Abstract
This paper describes a method for the quantitative detection of cells expressing BlaC, a β-lactamase naturally expressed by Mycobacterium tuberculosis, intended for the diagnosis of tuberculosis. The method is based on the compartmentalization of bacteria in picoliter droplets at limiting dilutions such that each drop contains one or no cells. The co-encapsulation of a fluorogenic substrate probe for BlaC allows the quantification of bacteria by enumerating the number of fluorescent drops. Quantification of 10 colony forming units per milliliter is demonstrated. Furthermore, the encapsulation of single cell in drops maintains the specificity of the detection scheme even when the concentration of bacteria that do not express BlaC exceeds that expressing BlaC by one million-fold.
I. INTRODUCTION
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), remains one of the world's deadliest infectious diseases with over a million deaths a year.1 A key challenge lies in the diagnosis of TB.1 In recent years, a significant proportion of TB patients are co-infected with HIV.2 HIV-co-infected patients, as well as children infected with TB, tend to have reduced pulmonary bacterial loads.2 The front-line diagnostic test for TB based on acid-fast smear microscopy using sputum is not sufficiently sensitive for such cases: positive smear test requires the presence of at least 5000 colony forming units per milliliter (or cfu/ml),3 and its sensitivity varies from 20% to 80%.2 Isolation and culture of Mtb has a higher sensitivity—possessing a detection limit of 10–100 cells3,4—than acid-fast smear does and has thus remained the gold standard to diagnose TB. Cultures take 4–8 weeks to determine whether the sample is Mtb-positive,5 however. Culturing facilities are also poorly available in developing countries. Immunoassays based on antibody or antigen detection are available but have been shown to have poor sensitivities and specificities due to the large variability in immune response from different patients.6,7 Polymerase chain reaction (PCR)-based assays are rapid (∼2 h) with improved sensitivity of ∼100 cfu/ml. The technology remains expensive, however, and cannot distinguish patients with active TB from patients recently exposed to Mtb.6 These tests also cannot differentiate live from dead cells due to the persistence of DNA after cell death.8
In this work, we use BlaC as the specific enzyme marker for TB diagnostics. Mtb naturally expresses BlaC, an enzyme that belongs to the class A β-lactamase family.9,10 Complete genome sequencing of more than 54 Mtb strains from numerous geographical regions demonstrates that BlaC is highly conserved in all Mtb clinical isolates.11 BlaC should, therefore, be a useful diagnostic marker for TB. Based on the hydrolysis of β-lactam, a number of fluorogenic and bioluminogenic probes have been developed for the detection of β-lactamase activity in vitro, in living cells, and even in whole animals.12–16 Previous probes generally lack specificity for BlaC in Mtb. Other β-lactamases such as the common TEM-1 β-lactamase (TEM-1 Bla) in gram-negative bacteria can also hydrolyze these probes. This non-specificity reduces the probes' utility for TB diagnosis. We have recently reported a BlaC-specific fluorogenic probe (CDG-OMe) based on chemically modified cephalosporins by taking advantage of the unique flexibility of the BlaC substrate-specificity loop.17 This probe demonstrated 200-fold increase in fluorescence upon hydrolysis by BlaC. While it was simple to obtain a positive result in the presence of cells expressing BlaC, the quantification of cell concentration was time-dependent and less reliable.
Here, we describe the use of droplet microfluidics for the quantification of cells expressing BlaC ultimately intended for the detection of TB. The approach is based on the co-encapsulation of BlaC-specific fluorogenic probe CDG-OMe and bacteria sample in a large number (N = 108) of picoliter droplets. To our knowledge, no prior work has applied droplet microfluidics for the detection of TB.18–22 The bacterial sample is prepared at a limiting dilution such that each drop contains one or no cell. We use Escherichia coli (E. coli) expressing BlaC as a surrogate to characterize our method and validate some of our results using Bacillus Calmette–Guérin (BCG), a strain of attenuated Mycobacterium bovis. If cells expressing BlaC are present inside a drop, the probe is hydrolyzed to render the droplet fluorescent (Figure 1(a)). Enumerating drops that are fluorescent allows us to quantify the initial concentration of cells. Combined with an automated droplet counting scheme, we demonstrate the detection of cells at concentrations ranging from 10 to 107 cfu/ml. Our lower detection limit is thus comparable to that of the gold standard based on cultures, which has a detection limit of 10–100 cfu/ml but requires 4–8 weeks to turn positive.23 Furthermore, the encapsulation of single cell in drops maintains the specificity of the detection scheme even when the concentration of bacteria that do not express BlaC exceeds that expressing BlaC by one million-fold.
