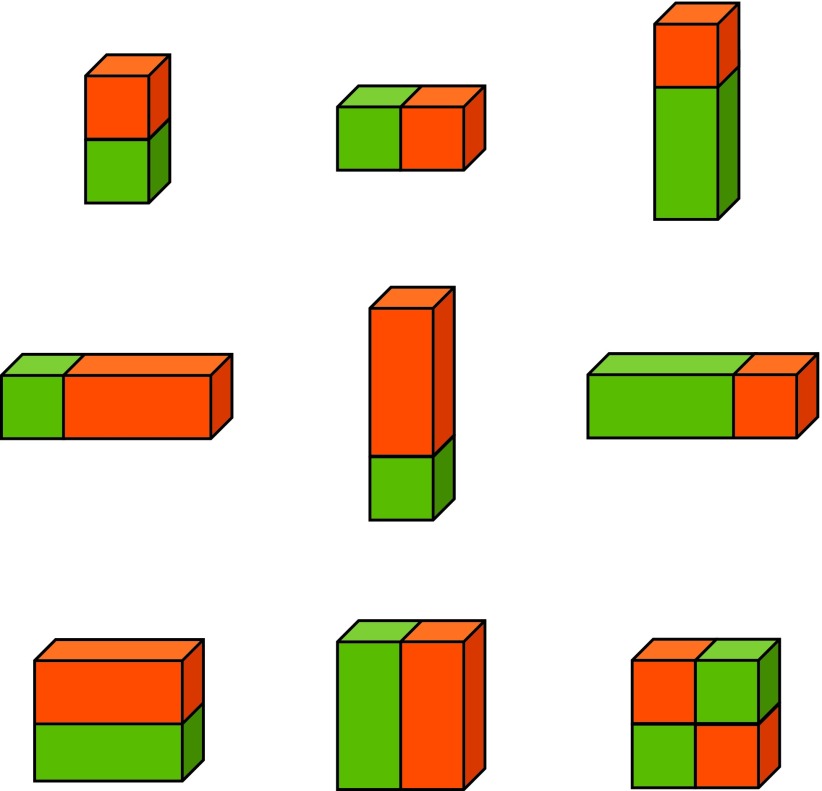Abstract
Purpose:
Positron emission tomography (PET) is a nuclear medical imaging technology that produces 3D images reflecting tissue metabolic activity in human body. PET has been widely used in various clinical applications, such as in diagnosis of brain disorders. High-quality PET images play an essential role in diagnosing brain diseases/disorders. In practice, in order to obtain high-quality PET images, a standard-dose radionuclide (tracer) needs to be used and injected into a living body. As a result, it will inevitably increase the patient’s exposure to radiation. One solution to solve this problem is predicting standard-dose PET images using low-dose PET images. As yet, no previous studies with this approach have been reported. Accordingly, in this paper, the authors propose a regression forest based framework for predicting a standard-dose brain [18F]FDG PET image by using a low-dose brain [18F]FDG PET image and its corresponding magnetic resonance imaging (MRI) image.
Methods:
The authors employ a regression forest for predicting the standard-dose brain [18F]FDG PET image by low-dose brain [18F]FDG PET and MRI images. Specifically, the proposed method consists of two main steps. First, based on the segmented brain tissues (i.e., cerebrospinal fluid, gray matter, and white matter) in the MRI image, the authors extract features for each patch in the brain image from both low-dose PET and MRI images to build tissue-specific models that can be used to initially predict standard-dose brain [18F]FDG PET images. Second, an iterative refinement strategy, via estimating the predicted image difference, is used to further improve the prediction accuracy.
Results:
The authors evaluated their algorithm on a brain dataset, consisting of 11 subjects with MRI, low-dose PET, and standard-dose PET images, using leave-one-out cross-validations. The proposed algorithm gives promising results with well-estimated standard-dose brain [18F]FDG PET image and substantially enhanced image quality of low-dose brain [18F]FDG PET image.
Conclusions:
In this paper, the authors propose a framework to generate standard-dose brain [18F]FDG PET image using low-dose brain [18F]FDG PET and MRI images. Both the visual and quantitative results indicate that the standard-dose brain [18F]FDG PET can be well-predicted using MRI and low-dose brain [18F]FDG PET.
Keywords: positron emission tomography (PET), [18F]FDG, brain [18F]FDG PET prediction, regression forest
1. INTRODUCTION
Positron emission tomography (PET) is a molecular imaging technique that produces 3D images reflecting tissue metabolic activity in the human body. Ever since the PET imaging scanner was built in the early 1970s,1,2 it has been widely used in diagnosing a variety of cancers3,4 and cardiovascular diseases.5,6 Moreover, PET has also been widely used for clinically diagnosing brain diseases/disorders.7–11
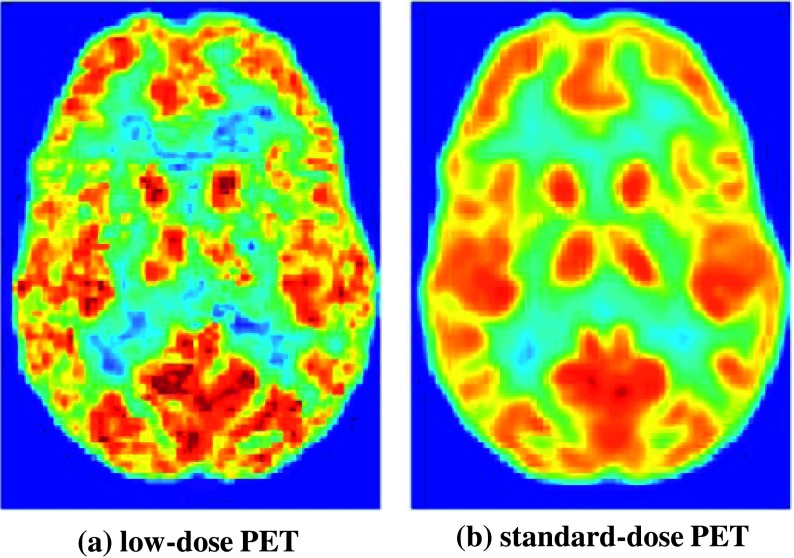
High-quality PET images play a crucial role in diagnosing brain diseases/disorders and are preferred in clinical practice for better diagnosis and assessment. Noise in PET images is mainly caused by the finite number of detected photons. Factors limiting photon counts include the amount of injected tracer, scan duration, and so on.4 Therefore, the total radioactivity injection is important to PET image acquisition, as higher radioactivity (and therefore higher radiation dose for the patient) will generate more detected events and thus obtain images with much higher quality as shown in Fig. 1. However, due to concerns about internal radiation exposure, it would be desirable to reduce the injected radioactivity, but image quality would be compromised as a result. Clearly, as shown in Fig. 1, the quality of a low-dose brain [18F] FDG PET image is inferior to that of a standard-dose brain [18F]FDG PET image. The quality of the low-dose PET image will be further decreased due to various unknown factors during the process of acquisition. Consequently, it will affect the accurate diagnosis of diseases/disorders. In practice, in order to obtain a high-quality brain PET image, a standard-dose radionuclide (tracer) needs to be injected into the living body. As a result, it will inevitably increase a patient’s exposure to radiation. Similar to computed tomography (CT) imaging, the total exposure dose to a patient should be considered,12 particularly when repetitive examinations are required for therapeutic monitoring. Therefore, reducing radiation exposure is very important for patients, especially children and younger patients.13
FIG. 1.
