Abstract
Malaria causes significant global morbidity and mortality. No routine vaccine is currently available. One of the major reasons for lack of a vaccine is the challenge of identifying suitable vaccine candidates. Malarial proteins expressed using prokaryotic and eukaryotic cell based expression systems are poorly glycosylated, generally insoluble and undergo improper folding leading to reduced immunogenicity. The wheat germ, rabbit reticulocyte lysate and Escherichia coli lysate cell free expression systems are currently used for expression of malarial proteins. However, the length of expression time and improper glycosylation of proteins still remains a challenge. We demonstrate expression of Plasmodium proteins in vitro using HeLa based cell free expression systems, termed “in vitro human cell free expression systems”. The 2 HeLa based cell free expression systems transcribe mRNA in 75 min and 3 µl of transcribed mRNA is sufficient to translate proteins in 90 min. The 1-step expression system is a transcription and translation coupled expression system; the transcription and co-translation occurs in 3 hr. The process can also be extended for 6 hr by providing additional energy. In the 2-step expression system, mRNA is first transcribed and then added to the translation mix for protein expression. We describe how to express malaria proteins; a hydrophobic PF3D7_0114100 Maurer’s Cleft – 2 transmembrane (PfMC-2TM) protein, a hydrophilic PF3D7_0925900 protein and an armadillo repeats containing protein PF3D7_1361800, using the HeLa based cell free expression system. The proteins are expressed in micro volumes employing 2-step and 1-step expression strategies. An affinity purification method to purify 25 µl of proteins expressed using the in vitro human cell free expression system is also described. Protein yield is determined by Bradford’s assay and the expressed and purified proteins can be confirmed by western blotting analysis. Expressed recombinant proteins can be used for immunizations, immunoassays and protein sequencing.
Keywords: Biochemistry, Issue 100, Cell free in vitro transcription-translation, HeLa cell free expression, rhoptry proteins, mammalian cell free expression system, Plasmodium falciparum, Pro Bond affinity purification
Introduction
In malaria research, expression of immunogenic proteins using prokaryotic or eukaryotic cell based systems remains a challenge. The A-T richness of the Plasmodium genome and unknown post translational mechanisms14,7, contribute to the difficulties associated with obtaining properly folded and immunogenic proteins for antibody production and vaccine studies. Prokaryotic systems such as Escherichia coli have been used for recombinant protein expression. Prokaryotic systems are low cost, effective, produce high yields of recombinant proteinand have multiple cloning vectors. Host E. coli cells are easy to transform and cells grow rapidly. However, prokaryotic systems have limitations such as lack of amino acid substitution, post-translational modification, risk of contamination, heterogeneous products and accumulation of recombinant proteins within inclusion bodies24.
In a study by Mehlin et al. (2006), 1000 open reading frames (ORF) were expressed. Approximately 7% of the expressed proteins were soluble14. The insolubility observed is due to the biased nature of the genes and the high frequency of codons that are used ideally by the Plasmodium A-T rich genome14. An alternative strategy to overcome this problem has been developed by using plasmids or host cells containing tRNAs that recognize rare codons or codons that match the frequencies3. Even after performing these optimizations, very small portions of proteins are expressed as soluble, active and immunogenic14. The cell-free expression systems contain all the components necessary for transcription and translation such as ribosomes, initiation factors, elongation factors (translations factors), tRNA and aminoacyl-tRNA synthetases. Both transcription and translation reactions are coupled in one-step procedure11,29. The transcription reaction is performed in a tube before appropriate amount of mRNA is incubated with translation machinery in a different tube11,29. Although these methods are successful in Plasmodium sp. protein expression, a major drawback is the length of time for protein expression, which is approximately 22 hr29. In addition, the high cost of supplies for inclusion in the protocols29, the labor-intensive preparations of the cell lysate and inconsistencies in component preparations, make these systems unattainable. The main focus of researchers for development of a cell free expression system depends on factors such as rapid genetic modification, fast yields with high concentrations and straight forward lysate preparations1.
Eukaryotic systems such as yeast, mammalian cell lines, baculovirus mediated expression system, Tetrahymena thermophilia, Dictyostelium discoideum and parasitic expression systems such as Leishmania have been used for recombinant protein expression7. Eukaryotic systems share phylogenetic relationship and therefore also share characteristics such as glycosylation, acylation, disulphide bond formation, chaperone interaction, proteolysis and sub-cellular compartmentalization. Protein secretion events in eukaryotes prevent accumulation and decreases toxicity for expression systems13. The use of yeast systems is preferred as they are suitable for protein expression in large scale and for obtaining high yields4. Two P. falciparum proteins PfCP-29 and Pvs25 were produced using the yeast system4. However, a major drawback in the synthesis of most of the proteins was irregular N and O-glycosylation patterns, improper folding and truncation of proteins from their native form4.
The use of mammalian cells for synthesis of recombinant proteins is labor-intensive and it is expensive to maintain stable recombinant cell lines4. Therefore, mammalian cells have been limited for the analysis of protein signaling, protein interactions and parasite-host interactions and also for testing DNA vaccines4. The Human DNA and mRNA in vitro protein expression cell-free system described herein is a HeLa cell-derived mammalian-based system that expresses proteins in 3 hr. Proteins expressed using pT7CFE1-cHis expression vector have a C-terminal tag facilitating identification and purification. The HeLa based system is supplemented with translation initiation factors and a translation regulator, thereby enhancing the efficiency of the translation system. Proteins can be rapidly translated, screened, verified and processed for use in various immunologic and structural applications15.