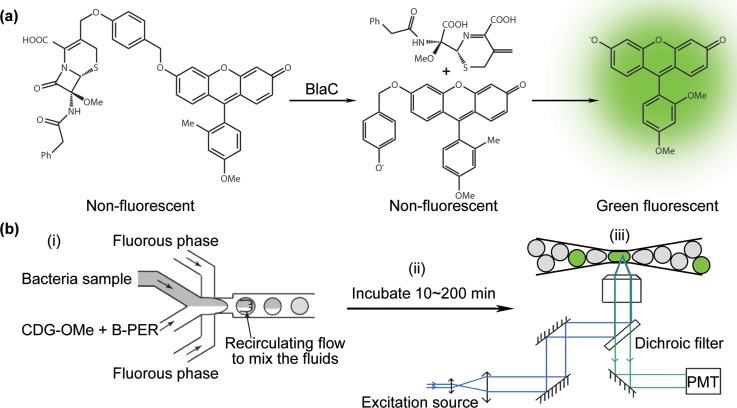
FIG. 1.
(a) Chemical structure of the fluorogenic probe CDG-OMe. (b) Scheme of the microfluidic process flow consisting of 3 parts: (i) Droplet generation and compartmentalization of the sample with probe CDG-OMe and lysis buffer B-PER. (ii) Incubation of the drops for fluorescence to turn on. (iii) Detection of fluorescence from the drops.
II. MATERIALS AND METHODS
A. Microfluidic systems
We used methods in soft lithography to fabricate microchannels in poly(dimethylsiloxane) (PDMS).24,25 The microchannels were rendered hydrophobic by treatment with Aquapel (Pittsburgh, PA) to avoid droplet wetting of the wall. We generated monodisperse droplets using flow-focusing nozzles with a dispersity <1%.26 The continuous phase was a hydrofluoroether HFE-7500 (3M, St. Paul, MN) containing a biocompatible EA-surfactant (RainDance Technologies, Lexington, MA) (2% w/w), a PEG-PFPE amphiphilic block copolymer,27 to stabilize the drops against coalescence. HFE-7500 was inert and permeable to gases and had been shown to be compatible with cell cultures in drops. We used two separate inlets for the disperse phase: one for the bacteria sample in 2-(N-morpholino)ethanesulfonic acid (MES) buffer (0.1M, pH 6.6), and one for a mixture of the probe CDG-OMe (at 20 μM in MES buffer) and lysis buffer “B-PER” (B-PER™ II Bacterial Protein Extraction Reagent (2X), Product No. 78260, Thermo Scientific) at 96% of as-purchased bottle concentration. The flow rate ratio of the fluids pumped into the two inlets was held at 1:1. The final concentration of the probe in a droplet was thus at 10 μM and that of B-PER was at 48% of the as-purchased concentration. The B-PER lysis buffer was needed for improving the efficiency of fluorescence detection (more details in Sec. III. Results and Discussion). The two inlets merged only at the flow-focusing nozzle (Figure 1(b)). All experiments were performed at 20 °C. We generated drops using a standard flow-focusing nozzle (Figure S1 in the supplementary material29). For experiments in Figures 3 and 4, droplet size of ∼30 pl was used. The droplet generation rate was ∼1400 drops/s for a single device. The total sample volume used was 0.2 ml to 1 ml. We collected the drops generated from the flow-focusing nozzles into Eppendorf tubes and incubated them at 20 °C off-chip for 5–200 min.
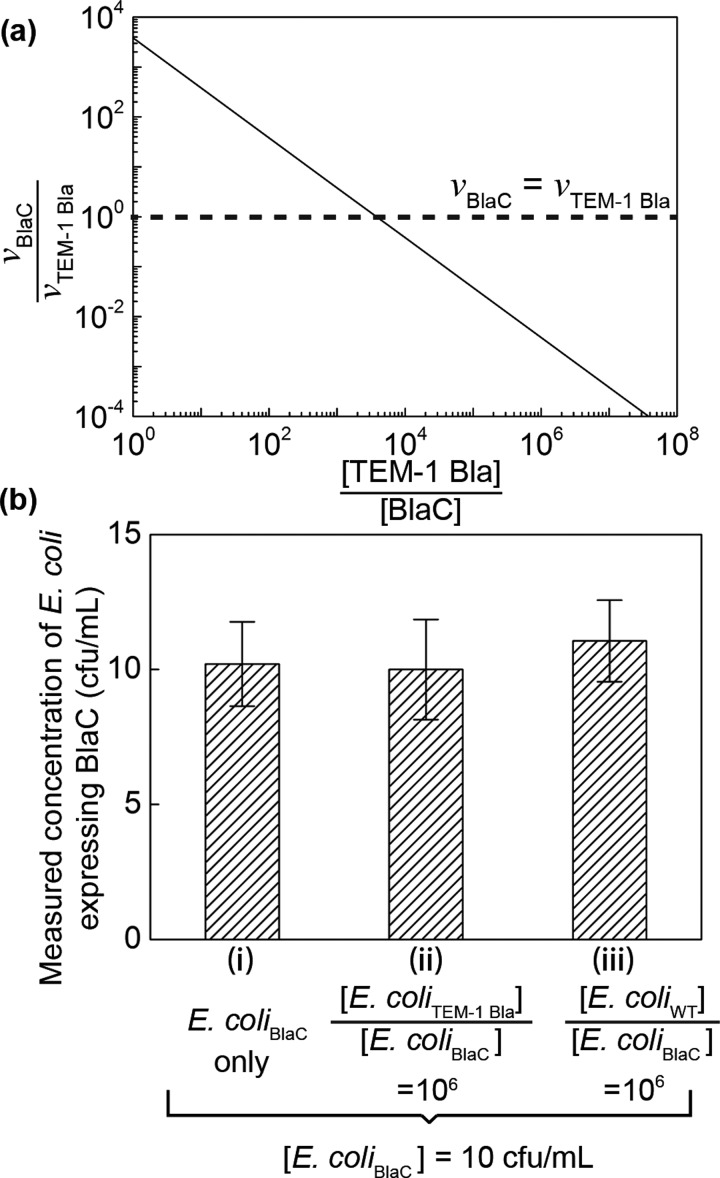
FIG. 3.
