Abstract
The zebrafish has become a very important model organism for studying vertebrate development, physiology, disease, and tissue regeneration. A thorough understanding of the molecular and cellular mechanisms involved requires experimental tools that allow for inducible, tissue-specific manipulation of gene expression or signaling pathways. Therefore, we and others have recently adapted the TetON system for use in zebrafish. The TetON system facilitates temporally and spatially-controlled gene expression and we have recently used this tool to probe for tissue-specific functions of Wnt/beta–catenin signaling during zebrafish tail fin regeneration. Here we describe the workflow for using the TetON system to achieve inducible, tissue-specific gene expression in the adult regenerating zebrafish tail fin. This includes the generation of stable transgenic TetActivator and TetResponder lines, transgene induction and techniques for verification of tissue-specific gene expression in the fin regenerate. Thus, this protocol serves as blueprint for setting up a functional TetON system in zebrafish and its subsequent use, in particular for studying fin regeneration.
Keywords: Developmental Biology, Issue 100, Tetracycline-controlled transcriptional activation, TetON, zebrafish, Regeneration, fin, tissue-specific gene expression, doxycycline, cryosectioning, transgenic, Tol2, I-SceI, anesthesia
Introduction
The zebrafish is a well-established vertebrate model organism to study many aspects of development, physiology, disease, and regeneration. With the growing adoption of zebrafish as a model for post-embryonic biological processes, experimental tools for inducible, tissue-specific manipulation of gene expression or signaling pathways have become increasingly important. Particularly, studies into organ and appendage regeneration in adult zebrafish have suffered from a lack of tools for dissection of the spatio-temporal requirements of signaling pathways during these regenerative processes.
Currently, three different systems have been used to achieve conditional, tissue-specific gene expression in regenerating organs of adult zebrafish: the Cre-lox system, mosaic expression of heat-shock inducible transgenes using transposon-mediated somatic transgenesis, and the TetON system1-3. TetON refers to a variant of a tetracycline-controlled transcriptional activation system, where expression is activated in the presence of the antibiotic tetracycline or a derivative, e.g. doxycycline. The Cre-lox system, as it has so far been used in adult fish, relies on a Tamoxifen-controlled Cre recombinase (CreERT2), whose expression is spatially restricted by tissue-specific regulatory elements. Cre-driven removal of a STOP cassette facilitates expression of the gene of interest driven by a promoter that should be active in all cell types1. Transposon-mediated creation of mosaically expressed somatic transgenes provides a system for inducible transgene expression in individual cell lineages. Injection of zebrafish embryos with a Tol2 transposon carrying a gene of interest under transcriptional control of a heat shock promoter results in chimeric individuals typically carrying the transgene only in discrete cell lineages of a regenerating organ2. While both systems allow for conditional tissue-specific gene expression, the Cre-lox system is not reversible, and the strategy using transposon-based clonal labeling suffers from its stochastic nature. Thus, we and others have recently adapted transgenic TetON systems for use in zebrafish, which facilitate temporally and spatially-controlled gene expression that is in addition tunable and reversible3-5.
The TetON system used here comprises a transgenic driver line (TetActivator) in which a Doxycycline (DOX)-inducible transcriptional activator (improved reverse tetracycline transactivator, irtTA, short TetA) is under control of tissue-specific genomic regulatory sequences. Secondly, it requires a transgenic responder line (TetResponder) that harbors a gene of interest under transcriptional control of the Tetracycline operator (Tet response element; TetRE) (Figure 1A). Thus, the use of specific combinations of TetActivator and TetResponder lines allows for conditional tissue-specific manipulation of gene expression.
