Abstract
The adenovirus late genes are organized in a complex transcription unit in which the production of multiple RNAs is controlled by specific RNA processing events. Adding to the complexity is the fact that the early E3 transcription unit is wholly contained within the late transcription unit. Within this overlapping region there are splicing events and poly(A) site choices specific to each transcription unit. Early in infection, there is near exclusive use of the E3 splice sites with little L4 poly(A) site use whereas the reverse is true late in infection. Using a plasmid containing an intact E3 transcription unit, we demonstrate that the L4 poly(A) site, located in the first E3 intron, is not used either in a mock-infected cell or a late infected cell; nor is it used if the same transcription unit is driven by the major late promoter. In addition, the nature of the poly(A) site appeared to be unimportant since the E3 poly(A) site was also not used in the intron if inserted in place of the L4 poly(A) site. Thus, splice site selection appears to dominate over poly(A) site choice. This conclusion was confirmed by the observation that deletion of either the E3 splice donor (5'ss) or the E3 splice acceptor (3'ss) allowed the use of the L4 poly(A) site. Finally, the L4 poly(A) site within the E3 intron was also used when additional sequence, including splicing signals from the L4 region and the tripartite leader, was inserted between the promoter and the E3 processing sites. It appears that splicing is the dominant event in governing production of E3/L4 RNAs.