An example of low-dose brain [18F]FDG PET and its corresponding standard-dose brain [18F]FDG PET.
Although many methods have been proposed to improve PET image quality, in the literature, most of them are mainly designed to overcome the limitation of PET imaging rather than to reduce dosage, such as partial volume correction,14,15 motion correction,16,17 and attenuation correction.18–20 To the best of our knowledge, no previous studies have been reported to predict a standard-dose PET image using a low-dose PET image or the combination of low-dose PET and magnetic resonance imaging (MRI) images.
In the process of acquiring PET images, image reconstruction will benefit from the use of anatomical information obtained from other imaging modalities, such as MRI.21 Recently, the combined PET/MRI imaging system has been developed as an alternative to PET/CT, which has matured into an important diagnostic tool.13 The major advantage of this dual-modality imaging system over the stand-alone PET is that, in a PET/MRI imaging system, the excellent soft-tissue contrast of MRI complements the molecular information of PET. Therefore, the combined PET/MRI system provides benefits for predicting standard-dose brain [18F]FDG PET images using low-dose brain [18F] FDG PET and MRI images.
Random forest, an ensemble method,22 has been proven to be one of the most powerful tools in the machine learning community, and has recently gained significantly more popularity for both classification and regression problems, such as remote sensing image classification,23,24 medical image segmentation,25–27 human diseases/disorders diagnosis,28–30 facial analysis,31 and so on. Note that, when applied to the nonlinear regression task, random forest is often called a regression forest (RF).
Accordingly, in this paper, we propose a regression forest-based framework to predict a standard-dose brain [18F]FDG PET image by using both low-dose brain [18F]FDG PET and MRI images. Our method consists of two major steps: (1) predicting an initial standard-dose brain [18F]FDG PET image by brain-tissue-specific regression forest models with image appearance features extracted from both low-dose brain [18F]FDG PET and MRI images and (2) incrementally refining the predicted results by iteratively estimating the image difference between the current prediction and the target standard-dose brain [18F]FDG PET (ground truth). By incrementally adding the estimated image difference onto the previously predicted standard-dose brain [18F]FDG PET, our proposed method is able to gradually improve the quality of predicted standard-dose brain [18F]FDG PET.
2. METHODS
2.A. Datasets and preprocessing
2.A.1. Data description
This study has been approved by the UNC Institutional Review Board. Written informed consent forms were also obtained from all participating patients. The dataset we used consists of 11 subjects. All data were acquired on a Siemens Biograph mMR MR-PET system. Seven subjects were observed to have normal FDG uptake where one subject had a regional uptake deficit in the left frontal lobe, and other three subjects were observed to have mild cognitive impairment (MCI). Subjects were administered an average of 203 MBq (range: 191–229 MBq) of [18F]FDG. The first brain PET scan (the “standard-dose” scan) was performed for a full 12 min with 60 min of injection, according to standard protocols. Immediately after, a second PET dataset was acquired in list-mode for 12 min, which was broken up into separate 3-min sets (the “low-dose” scans). We use the terms standard-dose and low-dose to refer to standard activity administered (averaging 203 MBq and effective dose of 3.86 mSv32) and low activity administered (approximately 51 MBq and effective dose of 0.97 mSv) since effective doses can generally be considered proportional to administered activity.32 The standard-dose activity level is at the low end of the range (185–740 MBq) for FDG brain PET recommended by the Society of Nuclear Medicine and Molecular Imaging (SNMMI);32 thus, the low-dose activity level is significantly lower than the recommended range.
For all subjects, the low-dose PET image sets are completely separate acquisitions from the standard-dose PET image sets. Otherwise, if only the first standard-dose scan was performed, the low-dose PET sets would have to be acquired by breaking up the first scanned standard-dose PET into four 3-min subsets. In this way, the low-dose PET sets and the standard-dose PET sets would actually come from the same set of data, despite difference in dosage. Note that, we used reduced acquisition time at standard-dose as a surrogate for standard acquisition time at reduced dose. Additionally, in our study, because the low-dose datasets were always acquired after the standard-dose ones, some additional uptake in the brain occurred and so the observed count levels in the low-dose sets were 27%–30% those of the standard-dose sets, not the 25% that the change in acquisition time would imply.
PET reconstructions were performed on an offline version of the vendor’s PET-MRI clinical reconstruction software and by using the ordered subsets expectation maximization (OSEM) algorithm.33 Corrections for patient attenuation, scatter, and scanner uniformity were included using the vendor’s standard procedures. Also, a T1-weighted MPRAGE MRI sequence was scanned.