We have recently utilized the TetON system to probe for tissue-specific functions of Wnt/beta–catenin signaling in the adult regenerating zebrafish tail fin3. In the protocol outlined here we describe a work flow for set-up and use of the TetON system in zebrafish, in particular for studies of fin regeneration. This includes detailed instructions on how to generate stable transgenic TetActivator and TetResponder lines and a protocol for transgene induction in embryos and adult zebrafish. Furthermore, we describe techniques for verification of tissue-specific gene expression in the fin regenerate, including a protocol for the preparation of cryosections of adult zebrafish fins. Additionally, we discuss considerations for the design of the TetActivator transgene, the choice of a transgenesis method, and detection of TetResponder expression. Hence, the overall goal of this protocol is to serve as a blueprint for setting-up a functional TetON system in zebrafish to achieve conditional tissue-specific gene expression, which can be applied to any tissue of interest.
We have created a TetActivator vector allowing for I-SceI or Tol2-mediated generation of stable TetActivator lines using short genomic regulatory sequences (enhancer fragments; Weidinger lab plasmid database no. 1247; Figure 1B). This construct contains a TetActivator cassette consisting of the M2 mutant variant of the reverse Tet repressor domain fused with the Herpes simplex virus VP16 transactivation domain-derivative 3F [irtTAM2(3F)]. Expression of the TetActivator (TetA) can be easily monitored since it is co-expressed with the fluorophore AmCyan from the same open reading frame; a p2a peptide mediates ribosomal skipping, which should result in production of TetA and AmCyan as separate proteins at a 1:1 ratio5,6. The construct also contains a poylinker 5’ of the TetActivator cassette to facilitate insertion of genomic regulatory sequences of interest using conventional cloning methods.
Additionally, we have created a construct consisting of the above described TetActivator cassette plus a Kanamycin selection cassette (Weidinger lab plasmid database no. 1180; Figure 1C), which can be recombined into a bacterial artificial chromosome (BAC) containing a large genomic region (usually into the start codon of a gene whose expression pattern is to be mimicked by the transgene). Both constructs are available from the Weidinger lab upon request.
Protocol
1. Generation of Transgenic TetActivator Fish Lines
- Generation of TetActivator construct
- Clone regulatory sequences of interest upstream of the TetActivator cassette in vector #1247 using standard techniques. Alternatively, use recombination techniques to generate a BAC, in which the TetActivator cassette (vector #1180) is inserted into the first exon of the target gene, and where the Tol2 inverted repeats are introduced into the BAC backbone (for a detailed recombineering protocol see7,8).
- Prepare toxin-free plasmid DNA or BAC DNA preparations using commercially available kits.
- Generation of transgenic TetActivator fish lines
- Perform microinjection of the TetActivator construct according to standard procedures9.
- Inject 25-50 pg of the plasmid DNA or up to 250 pg BAC DNA into one-cell-stage embryos together with capped sense RNA coding for tol2 transposase for transposon-mediated transgenesis, or with I-SceI protein for I-SceI-mediated transgenesis. NOTE: Considerations on the choice of the transgenesis method can be found in the discussion section. NOTE: Inject preferably into the cytoplasm, not the yolk, since efficiency of transgenesis might decrease when the DNA is not delivered into the cytoplasm.
- Raise injected G0 embryos to adulthood. Optionally, screen G0 embryos for AmCyan fluorescence and preferentially grow embryos showing the desired expression pattern. This might increase the rate of germ-line transmission, in particular for Tol2-mediated transgenesis.
- Assess successful germ-line integration by outcrossing individual G0 fish to wild-type fish and appearance of AmCyan fluorescence in the expected expression pattern in F1 embryos or larvae using a fluorescent stereomicroscope (example is shown in Figure 1D).
- Raise fluorescence-positive F1 embryos to adulthood and mate them with wild-type fish to obtain stable F2 transgenic fish.
- Raise and characterize at least 3 different sublines derived from different G0 founder fish, since individual lines might differ in expression levels, expression pattern, ability to induce TetResponder lines, leakiness, and their susceptibility to silencing. NOTE: Many transgenes that are robustly expressed in embryos are either partially or completely silenced in late-stage larvae or adult fish. Thus, if lines are meant to be used in adult fish, characterize several independent lines for their TetActivator expression and function during adulthood. NOTE: We have generated a panel of TetActivator lines for tissue-specific expression of the TetActivator in both embryos and adult fish, which are listed in Table 1. These lines are available from the Weidinger lab upon request.