Eukaryotic cell-free expression systems such as wheat germ and rabbit reticulocyte are currently used for the expression of various eukaryotic proteins including Plasmodium proteins11,29. In addition, E. coli based cell-free systems are also employed for eukaryotic protein expression11. Table 1 summarizes different cell free expression systems by comparing features of the systems as well as the ease of using the systems for protein expression. The human cell-free system in comparison to the others has the ability to perform post and co-translational modifications and is less expensive. Codon optimization is possible and all the reactions can be performed in 3 hr. The HeLa system is an ideal translation system for protein synthesis in the laboratory setting.
A major advantage of human cell free expression systems over other cell free systems is the availability of several different cell lines derived from different organs and tissues. Several varieties of cell free systems can be designed depending on the cell line. The extracts derived from mammalian cells have higher efficiency to synthesize large proteins than any other cell free expression systems. These cell free expression systems can also be used in diagnostic and protein characterization applications such as micro arrays30. The popular HeLa cell lines are most widely used to carry out the expression of proteins33. The endoplasmic reticulum in the HeLa cell lines is underdeveloped, leading to the absence of post-translational glycosylation modification activity33. However, this system is believed to be advantageous for Plasmodium protein synthesis as Plasmodium also lacks glycosylation post translation modification29. The HeLa cell free transcription-translation protocol is easy to perform, inexpensive and proteins can be expressed in 90 min, ready for purification and further downstream applications.
Protocol
1. Preparation of Parasite Cell Cultures, Collection of Parasite Pellets
Prepare RPMI-1640-HEPES media by adding 10 g of RPMI-1640 powder, 66 mg of gentamycin sulphate, 2 g sodium bicarbonate, 5.9 g HEPES buffer, 50 mg hypoxanthine and 3.8 g dextrose in 1 L sterile dH2O. Mix using table magnetic stirrer until all the powder dissolves in 1 L distilled water. Perform micro filtration of media using 0.22 µm filter. Store the media in sterile bottles.
Wash uninfected (fresh) type A+ erythrocytes using RPMI. Prepare 5% hematocrit of erythrocytes (0.5 ml of washed fresh erythrocytes in 9.5 ml 10% RPMI + human serum).
Make 15% human serum + RPMI (15 ml of human serum and 85 ml of RPMI) to initiate the cultivation of parasites. Make 10% human serum and RPMI (10 ml of human serum and 90 ml of RPMI) to continue culturing of parasites in red blood cells.
Culture P. falciparum (3D7 and FCR-3 strains) in type A+ human erythrocytes at 5% hematocrit and 20% parasitemia in petri dish or a 75 cm3 culture flask. Maintain culture in vitro according to the method of Trager and Jensen candle jar26. Alternately, maintain cultures are in incubator at 37 °C maintaining 9.5% CO2 and 3.4% N2 gas. Culture P. falciparum in RPMI 1640-HEPES media supplemented with 10% human serum.
Synchronize P. falciparum culture by adding 65% percoll (65 ml of percoll + 35 ml of RPMI-1640 without human serum) to the culture concentrate27.
Centrifuge the culture concentrate with percoll at 2500 x g for 5 min which results in 3 layers. Carefully, using a transfer pipette isolate schizonts from the middle layer transfer into a separate tube under aseptic conditions27.
Wash the isolated cultures with 10% human serum + RPMI-1640 twice to remove excess percoll by centrifuging at 7500 x g for 5 min. Discard the supernatant each time under aseptic conditions27.
Continue culturing the synchronized P. falciparum schizonts in 10% human serum + RPMI-164027.
Collect schizonts by treating Plasmodium-infected erythrocytes with 10 mM Tris pH 8.8 and centrifuging at 15000 x g for 15 min23. Store parasite pellets at -70 °C following addition of 5 µl of protease inhibitor (aprotinin) to the pellets. Use parasite pellets for protein extraction, genomic DNA isolation and RNA isolation.
2. Preparing Plasmid Vector
Isolate genomic DNA from P. falciparum schizont pellets prepared from cultures of approximately 20% parasitemia.
Add 600 µl of 10mM Tris-HCl of pH 7.6, 50 mM EDTA pH 8.0, 0.1% SDS and 1 mg/ml proteinase K, to pellets in a microcentrifuge tube, homogenize pellet using a 1 ml syringe attached to a 26 G needle. Alternate filling the syringe with the pellet mixture and emptying into a microcentrifuge tube 20 times. Incubate tube containing homogenate O/N in a 50 °C water bath.
Perform phenol – chloroform (P+C) extraction by adding 600 µl of P+C (300 µl of phenol and 300 µl of chloroform) to homogenized pellet. Cap tube tightly and shake the mixture vigorously by inverting tube end-to-end. Centrifuge tube at 14,000 x g for 2 min and collect upper aqueous solution using a micropipette into a new microcentrifuge tube. Repeat this step twice.
Add 600 µl of chloroform to the aqueous layer in the new tube from step 2.1.2. Vortex the tube for 10 sec and centrifuge at 14,000 x g for 2 min. Measure the upper aqueous layer with a micropipette and transfer it to a new microcentrifuge tube.
Add 0.1 vol of 5 M sodium acetate pH 5.5 and 1 vol of isopropanol (if aqueous layer is 300 µl add 30 µl of 5 M sodium acetate pH 5.5 + 330 µl of isopropanol) to aqueous layer from step 2.1.3. Incubate the tube at RT for 15 min.
Centrifuge the tube at 14,000 x g for 10 min to precipitate DNA pellet. Discard the supernatant and wash pellet with 100 µl of 70% ethanol (70 µl of ethanol and 30 µl of dH2O) to remove excess salt from the DNA. Store DNA in 50 µl of dH2O at -20 °C.