(a) The calculated ratio of fluorescence turn-on rate of probe CDG-OMe by BlaC (vBlaC) and by TEM-1 Bla (vTEM-1 Bla), when BlaC and TEM-1 Bla are mixed at different ratios ([TEM-1 Bla]/[BlaC]). When [TEM-1 Bla]/[BlaC] ∼ 103, the turn-on rate of CDG-OMe by BlaC equals that by TEM-1 Bla. The graph is calculated based on kinetic parameters of CDG-OMe by BlaC and TEM-1 Bla, respectively. See text for details. (b) Measured concentration of E. coli expressing BlaC using droplet microfluidics in three separate experiments: (i) 10 cfu/ml E. coli expressing BlaC (E. coliBlaC) in MES buffer, (ii) 10 cfu/ml E. coli expressing BlaC mixed with 107 cfu/ml E. coli expressing TEM-1 Bla (E. coliTEM-1 Bla), and (iii) 10 cfu/ml E. coli expressing BlaC mixed with 107 cfu/ml wild-type E. coli (E. coliWT). The height of each bar represents the mean concentration collected from at least 3 experiments. The height of the error bars represents one standard deviation from the mean.
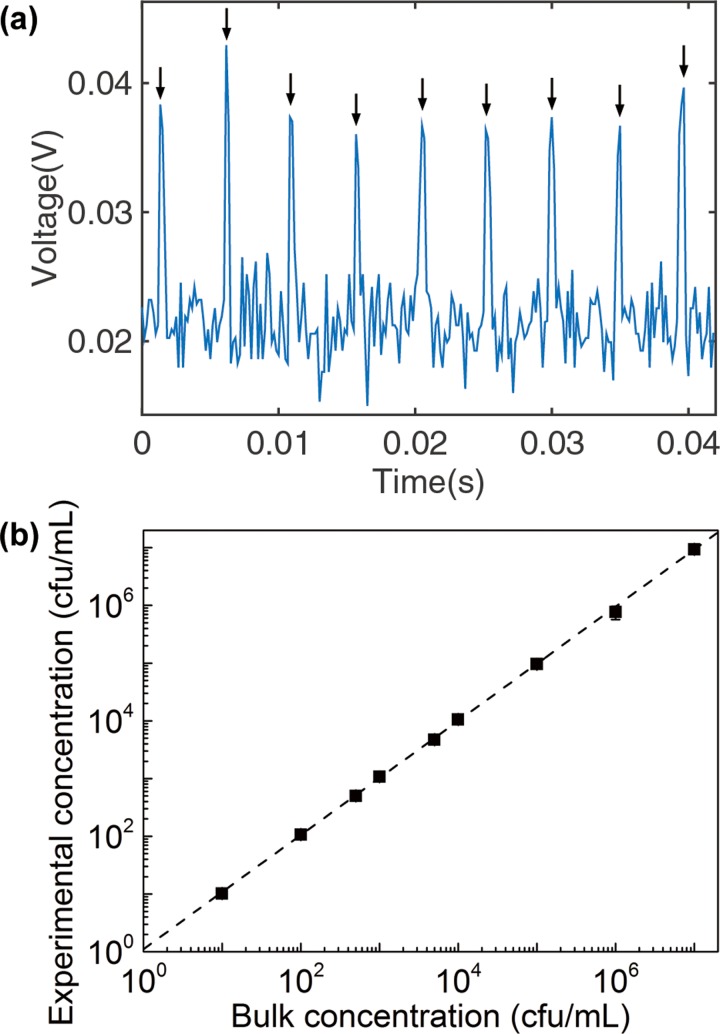
FIG. 4.
(a) Fluorescence signal collected from drops generated from a sample containing E. coli expressing BlaC at a concentration of 106 cfu/ml after 3 h of incubation. The 9 arrows indicate the fluorescent peaks that correspond to 9 fluorescent drops detected. The signal was obtained from a photomultiplier tube using the setup outlined in Figure 1(b). (b) Linear relationship between the experimentally measured bacteria concentration and the input bacteria concentration. Each data point represents the mean value collected from at least 3 experiments. The height of the error bars is less than that of the data point markers (see Table S1 in the supplementary material for details29).