2.A.2. Preprocessing
In our study, we used four images for each subject: MRI, two low-dose PETs (the first 3-min set, low-dose PET 1; and the second 3-min set, low-dose PET 2) and one standard-dose PET. Each image has the size of 128 × 128 × 128, and the voxel size of 2.09 × 2.09 × 2.03 mm3. The MRI data used in our study are not simultaneously scanned with PET data. All images were preprocessed with the following steps. (1) Image alignment: four images of each subject were aligned onto a common space by the algorithm.34 (2) Skull stripping: nonbrain tissue parts were removed from the aligned images.35 (3) Intensity normalization: each modality image was normalized via histogram matching. (4) Tissue segmentation: white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) were segmented from skull-stripped MRI brain image by the algorithm.36
2.B. Overview
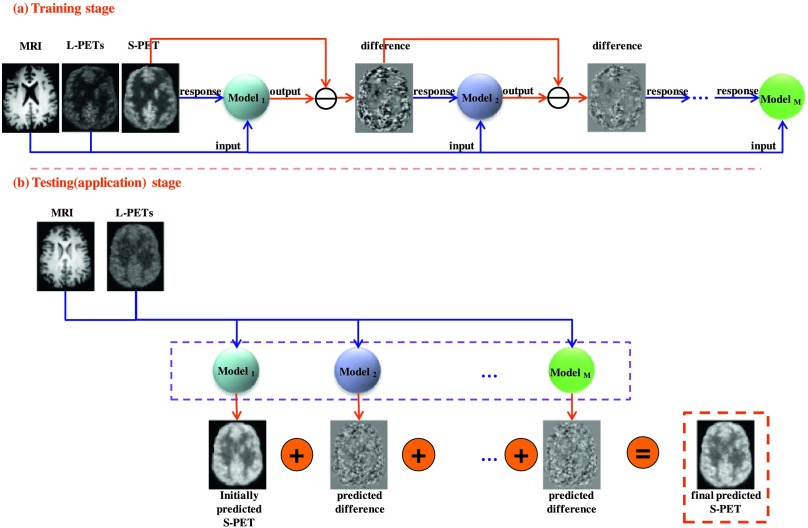
The main goal of this work is to estimate the intensity value of each voxel v ∈ ℝ3 in a standard-dose brain [18F]FDG PET for a new subject. In particular, the random forest22 is employed to estimate the standard-dose brain PET image by voxelwise prediction. Figure 2 shows the overview of our proposed algorithm that consists of training and application stages, where L-PETs stands for the low-dose PETs (low-dose PET 1 and 2), and S-PET stands for the standard-dose PET.
FIG. 2.
Schematic illustration of our proposed standard-dose brain [18F]FDG PET prediction framework, which includes a training stage (a) and an application stage (b). In the training stage (a), M models are sequentially trained by regression forest. In the application stage (b), the constructed M models were simultaneously applied to the new images (low-dose PET and MRI) to produce the final prediction result by adding up the initial prediction result and the predicted difference maps.
Assume we have a set of “pairs” of T1-weighted MRI image, low-dose PETs (low-dose PET 1 and low-dose PET 2), and standard-dose PET (ground truth) training images. We further suppose that the training data have been preprocessed with the procedures described in Sec. 2.A.2. In the training stage, we first extract appearance features (which will be detailed in Sec. 2.D) from each pair of MRI and low-dose PET images, and then use the extracted features as input [while using the corresponding standard-dose PET image (ground truth) as response] to train an initial regression forest (Model1 in Fig. 2). We then use the constructed forest (Model1) to predict the standard-dose PET image for each pair of MRI and low-dose PET images in the training set. Next, in the process of training the later regression forests/models Modelm(m = 2, …, M), we first compute the image difference between the “ground truth” and the standard-dose PET predicted using the Modelm−1 in the training set, and then take the image difference as regression response and the features extracted from the MRI and low-dose PET images as input to train the Modelm(m = 2, …, M). In the testing stage, we extract features from the new MRI and low-dose PET images and simultaneously feed them into the trained Modelm(m = 1, 2, …, M) in order to produce the initial prediction result and the difference maps, respectively. Then, we add up the prediction results (initial prediction result and difference maps) to obtain the final predicted standard-dose brain PET image.
2.C. Multisource integrated tissue-specific prediction with incremental refinement
As shown in Fig. 2, our proposed algorithm involves two major steps: the initial standard-dose PET prediction using Model1 and the incremental refinement using Modelm(m = 2, …, M).
2.C.1. Initial prediction of standard-dose PET by multisource integrated tissue-specific regression forests
Due to the large number of voxels in a human brain image, it is intractable to learn a global regression model for predicting a standard-dose PET image over the entire brain. Many studies37,38 have shown that learning multiple local models would help improve the prediction performance, compared to a single global model. Therefore, in this paper, we learn one regression forest for each brain tissue, i.e., WM, GM, and CSF.
Suppose that the training data have been preprocessed with the procedures described in Sec. 2.A.2. To train the regression forest for each tissue type, we first randomly sample a set of training samples/points within this tissue region for each training subject. Specifically, for the ith sample/point at position v ∈ ℝ3, we extract the patch-based features fi (i = 1, …, N) (which will be detailed in Sec. 2.D) centered at position v from both MRI and low-dose PET images (low-dose PET 1 and low-dose PET 2) for serving as the input. The voxel intensity yi(i = 1, …, N) of the corresponding standard-dose PET image at position v is taken as the regression target/response.
Next, each binary decision tree in a regression forest is trained independently by splitting the training voxels at each split node based on their features and then passing the voxels to either the left or right child node for further splitting. Specifically, for each voxel point ν, a binary decision is performed: , where πj(f) indicates the response of the jth feature in the input feature vector f, and θ is a threshold. According to the result of the decision function, the training voxel ν will be dispatched to either its left or right child node. The purpose of training is to learn the optimal combination of j and θ for each split node by maximizing the average variance decrease in each dimension of the regression response after splitting. Finally, the optimal combination of j and θ is stored at the node for testing. The training of a binary decision tree starts with finding the optimal combination at the root node, and recursively proceeds to child nodes until either the maximum tree depth is reached or the number of training samples is too small to split. Finally, the leaf node stores the average regression target/response of training samples falling into this node. The final prediction of a random forest is the average over the predictions of all its individual trees.