Design forward and reverse primers using software or manually for amplification of P. falciparum genes PF3D7_1361800, PF3D7_0925900, PF3D7_1436300 and PF3D7_0114100 previously identified in proteome studies16,18 with appropriate restriction sites to permit cloning of the gene sequences into the multiple cloning sites of expression plasmid pT7CFE1-CHis as shown in Table 2.
Make a 50 µl reaction mixture by adding; 5 µl of isolated P. falciparum genomic DNA, 5 µl of 10x buffer, 5µl of MgCl2, 1.5 µl of forward primer, 1.5 µl of reverse primer, 0.5 µl of T7 DNA polymerase and 31.5 µl of dH2O and perform polymerase chain reaction (PCR) at 94 °C for 8 min for denaturation, 50 °C for 1 min 30 sec for extension and 72 °C for 9 min for annealing and renaturation for amplifying genes shown in Table 1.
Separate PCR products on 1% agarose gel (1 g of agarose and 100 ml of Tris acetate EDTA (TAE) buffer)32.
Cut the PCR amplified DNA bands from the agarose gel, place the gel slices containing DNA in a DNA gel extraction spin column and freeze at -20 °C for 5 min. Add 100 µl of isopropanol to the frozen gel slices and centrifuge at 14,000 x g for 5 min.
Measure the aqueous flow-through filtered in the collection tube from step 2.5, using a micropipette and transfer to a new tube. Add 0.1 vol of 5 M sodium acetate and centrifuge at 14,000 x g for 5 min. Discard the supernatant and add 10 µl of dH2O to the DNA pellet.
Digest the pT7CFE1-CHis expression vector and purified DNA fragments (from 2.6) in two separate tubes by adding 3 µl of plasmid and 5 µl of PCR gene products to each tube. Add to each tube, 1 µl of specific restriction enzymes, 2 µl of reaction buffer and 14 µl of dH2O.
Add 3 µl of digested plasmid, 5 µl of digested DNA fragments, 2 µl of 10x ligase buffer, 1 µl of ligase and 9 µl of dH2O in an microcentrifuge tube. Incubate at 12 °C O/N.
Add 0.1 vol of 3 M sodium acetate at pH 5.5 and 2 volumes of ethanol (if your ligation tubes volume is 20 µl add 2 µl of 3 M sodium acetate + 44 µl of ethanol) to the tube from step 2.14. Mix well and incubate at -20 °C for 15 min.
Centrifuge the tube at 14,000 x g for 15 min. Carefully remove the supernatant using a micro pipette without disturbing the pellet.
Add 100 µl of 70% ethanol to the pellet from step 2.10. Centrifuge the tube at 14,000 x g for 5 min. Remove and discard the supernatant. Add 10 µl of nuclease-free water to the DNA pellet.
3. Protein Expression Using in Vitro Human Cell Free Expression System
Prepare 20 µl of transcription mixture by measuring 2 µl of the recombinant pT7CFE1-CHis plasmid DNA, 4 µl of 5x transcription buffer, 4 µl of NTP mix, 2 µl of T7 RNA Polymerase and 8 µl of nuclease-free water for 2-step Human in Vitro Protein Expression using DNA templates. Mix components in a microcentrifuge tube and incubate for 75 min at 30 °C in a water bath.
Take 2 µl of the transcription mixture and add it to 23 µl of translation mixture containing 12.5 µl of HeLa cell lysate, 2.5 µl of accessory proteins, 1 µl of salt solution A, 0.5 µl of amino acids –Met, 0.5 µl of amino acids –Leu, 1 µl of RNAse inhibitor, 1.25 µl of energy mix and 3.75 µl of nuclease-free water.
Incubate the translation mixture from step 3.2 at 30 °C for 90 min. Store translated products at -20 °C.
Prepare 25 µl of transcription and translation mixture by adding 3 µl of the recombinant pT7CFE1-CHis plasmid DNA, 2 µl of nuclease-free water, 5 µl of reaction mix, 12.5 µl of HeLa lysate and 2.5 µl of accessory proteins for 1-step Human Coupled IVT Protein Expression for DNA templates.
Incubate the reaction mixture from step 3.4 for 90 min to 6 hr at 30 °C. Store the translation products at -20°C.
4. Purification of Expressed Recombinant Proteins
Add 75 µl of 1x purification buffer to 25 µl of translation product to make 100 µl of purification mixture.
Add 100 µl of nickel chelating resin to 100 µl of purification mixture. Wash Ni-resin twice by adding 300 µl of dH2O and 300 µl of the binding buffer. Re-suspend resin and centrifuge at 14,000 x g for 1 min to remove excess ethanol.
Add 100 µl of purification mixture from step 4.2 to 100 µl of nickel chelating resin and incubate the mixture for 60 min at RT.
Centrifuge the mixture at 14,000 x g for 1 min and collect supernatant into a fresh tube labeled as “flow through”.
Wash resin with 100 µl of 1x wash buffer (20 mM imidazole, 250 mM NaH2PO4 at pH 8, 2.5 M NaCl and dH2O). Centrifuge at 14,000 x g for 1 min and collect supernatant in fresh tube labeled “wash”. Repeat step 4.5 twice.
Add 100 µl of 1x elution buffer (100 mM Imidazole, 250 mM Na2PO4 at pH 8.0, 2.5M NaCl and dH2O) to the resin and incubate for 15 min. Centrifuge at 14,000 x g for 1 min. Do the elution step twice. Each time collect supernatant into fresh microcentrifuge tubes and label as “eluate”.