B. Imaging of the drops
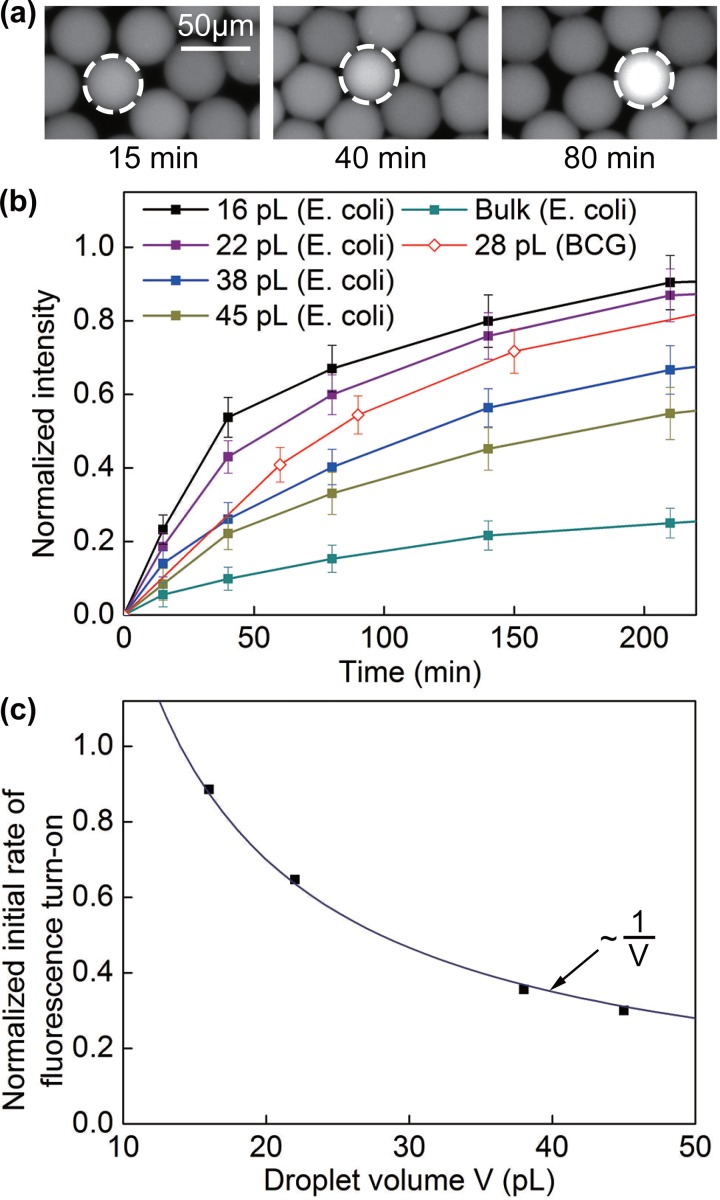
To image the drops (Figure 2), we injected the drops into a wide microfluidic channel and imaged them using an inverted optical microscope and an Electron Multiplying Charge Coupled Device (EMCCD) camera (Andor Technology, South Windsor, CT). For Figure 2(b), we normalized all intensity measurements (I) using this equation: Inorm = (I − Imin)/(Imax − Imin). The maximum intensity (Imax) was defined as the intensity when the enzymatic reaction was complete (i.e., when the intensity was saturated) at t = 30 h (see Figure S6 in the supplementary material29). Note this maximum intensity was the same for all drop sizes, as we used the same channel for intensity measurements and the path length for fluorescence measurement was fixed. The minimum intensity (Imin) was defined as the average intensity in negative drops at t = 15 min. For Figure 2(c), we calculated the initial rate of fluorescence increase by finding the slope of the initial linear portion of the curve.
FIG. 2.
(a) Images of selected droplets containing E. coli expressing BlaC with CDG-OMe and B-PER, after an incubation period of 15 min, 40 min, and 80 min, respectively. (b) Fluorescence signal from drops of various volumes containing a single cell of E. coli expressing BlaC and drops of 28 pl containing BCG. Each data point represents the mean intensity value collected from at least 10 drops. The height of the error bars represents one standard deviation from the mean. The intensities were normalized to the saturating intensity when the enzymatic reactions were complete (see supplementary material and Figure S6).29 (c) Initial rate of fluorescence turn-on as a function of droplet volume for the data presented in (b). The line is a fitted curve to the data.
C. Enumeration of fluorescent drops
For Figure 4, we prepared bacteria samples at concentrations of 10–107 cfu/ml by serial dilution from a concentrated bacteria culture (at 108 cfu/ml, verified by measuring the optical density at 600 nm (OD600) using Nanodrop). We then generated 30 pl drops from these samples (0.2–1 ml) following the methods described above. To count the number of fluorescent drops, we injected the drops into a funnel-shaped channel consisting of a narrow constriction with a cross section of 30 μm × 30 μm, less than one droplet diameter (Figure S2 in the supplementary material29). The reinjection rate was 1 ml/h, and the volume fraction of the drops upon reinjection was about 80%.