2.C.2. Incremental refinement by estimating image difference
The initially predicted standard-dose PET image using regression forest in the first step should appear to be very similar to the target standard-dose PET image (ground truth). However, due to the existence of fundamental differences among the MRI, the low-dose PET, and the target standard-dose PET, especially the difference between the MRI and the target standard-dose PET, it is difficult to capture the true relationship between the MRI and the target standard-dose PET and also between the low-dose PETs and the target standard-dose PET. Therefore, the inevitable discrepancies between the predicted standard-dose PET and the ground truth may still exist after the initial predicting.
To solve the above problem, motivated by the success of ensemble models,39–43 we propose an incremental refinement scheme to iteratively improve the quality of the predicted standard-dose PET image. To accomplish this, we learn a sequence of tissue-specific regression forests for gradually minimizing the image difference between the predicted and the target standard-dose PET images during the training stage. In particular, the tissue-specific regression forests at iteration m aim to estimate the image difference between the predicted standard-dose PET image by the previous m − 1 iterations and the target standard-dose PET image. Specifically, similar to training tissue-specific regression forests (as described in Sec. 2.C.1) for predicting initial standard-dose PET image, we will first randomly sample a set of training samples/points within each tissue region for each training subject, where is the features extracted from MRI and low-dose PETs, and the is the regression target/response which is the real difference between the ground truth and the standard-dose PET image predicted in the previous step. Next, the training of each decision tree in the regression forest is similar to the training procedure of the first iteration described in Sec. 2.C.1. The only difference is that during forest training of the refinement step, the response is not the voxel intensity yi(i = 1, …, N) of the ground truth, but the real difference computed between the ground truth and the previous predicted standard-dose PET image. In this way, we can learn a sequence of tissue-specific regression forests during the training stage, which will be in charge of estimating the image differences within their respective tissue region.
During testing/application stage, given a new subject with MRI and low-dose PET images, the learned tissue-specific regression forests [Modelm(m = 1, 2, …, M) as shown in Fig. 2] can be applied simultaneously to obtain a final predicted standard-dose PET image. Specifically, in the first iteration, only the technique described in Sec. 2.C.1 is adopted to predict an initial standard-dose PET image. Starting from the second iteration, the procedures of prediction work as follows: (1) for each voxel in the new subject, extract the patch-based appearance features fnew from both low-dose PET and MRI images; (2) based on the segmented tissue labels, simultaneously apply the corresponding constructed tissue-specific regression forests [Modelm(m = 2, …, M)] on fnew to get the difference estimations; (3) add all the estimated image differences onto the initially predicted standard-dose PET to obtain the final prediction.
2.D. Appearance features
The appearance features capture the local information around each voxel in the image. Haar features44 have been found effective in a wide range of applications and can be calculated efficiently at different scales by using integral images. In this paper, the appearance features we used include voxel intensities and 3D Haar-like features45 as shown in Fig. 3. For each training voxel, we compute the 3D Haar-like features at specific locations in a detection window by summing up the voxel intensities within green and orange cuboids, respectively, and then computing the difference between them. Finally, all these calculated features, together with voxel intensities, are concatenated to form a feature representation for the voxel.
FIG. 3.
3D Haar-like feature templates. Orange and green blocks indicate positive and negative blocks in Haar-like features.
3. EXPERIMENTS AND RESULTS
In this section, we will evaluate the performance of our proposed approach using a dataset consisting of 11 subjects. Leave-one-out cross-validation (LOOCV), which has been adopted in numerous papers,46–48 will be used for evaluating the performance of our approach. The dataset and preprocessing steps were described in detail in Sec. 2.A. The selection of parameter values is based on the cross-validation mentioned in Sec. 3.B.
3.A. Quantitative metrics for validating the predicted results
In order to quantitatively validate the performance of our proposed approach, we employ the commonly used metric, i.e., the mean standardized uptake value [SUV(mean)] within the region of interests (ROIs), to evaluate brain [18F]FDG PET activity. The SUV was calculated by normalizing the radioactive concentration to the injected dose corrected for patient weight,49
| (1) |
In our study, we selected eight ROIs and then computed the mean and standard deviation of SUVs in each region for each subject, and compared those values between the predicted standard-dose PET and the ground truth. The better prediction result is indicated by the smaller mean SUV difference [SUV(mean)_Diff] between the prediction result and the ground truth. The selected ROIs are the left frontal, right frontal, left parietal, right parietal, left temporal, right temporal, left occipital, and right occipital, respectively.
3.B. Influence of the parameters
In our proposed method, the parameter settings (i.e., the number of trees, maximal depth of trees, minimal number of samples at leaf node, and patch size) were determined via leave-one-out cross-validation on all training data. In the process of optimizing parameters, other parameters were set to their fixed values while optimizing a certain parameter. For instance, as shown in Fig. 4(a), we first study the influence of the number of trees on the prediction performance. We set the maximal depth of trees to 15, the minimal number of samples at leaf node to 5, and the patch size to 9 × 9 × 9, respectively. As shown in Fig. 4(a), we can see that “15” (15 trees) generate the best performance [i.e., with the smallest SUV(mean)_Diff] in our study.
FIG. 4.

Influence of parameter values on prediction performance.
The techniques for optimizing other parameters are similar to those for optimizing the number of trees. Figure 4(b) shows the influence of the maximal tree depth on prediction results. In our study, we note that the performance is significantly improved from depth of 5 to 15, slightly improved from depth of 15 to 25, and stably kept while the depth of trees is beyond 20. Figure 4(c) shows the influence of the minimal number of samples at the leaf node. We can see that the best performance is obtained when the minimal number of samples at leaf node equals to 2. Figure 4(d) shows the influence of patch size on the prediction performance. As we observed, increasing the patch size gradually improves the prediction performance. In addition, we also note that the prediction performance will not be significantly improved, when the patch size is beyond 11 × 11 × 11. Hence, in this paper, we select the patch size as 11 × 11 × 11, which is suitable for estimating reasonable predictions.
Finally, in this paper, we choose the following parameters in all experiments—the number of trees: 15; the depth of trees: 20; the minimal number of samples at leaf node: 2; the patch size: 11 × 11 × 11.