After elution, wash resin twice as described in step 4.5. Store flow through, washes and elution samples at -20 °C or lower. Resin must be stored at 4 °C. Use 30 µl of each sample for SDS-PAGE and immunoblotting analysis.
5. Coomassie (Bradford) Protein Assay 34
Prepare standard bovine serum albumin (BSA) solutions with the concentration 20 µg/ml (mix 10 µl of BSA (2 mg/ml) and 90 µl of dH2O), 40 µg/ml (mix 20 µl of BSA (2 mg/ml) and 80 µl of dH2O), 60 µg/ml (mix 30 µl of BSA (2 mg/ml) and 70 µl of dH2O), 80 µg/ml (mix 40 µl of BSA (2 mg/ml) and 60 µl of dH2O), 100 µg/ml (mix 50 µl of BSA (2 mg/ml) and 50 µl of dH2O), 160 µg/ml (mix 80 µl of BSA (2 mg/ml) and 20 µl of dH2O) and 200 µg/ml (100 µl of BSA (2 mg/ml).
Measure 5 µl of flow through, washes and eluate samples from step 4.7. Add 95 µl of dH2O.
Add 5 ml of Coomassie blue reagent to the mixtures from steps 5.1 and 5.2. Cover the tubes with parafilm and vortex for 10 sec then incubate for 20 min at RT.
Using a spectrophotometer, measure optical density (OD) at wavelength of 650 nm for the protein tubes from step 5.3. Plot concentration vs. OD reading to determine the protein concentrations34.
6. Western Blotting
Add 5 µl of schizont extracts from P. falciparum, 2 µl of Escherichia coli expressed recombinant Rhop-3 proteins31, 5 µl of recombinant proteins expressed using 2-step and 1-step in vitro human expression systems and 10 µl of protein products purified using affinity method to separate tubes.
Add electrophoresis sample buffer containing mercaptoethanol to each tube for a final volume of 20 µl to make solubilized protein samples. Boil tubes for 2 min.
Load solubilized protein samples onto 10% SDS-PAGE gels to separate the proteins.
Transfer separated proteins from SDS-PAGE gels to nitrocellulose paper (NCP) by electrophoresing at 35 mA current per gel in a semi-dry western blotting chamber for 2 hr.
Block NCPs following transfer with 2% nonfat milk.
Incubate blocked NCPs with the following polyclonal antibodies diluted 1:100 in 2% milk to perform western blotting; rabbit antibody #67621 specific for P. falciparum merozoite rhoptries, mouse antibodies20 specific for P. yoelii and P. berghei merozoite rhoptries, antisera #685 specific for the P. falciparum parasitophorous vacuole protein, SERA (serine rich antigen)22 and antibodies specific for the Maurer’s cleft protein PfMC-2TM28.
Incubate NCPs also in 1:100 diluted normal mouse and rabbit serum and spent culture supernatant (SCS) from SP2 myeloma cells as negative controls.
Incubate NCPs with antibodies and controls at 4 ºC O/N.
Wash NCPs 4x with Blot buffer and incubate NCPs with species specific secondary antibodies conjugated to horseradish peroxidase (HRP) diluted 1:1000 in 2% milk.
Wash NCPs 4x with Blot buffer. Incubate washed NCPs with the color development solution A+B (Solution A: add 50ml of 1x Blot buffer and 30 µl of H2O2; Solution B: add 10 ml of methanol and 30 mg of 3-chloro-naphtol) for 30 min in dark. Analyze western blot results colorimetrically.
Representative Results
Validation of the expressed recombinant proteins through reactivity with specific antibodies is an important first step in confirming the proper folding of the expressed proteins. Recombinant malarial proteins were expressed using the one step and two step in vitro human cell free expression systems. The recombinant proteins are purified Ni-chelating affinity method. We then used antisera against whole merozoite rhoptries in western blotting of SDS-PAGE separated recombinant proteins.
Figure 1 shows PCR amplified gene fragments of PY17X_1139200, PY17X_1402200, PY17X_1366000, PY17X_0830000 and PF3D7_1436300 in a 1% agarose gel. DNA bands were excised and DNA purified by freezing in DNA extraction Spin Column.
Malaria genes successfully amplified (shown in Table 2) from genomic DNA were cloned into the pT7CFE-CHis expression vector. Re-amplification of the genes from the recombinant plasmids demonstrated the presence of the cloned gene fragments in the vector. The flow chart of expression systems employed for protein translation is shown in Figure 2. Figures 2A and 2B shows a step wise procedure of the 2-step in vitro human cell free expression system and the 1-step IVT coupled translation system. Successful expression of Plasmodium proteins using in vitro human cell free expression systems was confirmed by rhoptry specific rabbit antisera. As illustrated in Figure 3A, recombinant proteins were recognized by rhoptry specific rabbit antisera #676. As shown previously, expressed recombinant proteins were not recognized by antisera against parasitophorous vacuole proteins from P. falciparum and P. yoelii demonstrating the specificity of rabbit antisera 676 against the expressed proteins32. Normal rabbit serum (NRS) did not react with recombinant proteins translated using 2-step in vitro human cell free expression system and 1-step IVT coupled translation system (Figure 3B). The successful expression of Plasmodium recombinant proteins was confirmed by different techniques such as In-gel histidine staining and Nickel-HRP staining32. Expressed proteins were purified by affinity purification system. Purification is performed in BATCH method (no columns used). The technique is optimized by changing the concentration of Imidazole from 60 mM to 100 mM. The optimum concentration for elution is 250 mM. Figure 4 shows successful purification of translated proteins from 25 µl of translated protein products. Figure 4A shows successful purification of Maurer’s cleft transmembrane protein18. Figure 4B shows successful purification of PF3D7_0925900, P. falciparum ortholog of Plasmodium yoelii gene PY17X_0830000, a hypothetical protein32. Figure 4C shows successful purification of armadillo repeats containing protein PY17X_1139200, a hypothetical protein using Ni resin beads and 100 mM imidazole in 1x elution buffer. The yield of protein after purification is 3.5 µg/25 µl.
