsychiatry.2008.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40(9):1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, Danoy P, Baltic S, Nyholt DR, Jenkins M, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378(9795):1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YH, Zillikens MC, Wilson SG, Farber CR, Demissie S, Soranzo N, Bianchi EN, Grundberg E, Liang L, Richards JB, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility loci for osteoporosis-related traits. PLoS Genet. 2010;6(6):e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YT, Vander Weele TJ, Lin X. Joint analysis of SNP and gene expression data in genetic association studies of complex diseases. Ann Appl Stat. 2014;34(1):162–178. doi: 10.1214/13-AOAS690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Chanock SJ. Genome-wide association studies and “the art of the soluble”. J Natl Cancer Inst. 2010;102(12):836–837. doi: 10.1093/jnci/djq197. [DOI] [PubMed] [Google Scholar]
- Jensen KP, Stein MB, Kranzler HR, Yang BZ, Farrer LA, Gelernter J. The alpha-endomannosidase gene (MANEA) is associated with panic disorder and social anxiety disorder. Transl Psychiatry. 2014;4:e353. doi: 10.1038/tp.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker E, Higdon R, Haynes W, Welch D, Broomall W, Lancet D, Stanberry L, Kolker N. MOPED: Model Organism Protein Expression Database. Nucleic Acids Res. 2012;40:D1093–D1099. doi: 10.1093/nar/gkr1177. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. Am J Hum Genet. 2008;82(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Howard TD, Moore WC, Ampleford EJ, Li H, Busse WW, Calhoun WJ, Castro M, Chung KF, Erzurum SC, et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127(6):1457–1465. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Morar N, Dixon AL, Lathrop GM, Abecasis GR, Moffatt MF, Cookson WO. A cross-platform analysis of 14177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013;23(4):716–726. doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Variance component testing in generalised linear models with random effects. Biometrika. 1997;84:309–326. [Google Scholar]
- Liu D, Ghosh D, Lin X. Estimation and testing for the effect of a genetic pathway on a disease outcome using logistic kernel machine regression via logistic mixed models. BMC Bioinformatics. 2008;9:292. doi: 10.1186/1471-2105-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon D. Introduction to Statistical Mediation Analysis. New York: Taylor and Francis; 2008. [Google Scholar]
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, vonBerg A, Bufe A, Rietschel E, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan F, Yandell BS, Raynor WJ., Jr Automatic smoothing of regression functions in generalized linear models. J Am Stat Assoc. 1986;81:96–103. [Google Scholar]
- Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358(16):1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parzen M, Wei L, Ying Z. A resampling method based on pivotal functions. Biometrika. 1994;81:341–350. [Google Scholar]
- Robins JM. In: Semantics of Causal DAG Models and the Identification of Direct and Indirect Effects. Geen P, Hjort NL, Richardson S, editors. New York, NY: Oxford University Press; 2003. [Google Scholar]
- Robins JM, Greenland S. Identifiability and exchangeability for direct and indirect effects. Epidemiology. 1992;3(2):143–155. doi: 10.1097/00001648-199203000-00013. [DOI] [PubMed] [Google Scholar]
- Rubin D. Bayesian inference of causal effects. Ann Stat. 1978;6:34–58. [Google Scholar]
- Schifano ED, Epstein MP, Bielak LF, Jhun MA, Kardia SL, Peyser PA, Lin X. SNP set association analysis for familial data. Genet Epidemiol. 2012;36(8):797–810. doi: 10.1002/gepi.21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, Himes BE, Levin AM, Mathias RA, Hancock DB, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marcano AC, Hajat C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82(1):139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ni L, Yu Q, Xiong J, Liu HC, Rosenwaks Z. Expression of the Lycat gene in the mouse cardiovascular and female reproductive systems. Dev Dyn. 2010;239(6):1827–1837. doi: 10.1002/dvdy.22300. [DOI] [PubMed] [Google Scholar]
- Wessels HJ, Vogel RO, Lightowlers RN, Spelbrink JN, Rodenburg RJ, vanden Heuvel LP, van Gool AJ, Gloerich J, Smeitink JA, Nijtmans LG. Analysis of 953 human proteins from a mitochondrial HEK293 fraction by complexome profiling. PLoS One. 2013;8(7):e68340. doi: 10.1371/journal.pone.0068340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86(6):929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Ancona N, Hauser ER, Mukherjee S, Furey TS. Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets. Genome Res. 2012;22(2):386–397. doi: 10.1101/gr.124370.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Kranzler HR, Panhuysen C, Weiss RD, Poling J, Farrer LA, Gelernter J. Substance dependence low-density whole genome association study in two distinct American populations. Hum Genet. 2008;123(5):495–506. doi: 10.1007/s00439-008-0501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapater JL, Colley KJ. Sequences prior to conserved catalytic motifs of polysialyltransferase ST8Sia IV are required for substrate recognition. J Biol Chem. 2012;287(9):6441–6453. doi: 10.1074/jbc.M111.322024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liang L, Morar N, Dixon AL, Lathrop GM, Ding J, Moffatt MF, Cookson WO, Kraft P, Qureshi AA, et al. Integrating pathway analysis and genetics of gene expression for genome-wide association study of basal cell carcinoma. Hum Genet. 2012;131(4):615–623. doi: 10.1007/s00439-011-1107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6(5):e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.