3.C. Predicted results with visual and statistical analysis
In this subsection, we will visually and statistically demonstrate the prediction results using our proposed method, i.e., multisource (low-dose PET 1, low-dose PET 2, and MRI) integrated, tissue-specific, two-step (initial prediction and incremental refinement) models.
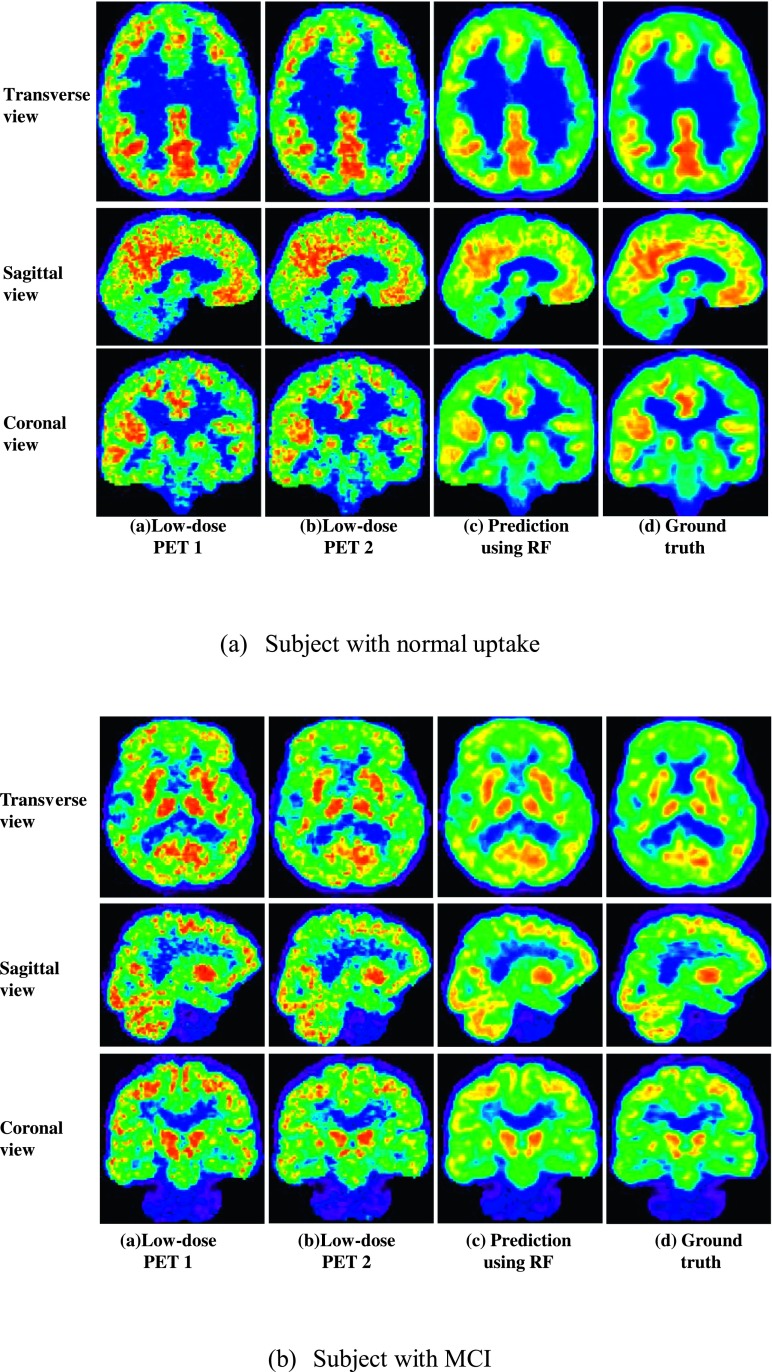
Figure 5 shows the qualitative prediction results on two different subjects using our proposed method. Here, for simplicity, we denote our proposed method as RF.
FIG. 5.
Prediction results on two different subjects using our proposed method (RF).
From Fig. 5, we can see that, compared to two low-dose PETs, the image qualities of the predicted standard-dose brain [18F]FDG PETs were substantially improved. Additionally, we also note that, both for the subject with normal uptake and for the subject with MCI, the predicted standard-dose PET images look very similar in appearance with the corresponding ground truth (i.e., the original standard-dose brain PET). All these indicate that our proposed method achieves desirable predictions.
In order to quantitatively compare the brain [18F]FDG PET activity between the predicted standard-dose PET and the ground truth, Table I tabulates the results in terms of SUV(mean) and its corresponding standard deviation within the selected ROIs.
TABLE I.
Mean SUV within each of the eight ROIs.
| Prediction result | Ground truth | SUV difference | |
|---|---|---|---|
| ROI 1 | 5.72 ± 1.51 | 5.74 ± 1.67 | 0.02 |
| ROI 2 | 5.80 ± 1.53 | 5.81 ± 1.77 | 0.01 |
| ROI 3 | 5.91 ± 1.73 | 5.96 ± 1.89 | 0.04 |
| ROI 4 | 5.90 ± 1.78 | 5.94 ± 1.99 | 0.04 |
| ROI 5 | 6.31 ± 2.01 | 6.35 ± 2.17 | 0.03 |
| ROI 6 | 6.23 ± 2.01 | 6.26 ± 2.20 | 0.02 |
| ROI 7 | 6.84 ± 2.03 | 6.87 ± 2.17 | 0.03 |
| ROI 8 | 6.99 ± 2.06 | 7.04 ± 2.22 | 0.05 |
| Average | 6.21 ± 1.83 | 6.25 ± 2.01 | 0.03 |
Note that the value for each ROI in Table I is calculated by averaging the values within the same ROIs from the 11 subjects.
From Table I, we can see that the predicted standard-dose PETs are close to the corresponding ground truths in terms of SUV(mean). It is clear that our method gives an unbiased estimate of the standard-dose image.
Moreover, in order to demonstrate the quality improvement of predicted standard-dose brain [18F]FDG PET image over the original low-dose brain [18F]FDG PET image, Table II tabulates a statistical comparison in terms of the coefficient of variation (CV, a kind of a noise-to-signal ratio) within ROI among the original low-dose PET 1 and low-dose PET 2, the prediction result, and the ground-truth, respectively. The CV is defined as CV = standard deviation/mean. The smaller value of CV means the better performance.