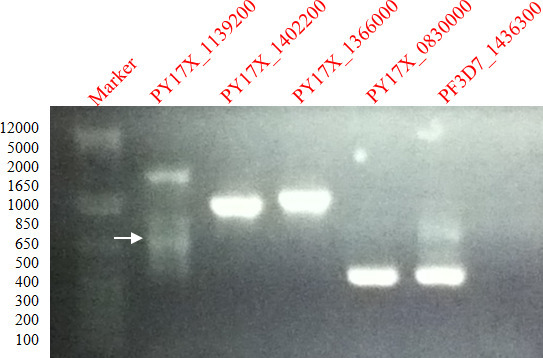 Figure 1. Amplification of P. falciparum and P. yoelii genes. 1% agarose gel showing ethidium bromide stained PCR products of gene fragments of PFc14_0344, PF3D7_0925900, PF3D7_1361800, PY07482 and PFA0680cw amplified from P. falciparum genomic DNA.
Figure 1. Amplification of P. falciparum and P. yoelii genes. 1% agarose gel showing ethidium bromide stained PCR products of gene fragments of PFc14_0344, PF3D7_0925900, PF3D7_1361800, PY07482 and PFA0680cw amplified from P. falciparum genomic DNA.
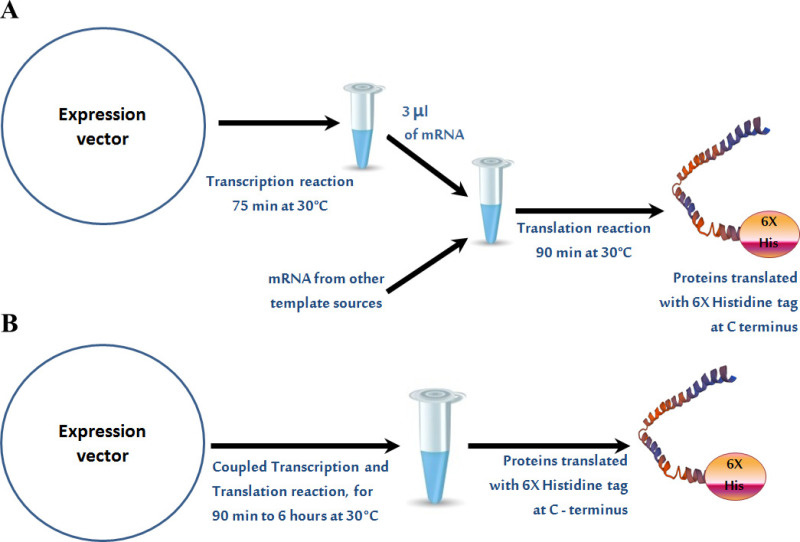 Figure 2. Flow chart of HeLa based cell free expression system. Schematic representation of both 2-step and 1-step in vitro human cell free expression systems. Please click here to view a larger version of this figure.
Figure 2. Flow chart of HeLa based cell free expression system. Schematic representation of both 2-step and 1-step in vitro human cell free expression systems. Please click here to view a larger version of this figure.
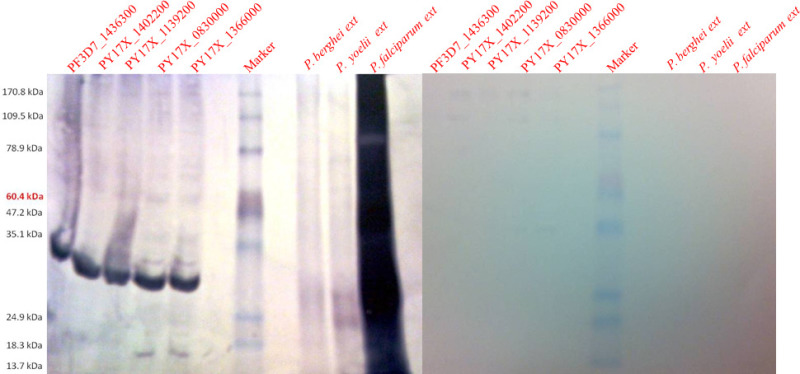 Figure 3. Immunoblotting confirmation of protein expression. Western blot analysis of expressed recombinant proteins using whole rhoptry specific rabbit anti-sera #676 and normal rabbit serum. (A) Reactivity of antisera 676 with expressed recombinant proteins. (B) Normal rabbit serum did not react with the expressed recombinant proteins. Parasitophorous vacuole protein did not react with recombinant proteins32. Please click here to view a larger version of this figure.
Figure 3. Immunoblotting confirmation of protein expression. Western blot analysis of expressed recombinant proteins using whole rhoptry specific rabbit anti-sera #676 and normal rabbit serum. (A) Reactivity of antisera 676 with expressed recombinant proteins. (B) Normal rabbit serum did not react with the expressed recombinant proteins. Parasitophorous vacuole protein did not react with recombinant proteins32. Please click here to view a larger version of this figure.