2. Generation of Transgenic TetResponder Fish Lines
NOTE: We have generated a TetResponder construct allowing for I-SceI or Tol2-mediated generation of stable TetResponder lines, which is available from the Weidinger lab upon request (Weidinger lab plasmid database no. 1444; Figure 1E). This construct contains a Tetracycline operator, followed by a polylinker region facilitating the insertion of coding sequences (CDS) of a gene of interest and the YFP-derivative YPet coding sequence. Thus, the construct is designed for TetA-mediated expression of a C-terminal fusion of the protein of interest with YPet. If expression of a tagged fusion protein has to be avoided, a p2a or t2a peptide can be introduced with the gene of interest CDS, which facilitates co-expression of the protein of interest and YPet as separate polypeptides6,10.
- Generation of TetResponder construct
- Clone CDS of the gene of interest 3’ of the Tetracycline operator and 5’ of the YPet CDS according to standard procedures. Ensure that the stop codon has been removed from the gene of interest CDS.
- Prepare toxin-free plasmid DNA preparations using commercially available kits.
- Generation of transgenic TetResponder fish lines
- Perform microinjection of TetResponder construct according to standard procedures9.
- Inject 25-50 pg of the plasmid DNA into the cytoplasm of one-cell-stage embryos together with capped sense RNA coding for tol2 transposase for transposon-mediated transgenesis or with I-SceI enzyme for meganuclease-aided transgenesis. NOTE: Considerations on the choice of the transgenesis method can be found in the discussion section. NOTE: Inject preferably into the cytoplasm, not the yolk, since efficiency of transgenesis might decrease when the DNA is not delivered into the cytoplasm.
- Raise injected G0 embryos to adulthood.
- Assess successful germ-line integration and inducibility of the transgene by mating individual G0 fish with a previously established TetActivator line followed by Doxycycline treatment of F1 embryos or F1 larvae. NOTE: Usually we use a ubiquitin promoter-driven TetActivator line (ubiquitin:irtTAM2(3F)-p2a-AmCyanulm2, short ubiquitin:TetA AmCyan3) to screen for functional TetResponder carriers, as this line drives fairly ubiquitous expression in embryos and adult fish and thus is well suited to assess TetResponder inducibility in all tissues of interest. This line is available from the Weidinger lab upon request.
- Collect embryos of each clutch in a separate 10 cm Petri dish containing E3 embryo medium (see Materials table) and incubate dishes at 28.5 °C.
- Place G0 fish that gave embryos into individual numbered tanks (breeding boxes; see Materials table) until embryos have been screened. This allows for recovery of G0 fish that show germ line transmission of the transgene.
- If only half of the embryos are expected to carry the TetActivator transgene (in case a heterozygous TetActivator carrier was crossed to a TetResponder founder fish), select for embryos containing the TetActivator transgene by appearance of AmCyan fluorescence at the appropriate developmental stage.
- Induce TetActivator transcriptional activity by treating embryos with Doxycycline (DOX) as described in section 2.3.
- Screen F1 embryos for emergence of YPet fluorescence. Prefer lines (G0 fish) that produce homogenous YPet expression, that is embryos in the clutch should display little mosaicism within the expected expression domain and little variation between individual embryos. See arrowheads in Figure 1F and Figure 1G for representative examples.
- Mate G0 fish transmitting an inducible TetResponder transgene identified in step 2.2.8 with wild-type fish and raise all F1 embryos to adulthood.
- Identify adult F1 transgenic fish either by PCR-based genotyping (see section or by crossing individual F1 fish to a TetActivator line followed by DOX treatment of F2 embryos and emergence of YPet fluorescence (see section 2.5).