TABLE II.
CV within each of the eight ROIs.
| Low-dose PET 1 | Low-dose PET 2 | Prediction result | Ground truth | |
|---|---|---|---|---|
| ROI 1 | 0.47 | 0.48 | 0.43 | 0.38 |
| ROI 2 | 0.43 | 0.44 | 0.39 | 0.35 |
| ROI 3 | 0.44 | 0.45 | 0.40 | 0.35 |
| ROI 4 | 0.42 | 0.43 | 0.38 | 0.34 |
| ROI 5 | 0.34 | 0.34 | 0.29 | 0.28 |
| ROI 6 | 0.33 | 0.33 | 0.28 | 0.27 |
| ROI 7 | 0.32 | 0.31 | 0.25 | 0.27 |
| ROI 8 | 0.29 | 0.29 | 0.22 | 0.25 |
| Average | 0.38 | 0.38 | 0.33 | 0.31 |
From Table II, it can be seen that the CV is higher for the low-dose PET images, but the CV of the predicted standard-dose PET images is close to that of the ground truth images. Hence, compared with the image quality of low-dose brain [18F]FDG PETs, the quality of predicted standard-dose brain [18F]FDG PET using our proposed method is much better. Furthermore, based on the results listed in Table II, we performed the null hypothesis tests to demonstrate the statistical significance of the improvement. The calculated P-value between the low-dose PET 1 and the predicted standard-dose PET, the low-dose PET 2 and the predicted standard-dose PET, is well below 0.05.
Note that the value for each ROI in Table II is calculated by averaging the values within the same ROIs from the 11 subjects.
4. DISCUSSION AND CONCLUSION
In this paper, we propose a regression forest-based framework for generating standard-dose brain [18F]FDG PET images. Different from traditional techniques for acquiring the standard-dose brain [18F]FDG PET, the proposed method utilizes low-dose brain [18F]FDG PETs, combined with MRI image, to predict standard-dose brain [18F]FDG PET, thus relatively reducing the radionuclide dose. Specifically, tissue-specific models are built to separately predict standard-dose brain [18F]FDG PET in each brain tissue. In addition, an incremental refinement strategy is also employed for estimating the image difference. Experimental results show that our proposed method substantially improved the quality of low-dose brain [18F]FDG PETs and is able to achieve very promising predictions for standard-dose brain [18F]FDG PET. To the best of our knowledge, this is the first time, in the literature, showing that the standard-dose brain [18F]FDG PET can be well-predicted using MRI and low-dose brain [18F]FDG PET.
Although our proposed method demonstrates good performance from cross-validations, several limitations should be indicated for the present study. First, we currently perform only a voxelwise prediction for estimating standard-dose brain [18F]FDG PET, i.e., each voxel’s intensity in standard-dose brain [18F]FDG PET is predicted independently. Thus, the proposed method may overlook the intrinsic relationship between the central voxel and its neighboring voxels in a patch. In our future work, we plan to extend our work in a patchwise fashion. Second, we use a dataset consisting of 11 subjects to evaluate the performance of our proposed method. The number of subjects is limited, but we are currently collecting more subjects, which will be used to further test the generality of our proposed method. Also, while our population included one case of abnormal uptake and three MCI subjects, it certainly does not adequately represent the range of possible uptake states. Third, our current method mainly focuses on the brain, and only uses [18F]FDG to generate PET images. Future research will apply the same method to other anatomical targets as well as to the PET images generated by other kinds of tracers. In addition, the proposed method will be extended to brain perfusion studies as well. Finally, we note that all conclusions in this work are based on quantitative measures. They indicate that our method reduces noise while imparting minimal bias on regional SUV estimates. It is not yet known how the method performs in a clinical task, so no conclusions can be drawn about the clinical value of the method. Future plans include a formal expert-observer study to evaluate the method in a clinical task.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health Grant Nos. MH100217, AG042599, EB006733, EB008374, EB009634, NS055754, MH064065, and HD053000.
REFERENCES
- 1.Phelps M. E., Hoffman E. J., Mullani N. A., and Ter-Pogossian M. M., “Application of annihilation coincidence detection to transaxial reconstruction tomography,” J. Nucl. Med. 16, 210–224 (1975). [PubMed] [Google Scholar]
- 2.Ter-Pogossian M. M., Phelps M. E., Hoffman E. J., and Mullani N. A., “A positron-emission transaxial tomograph for nuclear imaging (PETT),” Radiology 114, 89–98 (1975). 10.1148/114.1.89 [DOI] [PubMed] [Google Scholar]
- 3.Pace L., Nicolai E., Luongo A., Aiello M., Catalano O. A., Soricelli A., and Salvatore M., “Comparison of whole-body PET/CT and PET/MRI in breast cancer patients: Lesion detection and quantitation of 18F-deoxyglucose uptake in lesions and in normal organ tissues,” Eur. J. Radiol. 83, 289–296 (2014). 10.1016/j.ejrad.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 4.Bai B., Bading J., and Conti P. S., “Tumor quantification in clinical positron emission tomography,” Theranostics 3, 787–801 (2013). 10.7150/thno.5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saboury B., Ziai P., and Alavi A., “Role of global disease assessment by combined PET-CT-MR imaging in examining cardiovascular disease,” PET Clin. 6, 421–429 (2011). 10.1016/j.cpet.2011.10.003 [DOI] [PubMed] [Google Scholar]
- 6.Schindler T. H., Schelbert H. R., Quercioli A., and Dilsizian V., “Cardiac PET imaging for the detection and monitoring of coronary artery disease and microvascular health,” J. Am. Coll. Cardiol. 3, 623–640 (2010). 10.1016/j.jcmg.2010.04.007 [DOI] [PubMed] [Google Scholar]