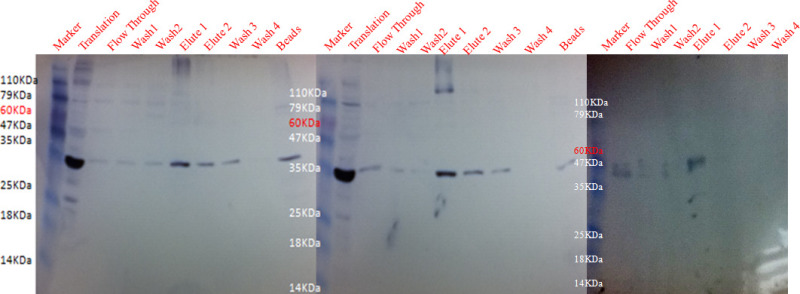 Figure 4. Purification of proteins from micro volumes of translation product. Purification of expressed recombinant proteins using Nickel chelating resin affinity method. Expressed proteins (translation) (A) PF3D7_0114100, (B) PY17X_0830000, and (C) PY17X_1139200 proteins were incubated with Nickel – chelating resins are purified with Nickel- chelating resin in the absence of imidazole in binding buffer and purification buffer. Following incubation, beads were centrifuged, unbound proteins were separated and the beads washed twice in wash buffer. Bound recombinant proteins were eluted twice followed by washing of the beads. Translated proteins, washes, eluates and beads were solubilized in electrophoresis sample buffer. Imidazole was used in the wash (20 mM of Imidazole) and elution (100 mM of Imidazole) buffers. Antisera #676 successfully recognized expressed and purified recombinant proteins. Normal rabbit serum did not react with expressed proteins as show in Figure 3B. Please click here to view a larger version of this figure.
Figure 4. Purification of proteins from micro volumes of translation product. Purification of expressed recombinant proteins using Nickel chelating resin affinity method. Expressed proteins (translation) (A) PF3D7_0114100, (B) PY17X_0830000, and (C) PY17X_1139200 proteins were incubated with Nickel – chelating resins are purified with Nickel- chelating resin in the absence of imidazole in binding buffer and purification buffer. Following incubation, beads were centrifuged, unbound proteins were separated and the beads washed twice in wash buffer. Bound recombinant proteins were eluted twice followed by washing of the beads. Translated proteins, washes, eluates and beads were solubilized in electrophoresis sample buffer. Imidazole was used in the wash (20 mM of Imidazole) and elution (100 mM of Imidazole) buffers. Antisera #676 successfully recognized expressed and purified recombinant proteins. Normal rabbit serum did not react with expressed proteins as show in Figure 3B. Please click here to view a larger version of this figure.
| Features | Human cell-Free expression system | Wheat germ cell free expression system | Rabbit reticulocyte expression system | E.coli lysate protein expression system |
| Time | Done transcription and translation in 3 hr | Transcription for 6 hr and translation for 10 - 20 hr | RNA has to be isolated translation takes 90 min | Done in 1 hr |
| Suitable vector | T7 vector can be | SP6 vector | T7 can be used | T7 can be used |
| Suitable scale | Suitable for laboratory synthesis | Used for the large scale protein synthesis | Suitable for laboratory and immunization studies | Suitable for laboratory and immunization studies |
| Extract | HeLa cell free extracts | Embryo of wheat extracts | Reticulocyte cell extracts | E. coli cell lysate extracts |
| Modification | Co-translational and post translational modification | Post translational modification | Co-translational modification | Post-translational modification |
| Size of protein translated | 8 KDa to 250 KDa | 220 KDa | 250 KDa | 200 KDa |
| Codon optimization | Tight | Tight | Loose | Tight |
| Disulphide bond formation | Yes | Yes | No | Yes |
| Yield | Low yield | High yield | Low Yield | Very high yield |
| Cost | $130.00 for 10 reactions | $173.00 for 10 reactions | $136.00 for 5 reactions | $159.00 for 8 reactions |
| Protein concentration | 3 to 5 µg per reaction | 100 µg/ml | 100 µg/ml | 500 µg/ml |
| References | 32, 30 | 7, 28, 29 | 7 | 7, 24, 25 |
Table 1. Cell free expression systems in malarial research. Comparison of cell free expression systems currently used in malarial research.
| Gene ID | Ortholog ID | Protein | Primer Sequence for PCR |
| PF3D7_1436300 | Translocon component | CTCGAGAATAATAACAATCATAATAATAAG | |
| GAATTCATTATCATCAGGTTTAGCTAATTTTC | |||
| PF3D7_0114100 | PfMC-2TM Maurer’s cleft protein | GGATCCATGTTAGGTCAAAAAAACACAAATA | |
| CTCGAGTGTTATTTGCTTTTTGTTTTGAAAA | |||
| PY17X_0830000 | PF3D7_0925900 | Hypothetical Protein | GGATCCATGAAATTTTTTAATATTCTCGCA |
| CTCGAGTCCTTGGACAACATATATACT | |||
| PY17X_1139200 | PF3D7_1361800 | Hypothetical Protein | GGATCCATGGGGTGCGACCCTGGGGTCAGCA |
| CTCGAGGCTCTTCAGATACTAAGCTACTAAT |
Table 2. Rhoptry genes in the study. Primers of different genes expressed in this study19.
| Name of Material | Company | Catalog number | Description |
| 2-step in vitro human cell free expression system32 | Thermo Fisher | 88856 | Discontinued. Always store at -80 °C. Do not thaw in warm water. |
| 1-step in vitro coupled translation system | Thermo Fisher | 88860 | Always store at -80 °C. Do not thaw in warm water. |
Table 3. Hela based cell free expression systems. Kits used for the expression of proteins.