Full text
PDF
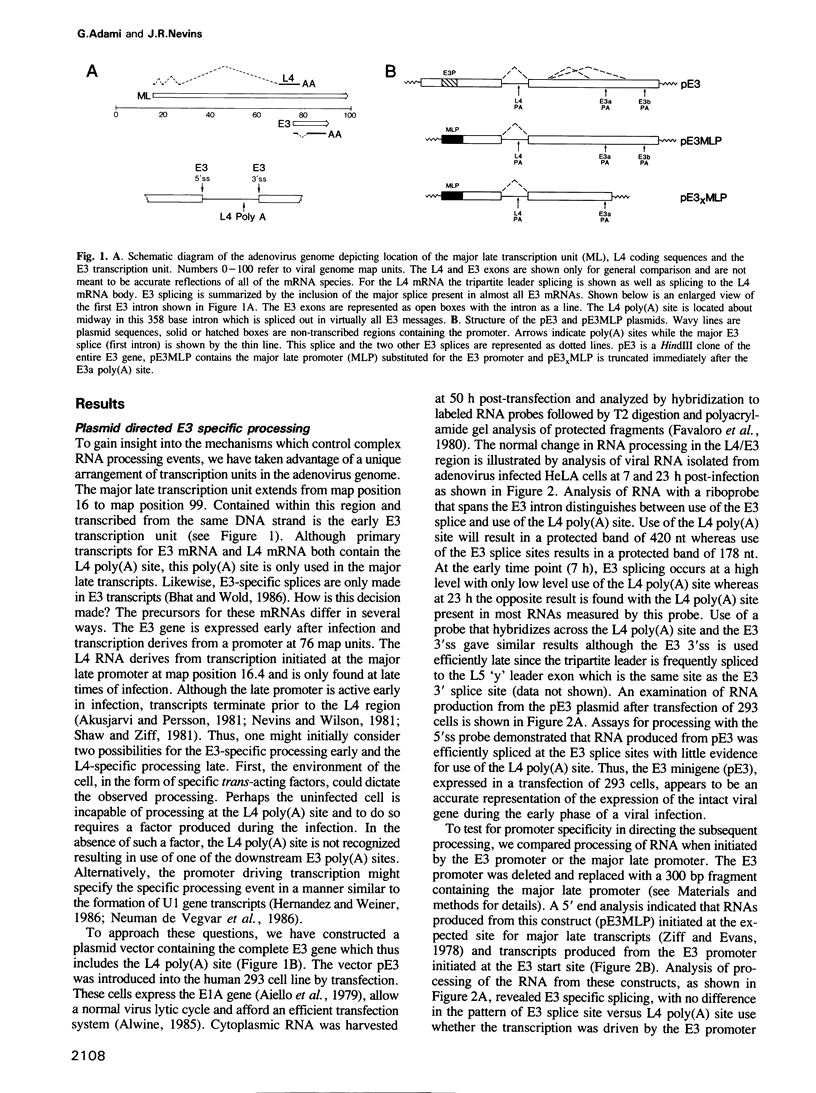
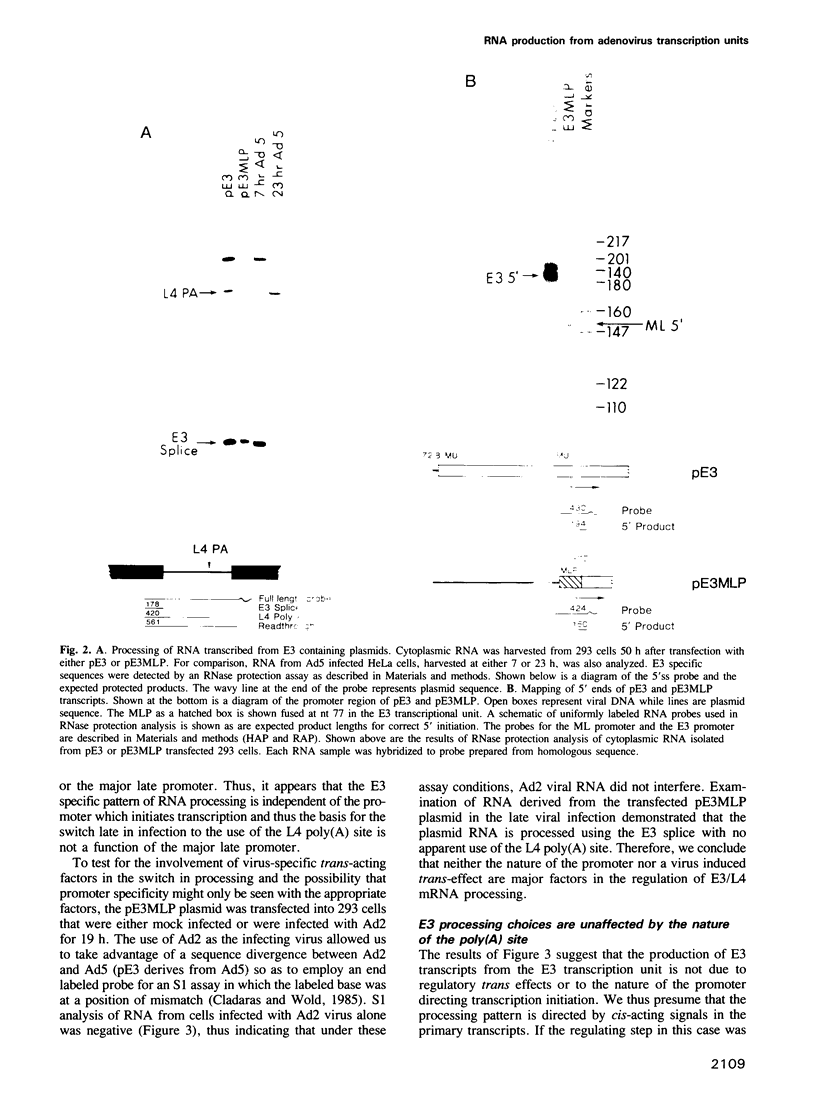


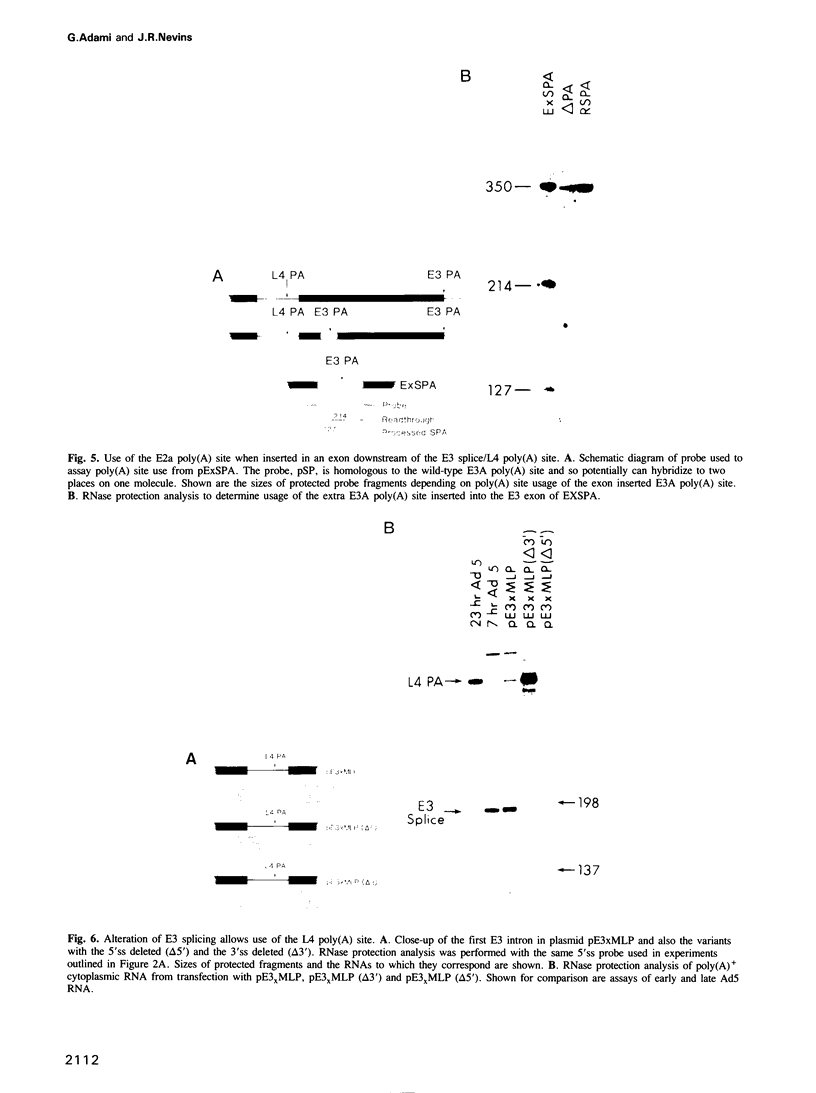
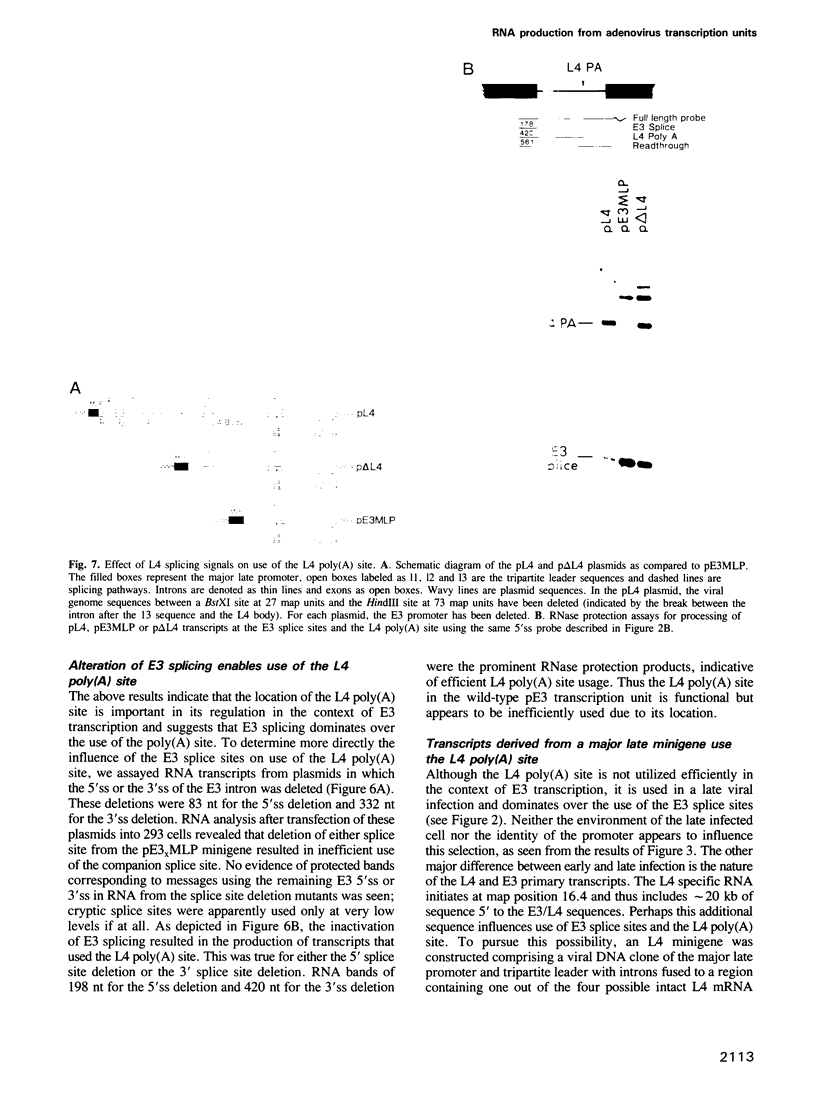



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiello L., Guilfoyle R., Huebner K., Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293). Virology. 1979 Apr 30;94(2):460–469. doi: 10.1016/0042-6822(79)90476-8. [DOI] [PubMed] [Google Scholar]
- Akusjärvi G., Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981 Jul 30;292(5822):420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Alwine J. C. Transient gene expression control: effects of transfected DNA stability and trans-activation by viral early proteins. Mol Cell Biol. 1985 May;5(5):1034–1042. doi: 10.1128/mcb.5.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara S. G., Evans R. M., Rosenfeld M. G. Calcitonin/calcitonin gene-related peptide transcription unit: tissue-specific expression involves selective use of alternative polyadenylation sites. Mol Cell Biol. 1984 Oct;4(10):2151–2160. doi: 10.1128/mcb.4.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. Genetic analysis of mRNA synthesis in adenovirus region E3 at different stages of productive infection by RNA-processing mutants. J Virol. 1986 Oct;60(1):54–63. doi: 10.1128/jvi.60.1.54-63.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Cladaras C., Wold W. S. DNA sequence of the early E3 transcription unit of adenovirus 5. Virology. 1985 Jan 15;140(1):28–43. doi: 10.1016/0042-6822(85)90443-x. [DOI] [PubMed] [Google Scholar]