- 7.Garraux G., Phillips C., Schrouff J., Kreisler A., Lemaire C., Degueldre C., Delcour C., Hustinx R., Luxen A., Destée A., and Salmon E., “Multiclass classification of FDG PET scans for the distinction between Parkinson’s disease and atypical parkinsonian syndromes,” NeuroImage: Clin. 2, 883–893 (2013). 10.1016/j.nicl.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F., Wee C. Y., Chen H., and Shen D., “Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s disease and mild cognitive impairment identification,” NeuroImage 84, 466–475 (2014). 10.1016/j.neuroimage.2013.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D., Wang Y., Zhou L., Yuan H., and Shen D., “Multimodal classification of Alzheimer’s disease and mild cognitive impairment,” NeuroImage 55, 856–867 (2011). 10.1016/j.neuroimage.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D., Shen D., and Alzheimer’s Disease Neuroimaging Initiative, “Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers,” PLoS One 7, e33182 (2012). 10.1371/journal.pone.0033182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Wolk D. A., Reddin J. S., Korczykowski M., Martinez P. M., Musiek E. S., Newberg A. B., Julin P., Arnold S. E., Greenberg J. H., and Detre J. A., “Voxel-level comparison of arterial spin-labeled perfusion MRI and FDG-PET in Alzheimer disease,” Neurology 77, 1977–1985 (2011). 10.1212/WNL.0b013e31823a0ef7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J., Huang J., Feng Q., Zhang H., Lu H., Liang Z., and Chen W., “Low-dose computed tomography image restoration using previous normal-dose scan,” Med. Phys. 38, 5713–5731 (2011). 10.1118/1.3638125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichler B. J., Kolb A., Nagele T., and Schlemmer H. P., “PET/MRI: Paving the way for the next generation of clinical multimodality imaging applications,” J. Nucl. Med. 51, 333–336 (2010). 10.2967/jnumed.109.061853 [DOI] [PubMed] [Google Scholar]
- 14.Coello C., Willoch F., Selnes P., Gjerstad L., Fladby T., and Skretting A., “Correction of partial volume effect in 18F-FDG PET brain studies using coregistered MR volumes: Voxel based analysis of tracer uptake in the white matter,” NeuroImage 72, 183–192 (2013). 10.1016/j.neuroimage.2013.01.043 [DOI] [PubMed] [Google Scholar]
- 15.Lehnert W., Gregoire M. C., Reilhac A., and Meikle S. R., “Characterization of partial volume effect and region-based correction in small animal positron emission tomography (PET) of the rat brain,” NeuroImage 60, 2144–2157 (2012). 10.1016/j.neuroimage.2012.02.032 [DOI] [PubMed] [Google Scholar]
- 16.Gigengack F., Ruthotto L., Burger M., Wolters C. H., Jiang X., and Schafers K. P., “Motion correction in dual gated cardiac PET using mass-preserving image registration,” IEEE Trans. Med. Imaging 31, 698–712 (2012). 10.1109/TMI.2011.2175402 [DOI] [PubMed] [Google Scholar]
- 17.Olesen O. V., Sullivan J. M., Mulnix T., Paulsen R. R., Hojgaard L., Roed B., Carson R. E., Morris E. D., and Larsen R., “List-mode PET motion correction using markerless head tracking: Proof-of-concept with scans of human subject,” IEEE Trans. Med. Imaging 32, 200–209 (2013). 10.1109/TMI.2012.2219693 [DOI] [PubMed] [Google Scholar]
- 18.Andersen F. L., Ladefoged C. N., Beyer T., Keller S. H., Hansen A. E., Højgaard L., Kjær A., Law I., and Holm S., “Combined PET/MR imaging in neurology: MR-based attenuation correction implies a strong spatial bias when ignoring bone,” NeuroImage 84, 206–216 (2014). 10.1016/j.neuroimage.2013.08.042 [DOI] [PubMed] [Google Scholar]
- 19.Bai W. and Brady M., “Motion correction and attenuation correction for respiratory gated PET images,” IEEE Trans. Med. Imaging 30, 351–365 (2011). 10.1109/tmi.2010.2078514 [DOI] [PubMed] [Google Scholar]
- 20.Keller S. H., Svarer C., and Sibomana M., “Attenuation correction for the HRRT PET-scanner using transmission scatter correction and total variation regularization,” IEEE Trans. Med. Imaging 32, 1611–1621 (2013). 10.1109/TMI.2013.2261313 [DOI] [PubMed] [Google Scholar]
- 21.Vunckx K., Atre A., Baete K., Reilhac A., Deroose C. M., Van Laere K., and Nuyts J., “Evaluation of three MRI-based anatomical priors for quantitative PET brain imaging,” IEEE Trans. Med. Imaging 31, 599–612 (2012). 10.1109/TMI.2011.2173766 [DOI] [PubMed] [Google Scholar]
- 22.Breiman L., “Random forests,” Mach. Learn. 45, 5–32 (2001). 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 23.Guo L., Chehata N., Mallet C., and Boukir S., “Relevance of airborne lidar and multispectral image data for urban scene classification using random forests,” ISPRS J. Photogramm. Remote Sens. 66, 56–66 (2011). 10.1016/j.isprsjprs.2010.08.007 [DOI] [Google Scholar]
- 24.Ham J., Chen Y., Crawford M. M., and Ghosh J., “Investigation of the random forest framework for classification of hyperspectral data,” IEEE Trans. Geosci. Remote Sens. 43, 492–501 (2005). 10.1109/TGRS.2004.842481 [DOI] [Google Scholar]
- 25.Lindner C., Thiagarajah S., Wilkinson J. M., arcOGEN Consortium, Wallis G. A., and Cootes T. F., “Fully automatic segmentation of the proximal femur using random forest regression voting,” IEEE Trans. Med. Imaging 32, 1462–1472 (2013). 10.1109/TMI.2013.2258030 [DOI] [PubMed] [Google Scholar]
- 26.McIntosh C., Svistoun I., and Purdie T. G., “Groupwise conditional random forests for automatic shape classification and contour quality assessment in radiotherapy planning,” IEEE Trans. Med. Imaging 32, 1043–1057 (2013). 10.1109/TMI.2013.2251421 [DOI] [PubMed] [Google Scholar]
- 27.Yaqub M., Javaid M. K., Cooper C., and Noble J. A., “Investigation of the role of feature selection and weighted voting in random forests for 3-D volumetric segmentation,” IEEE Trans. Med. Imaging 33, 258–271 (2014). 10.1109/TMI.2013.2284025 [DOI] [PubMed] [Google Scholar]
- 28.Azar A. T., Elshazly H. I., Hassanien A. E., and Elkorany A. M., “A random forest classifier for lymph diseases,” Comput. Methods Programs Biomed. 113, 465–473 (2014). 10.1016/j.cmpb.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 29.Gray K. R., Aljabar P., Heckemann R. A., Hammers A., Rueckert D., and Alzheimer’s Disease Neuroimaging Initiative, “Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease,” NeuroImage 65, 167–175 (2013). 10.1016/j.neuroimage.2012.09.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripoliti E. E., Fotiadis D. I., and Manis G., “Automated diagnosis of diseases based on classification: Dynamic determination of the number of trees in random forests algorithm,” IEEE Trans. Inf. Technol. Biomed. 16, 615–622 (2012). 10.1109/TITB.2011.2175938 [DOI] [PubMed] [Google Scholar]
- 31.Fanelli G., Dantone M., Gall J., Fossati A., and Gool L. V., “Random forests for real time 3D face analysis,” Int. J. Comput. Vision 101, 437–458 (2013). 10.1007/s11263-012-0549-0 [DOI] [Google Scholar]
- 32.Waxman A. D., Herholz K., Lewis D. H., Herscovitch P., Minoshima S., Ichise M., Drzezga A. E., Devous M. D., and Mountz J. M., Society of Nuclear Medicine Procedure Guideline for FDG PET Brain Imaging, Version 1.0, Society of Nuclear Medicine, 2009.