Discussion
The important steps for successful expression of proteins using two step in vitro human cell free expression system and purification are: 1) successful amplification and cloning of genes into pT7CFE-CHis plasmid vector; 2) transcribing RNA at 30oC; 3) translation of protein at 30 °C in the presence of RNAse inhibitor; 4) developing the affinity based purification protocol using resin beads coated with Ni and 5) analyzing results on western blot using polyclonal and monoclonal antibodies. The important steps for successful expression of proteins using one step IVT coupled protein translation system are: 1) successful amplification and cloning of genes into pT7CFE-CHis plasmid vector; 2) Transcription and translation of proteins at 30 °C; 3) developing the affinity based purification protocol using resin beads coated with Ni and 4) analyzing results on western blot using polyclonal and monoclonal antibodies.
Currently, cell free expression systems are used for expressing recombinant malarial proteins. The critical features of cell free expression systems to express malaria proteins is the ability to decode A-T rich genes, a lack of long translational pause, successful expression of repeated amino acid sequences, lack of glycosylation mechanisms and the ability of plasmids to control gene truncation. In this study, we demonstrate use of a two-step in vitro human cell free expression system32 and a one-step IVT coupled translation systems for expression of P. falciparum gene orthologs of P. yoelii merozoite rhoptry genes as shown in Table 1. A purification protocol was developed and optimized for purifying micro-volumes of recombinant proteins obtained from 1-step and 2-step in vitro human cell free expression systems. 4 P. falciparum genes, PF3D7_0114100 a transmembrane hydrophobic protein, a predicted hypothetical ARM repeat protein PF3D7_1361800 and a hydrophilic protein PF3D7_0925900, were expressed using a 2-step and 1-step in vitro human cell free expression system. Purified protein yields were determined. The protein PfMC-2TM was included in these studies because it is an integral membrane protein with two transmembrane domains and we were interested in determining whether the in vitro expression system could correctly express the transmembrane protein and allow for ease of purification.
The key difference in both the one step and two step expression systems is that the two step expression system is an mRNA dependent system, where a stable mRNA is transcribed and added to the translation reaction. In contrast, the one step in vitro coupled transcription/translation system involves a continuous transcription and supply of mRNA followed by the co-translation of protein during the reaction. Both 1-step IVT transcription/translation coupled expression system and 2-step in vitro human cell free expression system were tested for expression of high molecular weight proteins. Both systems were successful in expressing low molecular weight recombinant proteins. In the 1-step expression system, antibody cross reactivity with proteins in the translation reaction mixture was observed. The specific proteins cross reacting with the antibodies are unknown and need further investigation. Expression of high molecular weight proteins was unsuccessful using both the 1-step and 2-step systems. A purification protocol was developed for purifying microvolumes of recombinant proteins of different solubility properties expressed using both 1-step and 2-step in vitro human cell free expression systems. Proteins were successfully purified and the yield obtained was 3.5 µg per 25 µl when expressed using 2-step in vitro protein translation system and 1.5 µg per 25 µl when translated with the 1-step IVT protein expression system. Expression of low molecular weight proteins varying from 18 kDa to 37 kDa is reproducible and straight forward. However, high molecular weight expression results in protein truncations. Due to the microvolumes obtained for cloned DNA, PCR amplification was used to verify cloned products. Potential obstacles for expressing high molecular weight proteins using cell free expression systems could be constant phosphorylation of eIF2 alpha subunit due to high concentration of ATP in the reaction. This impairs the translation initiation in the system which potentially affects the elongation of the protein synthesis during synthesis of large proteins in cell free expression systems10.
The protocol described here is uninfluenced by optimization conditions for expression of proteins. Neither the gene codons nor the plasmid were optimized for expression of proteins, and protein refolding was not required. This system expresses proteins in 3 hr and the purification of the protein can be obtained in 2 hr, allowing for further downstream applications such as processing of the recombinant protein for immunizations or peptide sequencing. Multiple recombinant proteins can be expressed with the HeLa cell free system, using microarray formats for immune sera screening and identification of potential vaccine candidate molecules. Furthermore, potentially immunogenic proteins can be identified through screens and selected proteins obtained by scaling up expression.
Disclosures
We do not have any competing financial interests in this study.
Acknowledgments
This research was supported by Cleveland State University Faculty Development Funds.