- Costa R. H., Lai E., Darnell J. E., Jr Transcriptional control of the mouse prealbumin (transthyretin) gene: both promoter sequences and a distinct enhancer are cell specific. Mol Cell Biol. 1986 Dec;6(12):4697–4708. doi: 10.1128/mcb.6.12.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fraser N. W., Nevins J. R., Ziff E., Darnell J. E., Jr The major late adenovirus type-2 transcription unit: termination is downstream from the last poly(A) site. J Mol Biol. 1979 Apr 25;129(4):643–656. doi: 10.1016/0022-2836(79)90474-1. [DOI] [PubMed] [Google Scholar]
- Galli G., Guise J. W., McDevitt M. A., Tucker P. W., Nevins J. R. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted mu-chain expression during B-cell development. Genes Dev. 1987 Jul;1(5):471–481. doi: 10.1101/gad.1.5.471. [DOI] [PubMed] [Google Scholar]
- Galli G., Guise J., Tucker P. W., Nevins J. R. Poly(A) site choice rather than splice site choice governs the regulated production of IgM heavy-chain RNAs. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2439–2443. doi: 10.1073/pnas.85.8.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Ali H., Nevins J. R. Definition of essential sequences and functional equivalence of elements downstream of the adenovirus E2A and the early simian virus 40 polyadenylation sites. Mol Cell Biol. 1985 Nov;5(11):2975–2983. doi: 10.1128/mcb.5.11.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Chen-Kiang S. Processing of adenovirus nuclear RNA to mRNA. Adv Virus Res. 1981;26:1–35. doi: 10.1016/s0065-3527(08)60419-4. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. Processing of late adenovirus nuclear RNA to mRNA. Kinetics of formation of intermediates and demonstration that all events are nuclear. J Mol Biol. 1979 Jun 5;130(4):493–506. doi: 10.1016/0022-2836(79)90436-4. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Wilson M. C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981 Mar 12;290(5802):113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- Osheim Y. N., Miller O. L., Jr, Beyer A. L. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985 Nov;43(1):143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parker B. A., Stark G. R. Regulation of simian virus 40 transcription: sensitive analysis of the RNA species present early in infections by virus or viral DNA. J Virol. 1979 Aug;31(2):360–369. doi: 10.1128/jvi.31.2.360-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Early P., Carter C., Calame K., Bond M., Hood L., Wall R. Two mRNAs with different 3' ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980 Jun;20(2):303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- Shaw A. R., Ziff E. B. Transcripts from the adenovirus-2 major late promoter yield a single early family of 3' coterminal mRNAs and five late families. Cell. 1980 Dec;22(3):905–916. doi: 10.1016/0092-8674(80)90568-1. [DOI] [PubMed] [Google Scholar]
- Sittler A., Gallinaro H., Jacob M. In vivo splicing of the premRNAs from early region 3 of adenovirus-2: the products of cleavage at the 5' splice site of the common intron. Nucleic Acids Res. 1986 Feb 11;14(3):1187–1207. doi: 10.1093/nar/14.3.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]