We focused UV light from a mercury lamp past an excitation filter onto the constriction using a 20× microscope objective on an inverted microscope (Nikon Eclipse TE2000). We collected the fluorescence from the drops from the same objective through an emission filter into a photomultiplier tube (PMT) (Hamamatsu Product No. 56420001) (Figure 1(b)). A gain of 13.0 V was applied to the PMT, and the output of the PMT was measured in voltages: a high voltage corresponds to a high intensity value collected by the PMT. LabView and MATLAB scripts were used to automate the acquisition and recording of voltage values from the PMT as a function of time for a duration of 5 to 60 min. A peak in voltage value corresponded to the presence of a drop. Counting the number of voltage peaks allowed us to count the number of fluorescent drops. A voltage peak was identified according to the following criteria: we generated drops from a sample without bacteria (negative drops) and measured the output voltages using the PMT (Figure S3, right, supplementary material29). We calculated the mean (negative) and the standard deviation (σ) from these data. A threshold value Vth was set at Vth = negative + nσ. n = 6 was used such that less than of the negative drops had value exceeding Vth. A large number of n was used here, so we had less than one false positive drop when counting ∼3 × 107 drops for the sample at 10 cfu/ml. This threshold was then applied to the sample with bacteria. Voltage peaks with values V > Vth were counted as a drop containing E. coli expressing BlaC.
D. Expression of BlaC
In brief, pET28b-BlaC transformed BL21 strain was cultured in Luria-Bertani medium containing 50 μg/ml of kanamycin at 30 °C until the OD600 reached 0.6–0.8. Expression of the BlaC gene was induced for 20 h at 16 °C by the addition of 1 mM isopropyl β-D-thiogalactopyranoside (IPTG). Cells were harvested and proteins were then purified using BugBuster protein extraction reagent (Novagen) and Ni-NTA agarose bead (Qiagen) following the manufacturer's protocol (Wash buffer 1: 0.01M Phosphate-buffered saline (PBS buffer), 150 mM NaCl at pH 7.4. Wash buffer 2: 20 mM imidazole in 0.01M PBS buffer at pH 7.4; this washing step was repeated 3 times. Elution buffer: 250 mM imidazole in 0.01M PBS at pH 7.4). After purification, the eluted protein was further purified using PD-10 column (GE healthcare). Purity was determined by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Figure S8 in the supplementary material29), and the concentration was determined by Bradford assay kit (BioRad).
E. Cell culture
E. coli was cultured in Luria–Bertani medium until OD600 reached a value of 0.5–1. For testing the specificity of probe CDG-OMe, we also used E. coli that does not express BlaC (IDEXX-QC E. coli intended for water safety quality control, purchased from IDEXX Laboratories). We refer to this E. coli as “wild-type E. coli” in Sec. III. Results and Discussion. Mycobacterium Smegmatis was cultured in 7H9 medium with a 10% oleic acid albumin dextrose complex (OADC) and 0.25% Tween-80 until the culture reached the log phase (OD600 = 0.5–1). BCG was cultured in Middlebrook 7H9 Broth (Sigma, St. Louis, MO) supplemented with 10% albumin dextrose catalase (ADC) supplement (Sigma, St. Louis, MO) and 0.1% Tween-80 until the culture reached the log phase (OD600 = 0.8–1).
F. Probe synthesis
Fluorogenic probes CDG-1 and CDG-OMe were synthesized according to procedures described previously.28 Both probes contain Tokyo Green as the fluorophore. These probes have been confirmed for their activity towards recombinant TEM-1 Bla and BlaC, respectively, as reported previously.28
III. RESULTS AND DISCUSSION
A. Fluorescence detection in micro-droplets
To characterize the effect of droplet size on the rate of fluorescence increase, we used an initial bacteria concentration such that each drop contained one or zero cell. Drops were generated by a microfluidic flow-focusing device26,29 (Figure 1(b); see supplementary material for experimental design). The probe CDG-OMe and lysis buffer B-PER were co-encapsulated into each drop. The lysis buffer did not impair the hydrolysis of the probe by BlaC. On the contrary, the absolute intensity of fluorescence measured at a fixed time point in drops with B-PER was higher than that without B-PER for E. coli expressing BlaC (Figure S4 in the supplementary material29). B-PER enabled the mild extraction of proteins from bacteria and was effective in facilitating the transport of the probe to the enzyme here, although the probe was already designed to be cell-permeable. Furthermore, the enzyme extracted from bacteria allowed the fluorophore Tokyo Green to be formed outside the cell and render the entire droplet fluorescent. This ability was important in facilitating the counting of fluorescent drops using a simple optical setup. We have thus used B-PER in all our subsequent experiments. We verified that B-PER also lysed BCG (Figure S5 in the supplementary material29). Since BCG and Mtb are both mycobacteria, we expect that B-PER would also be effective in lysing Mtb.
Figure 2 shows the effect of droplet size on the rate of fluorescence increase. For drops that contained a cell, the rate of increase in fluorescence intensity increased with decreasing droplet size as expected. We measured the initial rate of fluorescence increase v by finding the slope of the initial linear portion of the curves. Figure 2(c) shows that v scaled nicely with the inverse of droplet volume V, i.e., v ∼ V−1. This scaling confirms that the primary effect of drop size was indeed that of increasing the concentration of the cell (and the enzymes) inside the drop. For drops that were 16 pl in volume, the fluorescence intensity reached 80% of its maximum value in about 150 min for E. coli expressing BlaC (Figure 2(b)). With our current imaging system, the detection of a positive fluorescence signal (defined to be at an intensity level that is 6 times the standard deviation of the noise level) can be achieved in less than 60 min. We further validated our method using BCG in 28-pl drops and found that the fluorescence turn-on rate was comparable with that of E. coli expressing BlaC (Figure 2(b)). The expression level of BlaC in BCG may be different from the endogenous BlaC level in Mtb. The actual detection time for Mtb may be different from that shown here under similar conditions but is still expected to be significantly shorter than the 4–8 weeks lag time required for the culture-based method.