References
- Alexandrov K, et al. Leishmania. cell-free expression system. Methods. 2011;55(1):58–64. doi: 10.1016/j.ymeth.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Alexandrov K, et al. Towards the construction of expressed proteomes using a Leishmania tarentolae. based cell-free expression system. PLOS One. 2010;5(12):e14388. doi: 10.1371/journal.pone.0014388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca AM, Hol WGJ. Overcoming codon bias: A method for high-level over-expression of Plasmodium. and other AT-rich parasite genes in Escherichia coli. Int J Parasitol. 2000;30(2):113–118. doi: 10.1016/s0020-7519(00)00019-9. [DOI] [PubMed] [Google Scholar]
- Birkholtz LM, et al. Heterologous expression of plasmodial proteins for structural studies and functional annotation. Malar. J. 2008;7:197. doi: 10.1186/1475-2875-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan DL, Mu Y, Unal B. Profiling humoral immune responses to P. falciparum. infection with protein microarrays. Proteomics. 2008;8(22):4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Sawasaki T. Cell-free expression systems for eukaryotic protein production. Curr. Opin. Biotechnol. 2006;17(4):373–380. doi: 10.1016/j.copbio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Fernández-Robledo JA, Vasta GR. Production of recombinant proteins from protozoan parasites. Trends Parasitol. 2010;26(5):244–254. doi: 10.1016/j.pt.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda DC, Davidson EA. Protein glycosylation in the malaria parasite. Parasitol. Today. 1999;15(4):147–152. doi: 10.1016/s0169-4758(99)01412-x. [DOI] [PubMed] [Google Scholar]
- Goshima N, et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods. 2008;5:1011–1017. doi: 10.1038/nmeth.1273. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. Vol. 5. Cold Spring Harbor, N.W: Cold Spring Harbor Laboratory; 2000. pp. 185–243. [Google Scholar]
- Hino M, et al. Efficiency of cell-free protein synthesis based on a crude cell extract from Escherichia coli., wheat germ, and rabbit reticulocytes. Journal of Biotechnology. 2008;133(2):183–189. doi: 10.1016/j.jbiotec.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301(5639):1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996;60(3):512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin C, et al. Heterologous expression of proteins from Plasmodium falciparum.: results from 1000 genes. Mol. Biochem. Parasitol. 2006;148(2):144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Mikami S, et al. A human cell-derived in vitro coupled transcription/translation system optimized for production of recombinant proteins. Protein Expr. Purif. 2008;62(2):190–198. doi: 10.1016/j.pep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe TY, et al. Proteome analysis of rhoptry: Enriched isolated from Plasmodium. merozoites. Journal of Proteome Research. 2004;3(5):995–1001. doi: 10.1021/pr049926m. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe TY, Fujioka H, Aikawa M, Hall T, Drazbar JA. Plasmodium falciparum. protein located in Maurer’s clefts underneath knobs and protein localization in association with Rhop-3 and SERA in the intracellular network of infected erythrocytes. Parasitol Res. 2001;87(3):173–185. doi: 10.1007/pl00008572. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe TY, et al. A Plasmodium. gene family encoding Maurer’s cleft membrane proteins: Structural properties and expression profiling. Genome Res. 2004;14(6):1052–1059. doi: 10.1101/gr.2126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam-Yellowe TY, Banks TL, Fujioka H, Drazba JA, Yadav SP. Plasmodium yoelii.: Novel rhoptry proteins identified within the body of merozoite rhoptries in rodent Plasmodium malaria. Exp Parasitol. 2008;120(1):113–117. doi: 10.1016/j.exppara.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe TY, Rio D, Fujioka H, Aikawa M, Yang JC, Yakubu Z. Isolation of merozoite rhoptries, identification of novel rhoptry associated proteins from P. yoelii., P. chabaudi., P. berghei. and conserved interspecies reactivity of organelles and proteins with P. falciparum. rhoptry-specific antibodies. Exp Parasitol. 1998;89(3):271–284. doi: 10.1006/expr.1998.4280. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe TY, Fujioka H, Masamichi A, Messineo DG. Plasmodium falciparum. rhoptry proteins of 140/130/110kd (Rhop-H) are located in an electron lucent compartment in the neck of the rhoptries. J Euk Microbial. 1995;42(3):224–231. doi: 10.1111/j.1550-7408.1995.tb01570.x. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe TY, Hishashi F, Masamichi A, Messineo DG, Leash AM, Hall T, Drazba JA, Ndengele MM. Molecular Organization and cross linking analysis of the Plasmodium falciparum erythrocyte binding proteins Rhop-H and SERA. J Protozool. Res. 2000;10(3):128–154. [Google Scholar]
- Sam-Yellowe TY, Shio H, Perkins M. Secretion of Plasmodium falciparum. rhoptry protein into plasma membrane of host erythrocytes. J Cell Biol. 1988;106(5):1507–1513. doi: 10.1083/jcb.106.5.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresh S, et al. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics. 2006;22(14):1760–1766. doi: 10.1093/bioinformatics/btl162. [DOI] [PubMed] [Google Scholar]
- Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2006;72(2):211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193(4254):673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Tsarukyanova I, Drazba JA, Fujioka H, Yadav S, Sam-Yellowe TY. Proteins of the Plasmodium falciparum. two transmembrane Maurer’s cleft protein family, PfMC-2TM and the 130 kDa Maurer’s cleft protein define different domains of the infected erythrocyte intramembranous network. Parasitol Res. 2009;104(4):875–891. doi: 10.1007/s00436-008-1270-3. [DOI] [PubMed] [Google Scholar]
- Tsuboi T, et al. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect. Immun. 2008;76(4):1702–1708. doi: 10.1128/IAI.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Takeo S, Sawasaki T, Torii M, Endo Y. An efficient approach to the production of vaccines against the malaria parasite. Methods in Mol. Biol. 2010;607:73–83. doi: 10.1007/978-1-60327-331-2_8. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. A versatile protein microarray platform enabling antibody profiling against denatured proteins. Proteomics Clin. Appl. 2013;7(5-6):378–383. doi: 10.1002/prca.201200062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Fujioka H, Drazba JA, Sam-Yellowe TY. Rhop-3 protein conservation among Plasmodium. species and induced protection against lethal. 2006;99(3):238–252. doi: 10.1007/s00436-006-0136-9. [DOI] [PubMed] [Google Scholar]
- Yadavalli R, Ledger C, Sam Yellowe TY. In vitro human cell free expression for synthesis of malaria proteins. Parasitol Res. 2012;111(6):2461–2465. doi: 10.1007/s00436-012-3014-7. [DOI] [PubMed] [Google Scholar]
- Machida K, Masutan M, Imataka H. Protein Synthesis in vitro.: Cell-Free Systems Derived from Human Cells. Intech. 2012.
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]