- 33.Hudson H. M. and Larkin R. S., “Accelerated image reconstruction using ordered subsets of projection data,” IEEE Trans. Med. Imaging 13, 601–609 (1994). 10.1109/42.363108 [DOI] [PubMed] [Google Scholar]
- 34.Wu G., Jia H., Wang Q., and Shen D., “SharpMean: Groupwise registration guided by sharp mean image and tree-based registration,” NeuroImage 56, 1968–1981 (2011). 10.1016/j.neuroimage.2011.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi F., Wang L., Dai Y., Gilmore J. H., Lin W., and Shen D., “LABEL: Pediatric brain extraction using learning-based meta-algorithm,” NeuroImage 62, 1975–1986 (2012). 10.1016/j.neuroimage.2012.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Brady M., and Smith S., “Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm,” IEEE Trans. Med. Imaging 20, 45–57 (2001). 10.1109/42.906424 [DOI] [PubMed] [Google Scholar]
- 37.Gao Y., Liao S., and Shen D., “Prostate segmentation by sparse representation based classification,” Med. Phys. 39, 6372–6387 (2012). 10.1118/1.4754304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao Y., Gao Y., Guo Y., Shi Y., Yang X., and Shen D., “Hierarchical lung field segmentation with joint shape and appearance sparse learning,” IEEE Trans. Med. Imaging 33, 1761–1780 (2014). 10.1109/TMI.2014.2305691 [DOI] [PubMed] [Google Scholar]
- 39.Natekin A. and Knoll A., “Gradient boosting machines, a tutorial,” Front. Neurorobotics 7, 1–21 (2013). 10.3389/fnbot.2013.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kazemi V. and Sullivan J., “One millisecond face alignment with an ensemble of regression trees,” in 2014 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Columbus, Ohio, 23-28 June (IEEE, New York, NY, 2014), pp. 1867–1874. 10.1109/CVPR.2014.241 [DOI] [Google Scholar]
- 41.Gao Y., Zhan Y., and Shen D., “Incremental learning with selective memory (ILSM): Towards fast prostate localization for image guided radiotherapy,” IEEE Trans. Med. Imaging 33, 518–534 (2014). 10.1109/tmi.2013.2291495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W., Liao S., Feng Q., Chen W., and Shen D., “Learning image context for segmentation of the prostate in CT-guided radiotherapy,” Phys. Med. Biol. 57, 1283–1308 (2012). 10.1088/0031-9155/57/5/1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tu Z. and Bai X., “Auto-context and its application to high-level vision tasks and 3D brain image segmentation,” IEEE Trans. Pattern Anal. Mach. Intell. 32, 1744–1757 (2010). 10.1109/tpami.2009.186 [DOI] [PubMed] [Google Scholar]
- 44.Viola P. and Jones M. J., “Robust real-time face detection,” Int. J. Comput. Vision 57, 137–154 (2004). 10.1023/B:VISI.0000013087.49260.fb [DOI] [Google Scholar]
- 45.Kim M., Wu G., Li W., Wang L., Son Y., Cho Z., and Shen D., “Automatic hippocampus segmentation of 7.0 Tesla MR images by combining multiple atlases and auto-context models,” NeuroImage 83, 335–345 (2013). 10.1016/j.neuroimage.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi F., Fan Y., Tang S., Gilmore J. H., Lin W., and Shen D., “Neonatal brain image segmentation in longitudinal MRI studies,” NeuroImage 49, 391–400 (2009). 10.1016/j.neuroimage.2009.07.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L., Shi F., Li G., Gao Y., Lin W., Gilmore J. H., and Shen D., “Segmentation of neonatal brain MR images using patch-driven level sets,” NeuroImage 84, 141–158 (2014). 10.1016/j.neuroimage.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L., Gao Y., Shi F., Li G., Gilmore J. H., Lin W., and Shen D., “LINKS: Learning-based multi-source IntegratioN frameworK for Segmentation of infant brain images,” NeuroImage 108, 160–172 (2015). 10.1016/j.neuroimage.2014.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higgins K. A., Hoang J. K., Roach M. C., Chino J., Yoo D. S., Turkington T. G., and Brizel D. M., “Analysis of pretreatment FDG-PET SUV parameters in head-and-neck cancer: Tumor SUVmean has superior prognostic value,” Int. J. Radiat. Oncol., Biol., Phys. 82, 548–553 (2012). 10.1016/j.ijrobp.2010.11.050 [DOI] [PubMed] [Google Scholar]