B. Specificity of the detection scheme
Contamination of patient samples with non-Mtb or environmental organisms can cause false positives in the diagnosis. It was recently shown that a healthy lung contains diverse microbial communities.28 In patients with pneumonia caused by the infection of Streptococcus pneumoniae, the concentration of S. pneumoniae can exceed 107 cfu/ml.28 In order to differentiate Mtb from other micro-organisms and to avoid false positives, it is critical that our method is specific to Mtb, or BlaC, only. Previously, we have shown the specificity and sensitivity of our probe CDG-OMe for BlaC over its close class A homologue TEM-1 Bla, as well as over β-lactamases produced by Pseudomonas, Staphylococcus, and the environmental mycobacterium M. smegmatis.17 The probe CDG-OMe has more than 1000-fold selectivity of BlaC over its close A homologue TEM-1 Bla, when the concentrations of BlaC and TEM-1 Bla are the same. In actual patient samples, the concentration of Mtb (and thus BlaC) can be many times smaller than that of other bacteria. The probe fluorescence intensity for 10 cfu of BCG was only 1.2 times higher than that of 105 cfu of other bacteria that expressed β-lactamase, including methicillin-resistant Staphylococcus aureus (MRSA).17 Based on the enzyme kinetic parameters for the hydrolysis of CDG-OMe by BlaC and TEM-1 Bla,17 we used a simple Michaelis-Menten model to estimate the concentration of TEM-1 Bla above which the fluorescence turn-on rate of CDG-OMe by TEM-1 Bla exceeds that by BlaC for a fixed probe concentration. Figure 3(a) shows that when BlaC is equally concentrated as TEM-1 Bla, the probe fluorescence turn-on rate is 3800-fold higher for BlaC than for TEM-1 Bla (i.e., , where and are the initial rates of product formation, or the rate of probe fluorescence turn-on, in the presence of BlaC and TEM-1 Bla, respectively). Such ratio diminishes to 1 when TEM-1 Bla is about 3800-fold more concentrated than BlaC, however.
This challenge can be overcome in droplet microfluidics when individual cells are compartmentalized into droplets. The effective concentration of a single cell in a drop is fixed and is determined by the volume of the drop only, although the bulk concentration of non-BlaC expressing cells is many times higher than that of BlaC-expressing cells. The use of droplets for single-cell compartmentalization thus allows the original selectivity of the probe to be preserved at . Figure 3(b) shows that when used with droplet microfluidics, the specificity was indeed maintained in each drop. We were able to measure the concentration of E. coli expressing BlaC accurately at 10 cfu/ml, when it was mixed with E. coli expressing TEM-1 Bla or wild-type E. coli present at 106 times higher concentration than E. coli expressing BlaC.
C. Dynamic range of our method
We counted the number of fluorescent drops (Figure 4(a)) and compared with the number of cells as detected by standard methods based on optical density (NanoDrop) and serial dilutions of the bacteria sample (see supplementary material for details).29 Figure 4(b) shows a linear relationship between the measured concentration of cells and the input concentration of cells over the range of 10 to 107 cfu/ml. In deriving the concentration of cells from the counted number of fluorescent drops, we have accounted for Poisson statistics in the encapsulation of cells in drops (see Note S1). The upper detection limit of our method is bounded by bacteria concentration at which all drops become occupied by cells. For droplet volume of 10 pl, the highest concentration of cells we can detect is approximately 108 cfu/ml, which should be much higher than that needed in practical applications.
The lower detection limit of our method is, in theory, one fluorescent drop or one cell. At very low concentrations of cells, the practicality of our method is limited by the speed of the droplet generation and interrogation process.30 For the sample at 10 cfu/ml, we used a sample volume of 1 ml. At the flow rates used for a single droplet generator, it took about 6 h to generate the drops, much longer than the incubation time needed for the fluorescence signal to turn on. To reduce the droplet generation time, current work is in progress to incorporate parallel droplet generators, which have been described by multiple groups where up to 512 parallel generators have been reported.28,31–35 The use of a single droplet generator here did not change the principle of our method, however. For the serial interrogation of the sample at 10 cfu/ml, we counted a total of 1 ml of drops, which took about 1 h. We have recently shown that it is possible to count drops in massively parallel format at a rate of ∼0.25 million drops/second.36 Given this rate, it would require only 2 min to interrogate 1 ml of sample. The assay would then be rate-limited by the kinetics of the probe and the enzyme. Current work is in progress to improve the light sensitivity of this parallel interrogation scheme for use with the BlaC assay described in this paper.
IV. CONCLUSIONS
We have demonstrated a droplet microfluidics-based method for the quantitative detection of cells expressing BlaC with intended application in the diagnosis of tuberculosis. The key advantages of our method are (1) the ability to enumerate the number of cells with a lower detection limit of 10 cfu/ml. This ability is useful not only for diagnosing TB but also for quantifying the efficacy of therapeutic methods and measuring drug resistance. (2) It does not rely on bacteria growth and is less susceptible to the risk of contamination by non-M. tuberculosis cells than culture-based methods are.37 (3) It maintains the specificity of the probe regardless of the concentration of non-Mtb cells present in the sample. The detection time can be further reduced if photodetectors with increased sensitivity are used and when combined with parallel droplet generation and interrogation schemes. While this work is not yet ready for point-of-care applications, we have demonstrated the feasibility of the concept and have established a process flow necessary for the subsequent design and development of an integrated system that combines sample handling, microfluidics, and optical detection. To apply our method to actual sputum samples, we intend to follow standard protocols suggested for the liquefaction of sputum, for example, by adding N-acetyl-L-cysteine (NALC) as a mucolytic agent.3 As our method is highly specific to bacteria generating BlaC only, we do not expect the addition of decontaminant agents—typically consisting of 1%–2% sodium hydroxide solution—to be necessary for rendering non-Mtb bacterial contaminants non-viable, as the use of decontaminants is known to kill a certain amount of Mtb.3 However, the detailed characterization of the effect of the pre-processing steps of sputum on our assay results is out of the scope of the current work. Finally, we expect our method to be applicable to other pathogen-induced diseases so long as a specific fluorogenic probe is available.38 Current work is in progress to test our method on clinical samples.
ACKNOWLEDGMENTS
Liat Rosenfeld is acknowledged for initial help with the work. We acknowledge partial support of the work by the Stanford Center for Innovation in Global Health and the Stanford Nano Shared Facilities Bio/Medical Mini Seed Grant. S.K.Y.T. acknowledges additional support from the 3M Non-tenured Faculty Award, and J.R. thanks the support from SCPKU Faculty Fellowship Program.
References
- 1.W. H. Organization, Global Tuberculosis Control: WHO Report 2010 ( World Health Organization, 2010). [Google Scholar]
- 2. Parsons L. M., Somoskövi A., Gutierrez C., Lee E., Paramasivan C. N., Abimiku A., Spector S., Roscigno G., and Nkengasong J., Clin. Microbiol. Rev. 24, 314 (2011). 10.1128/CMR.00059-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Thoracic Society and the Centers for Disease Control and Prevention (official statement), Am. J. Respir. Crit. Care Med. 161, 1376 (2000). 10.1164/ajrccm.161.4.16141 [DOI] [PubMed] [Google Scholar]
- 4. Yeager H., Lacy J., Smith L. R., and LeMaistre C. A., Am. Rev. Respir. Dis. 95, 998 (1967). [DOI] [PubMed] [Google Scholar]
- 5. Sommers H. and McClatchy J. K., Laboratory Diagnosis of the Mycobacterioses ( American Society for Microbiology, Washington D.C., 1983). [Google Scholar]
- 6. McNerney R. and Daley P., Nat. Rev. Microbiol. 9, 204 (2011). 10.1038/nrmicro2521 [DOI] [PubMed] [Google Scholar]
- 7. Steingart K. R., Henry M., Laal S., Hopewell P. C., Ramsay A., Menzies D., Cunningham J., Weldingh K., and Pai M., PLoS Med. 4, 1041 (2007). 10.1371/journal.pmed.0040202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miotto P., Bigoni S., Migliori G. B., Matteelli A., and Cirillo D. M., Eur. Respir. J. 39, 1269 (2012). 10.1183/09031936.00124711 [DOI] [PubMed] [Google Scholar]
- 9. Flores A. R., Parsons L. M., and Pavelka M. S., Microbiology 151, 521 (2005). 10.1099/mic.0.27629-0 [DOI] [PubMed] [Google Scholar]
- 10. Hugonnet J.-E., Tremblay L. W., Boshoff H. I., Barry C. E., and Blanchard J. S., Science 323, 1215 (2009). 10.1126/science.1167498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ioerger T. R., Feng Y., Chen X., Dobos K. M., Victor T. C., Streicher E. M., Warren R. M., Gey van Pittius N. C., Van Helden P. D., and Sacchettini J. C., BMC Genomics 11, 670 (2010). 10.1186/1471-2164-11-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zlokarnik G., Negulescu P. A., Knapp T. E., Mere L., Burres N., Feng L., Whitney M., Roemer K., and Tsien R. Y., Science 279, 84 (1998). 10.1126/science.279.5347.84 [DOI] [PubMed] [Google Scholar]
- 13. Gao W., Xing B., Tsien R. Y., and Rao J., J. Am. Chem. Soc. 125, 11146 (2003). 10.1021/ja036126o [DOI] [PubMed] [Google Scholar]
- 14. Xing B., Khanamiryan A., and Rao J., J. Am. Chem. Soc. 127, 4158 (2005). 10.1021/ja042829+ [DOI] [PubMed] [Google Scholar]
- 15. Yao H., So M., and Rao J., Angew. Chem. Int. Ed. Engl. 46, 7031 (2007). 10.1002/anie.200701931 [DOI] [PubMed] [Google Scholar]
- 16. Rukavishnikov A., Gee K. R., Johnson I., and Corry S., Anal. Biochem. 419, 9 (2011). 10.1016/j.ab.2011.07.020 [DOI] [PubMed] [Google Scholar]
- 17. Xie H., Mire J., Kong Y., Chang M., Hassounah H. A., Thornton C. N., Sacchettini J. C., Cirillo J. D., and Rao J., Nat. Chem. 4, 802 (2012). 10.1038/nchem.1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boedicker J. Q., Li L., Kline T. R., and Ismagilov R. F., Lab Chip 8, 1265 (2008). 10.1039/b804911d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Churski K., Kaminski T. S., Jakiela S., Kamysz W., Baranska-Rybak W., Weibel D. B., and Garstecki P., Lab Chip 12, 1629 (2012). 10.1039/c2lc21284f [DOI] [PubMed] [Google Scholar]
- 20. Marcoux P. R., Dupoy M., Mathey R., Novelli-Rousseau A., Heran V., Morales S., Rivera F., Joly P. L., Moy J.-P., and Mallard F., Colloids Surf., A 377, 54 (2011). 10.1016/j.colsurfa.2010.12.013 [DOI] [Google Scholar]
- 21. Rane T. D., Zec H. C., Puleo C., Lee A. P., and Wang T.-H., Lab Chip 12, 3341 (2012). 10.1039/c2lc40537g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang D.-K., Ali M. M., Zhang K., Huang S. S., Peterson E., Digman M. A., Gratton E., and Zhao W., Nat. Commun. 5, 5427 (2014). 10.1038/ncomms6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marlowe E. M., Novak-Weekley S. M., Cumpio J., Sharp S. E., Momeny M. A., Babst A., Carlson J. S., Kawamura M., and Pandori M., J. Clin. Microbiol. 49, 1621 (2011). 10.1128/JCM.02214-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brittain S., Paul K., Zhao X.-M., and Whitesides G., Phys. World 11, 31 (1998). [Google Scholar]
- 25. Siegel A. C., Tang S. K. Y., Nijhuis C. A., Hashimoto M., Phillips S. T., Dickey M. D., and Whitesides G. M., Acc. Chem. Res. 43, 518 (2010). 10.1021/ar900178k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anna S. L., Bontoux N., and Stone H. A., Appl. Phys. Lett. 82, 364 (2003). 10.1063/1.1537519 [DOI] [Google Scholar]
- 27. Holtze C., Rowat A. C., Agresti J. J., Hutchison J. B., Angilè F. E., Schmitz C. H. J., Köster S., Duan H., Humphry K. J., Scanga R. A., Johnson J. S., Pisignano D., and Weitz D. A., Lab Chip 8, 1632 (2008). 10.1039/b806706f [DOI] [PubMed] [Google Scholar]
- 28. Blainey P. C., Milla C. E., Cornfield D. N., and Quake S. R., Sci. Transl. Med. 4, 153ra130 (2012). 10.1126/scitranslmed.3004458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.See supplementary material at http://dx.doi.org/10.1063/1.4928879E-BIOMGB-9-027504 for experiment details and figures.
- 30. Rosenfeld L., Fan L., Chen Y., Swoboda R., and Tang S. K. Y., Soft Matter 10, 421 (2014). 10.1039/C3SM51843D [DOI] [PubMed] [Google Scholar]
- 31. Li W., Greener J., Voicu D., and Kumacheva E., Lab Chip 9, 2715 (2009). 10.1039/b906626h [DOI] [PubMed] [Google Scholar]
- 32. Romanowsky M. B., Abate A. R., Rotem A., Holtze C., and Weitz D. A., Lab Chip 12, 802 (2012). 10.1039/c2lc21033a [DOI] [PubMed] [Google Scholar]
- 33. Nisisako T. and Torii T., Lab Chip 8, 287 (2008). 10.1039/B713141K [DOI] [PubMed] [Google Scholar]
- 34. Nisisako T., Ando T., and Hatsuzawa T., Lab Chip 12, 3426 (2012). 10.1039/c2lc40245a [DOI] [PubMed] [Google Scholar]
- 35. Conchouso D., Castro D., Khan S. A., and Foulds I. G., Lab Chip 14, 3011 (2014). 10.1039/C4LC00379A [DOI] [PubMed] [Google Scholar]
- 36. Kim M., Pan M., Gai Y., Pang S., Han C., Yang C., and Tang S. K. Y., Lab Chip 15, 1417 (2015). 10.1039/C4LC01465K [DOI] [PubMed] [Google Scholar]
- 37. Tortoli E., Cichero P., Piersimoni C., Simonetti M. T., Gesu G., and Nista D., J. Clin. Microbiol. 37, 3578 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi H., Cheng Y., Lee K. H., Luo R. F., Banaei N., and Rao J., Angew. Chem. Int. Ed. 53, 8113 (2014). 10.1002/anie.201402012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4928879E-BIOMGB-9-027504 for experiment details and figures.