- Establish and characterize at least 3 different TetResponder sublines derived from different G0 founder fish, since inducibility might differ in individual lines. In particular achievable expression levels and whether the responder can be activated in all cell types might vary. In addition, susceptibility to silencing will differ between lines. Finally, in some cases it might be preferable to select lines that mediate only moderate expression levels of the protein of interest, in particular if leaky expression produced in combination with a TetActivator line in the absence of Doxycycline is expected to cause developmental defects or toxicity. NOTE: TetResponder lines that are robustly inducible in embryos may be either partially or completely silenced in late-stage larvae or adult fish. Thus, if lines are meant to be used in adult fish, several independent lines should be characterized for inducibility in adults. If induction of the gene of interest is compatible with further development, it has been proven useful to create double transgenic embryos (TetActivator x TetResponder), to induce GOI expression in embryos and to select and raise only the individuals showing the most robust and least mosaic induction.
- To maintain and propagate established TetResponder lines, genotype adult fish as described in section 2.5).
- Doxycycline treatment of embryos
- Prepare 2,000x Doxycycline (DOX) stocks. Solve DOX in 50% EtOH to achieve a concentration of 50 mg/ml (97 mM). Store DOX stocks protected from light at -20 °C.
- Decant the majority of E3 medium from embryos and replace it with E3 embryo medium containing DOX at a final concentration of 25 µg/ml DOX (2.5 µl of 2,000x DOX stock per 50 ml E3). Dechorionation is not required.
- Return embryos to 28.5 °C for a minimum of 6 hr before assessing expression of the tagged gene of interest by appearance of YPet fluorescence.
- Assess leakiness of the TetActivator/TetResponder combination by treating embryos with EtOH solvent only. NOTE: In situ hybridization (ISH) or RT-PCR to detect the TetResponder mRNA can be used as more sensitive measures of leakiness than fluorescence of the tagged gene of interest. The extent of leakiness will depend on the particular TetActivator/TetResponder line combination and it may also vary between tissues and developmental stages.
- Anesthesia of adult zebrafish
- Prepare 10 cm diameter Petri dish or small beaker filled with fish system water containing 1 mg/ml Tricaine (Ethyl 3-aminobenzoate methanesulfonate, MS-222).
- Transfer fish to container containing anesthetic and wait until it reaches level 4 of anesthesia as indicated by complete loss of equilibrium (fish lies on its side), no movements and no response to touching11.
- Carefully transfer fish using tweezers or a spoon onto a glass slide, lid of a plastic petri dish, or agarose-coated petri dish and perform fin amputation or imaging (see section 3.2).
- Return fish to a container containing a large volume of fresh fish system water and monitor it. If gill movements do not resume within 3 min, assist by blowing water over the gills using a plastic transfer pipet.
- PCR-based genotyping of TetResponder fish
- Perform partial amputation of the tail fin12. Place fish into individual, numbered tanks (breeding boxes; see Materials table). This allows for the recovery of transgenic fish following successful genotyping.
- Transfer fin tissues to individual wells of a 96-well PCR plate pre-filled with 20 µl DNase-free water for isolation of genomic DNA13.
- Add 120 µl 1 M NaOH to each well and incubate samples for 20 min at 95°C using a thermocycler.
- Chill samples to 4°C, add 14 µl of 1 M Tris-HCl (pH 8.0) to each well, and briefly vortex samples. Centrifuge samples for 5 min at 16,000 x g to pellet cell debris. Store genomic DNA at 4 °C until needed.
- Design primers to specifically amplify a fragment of the TetResponder transgene. Use 2 µl of isolated DNA in a standard PCR reaction and analyze PCR product on an agarose gel.
3. Tissue-specific Induction of Transgene Expression in the Adult Regenerating Zebrafish Tail Fin
- Establishment of TetActivator; TetResponder double transgenic fish
- Mate TetActivator carriers with TetResponder carriers.
- Select embryos carrying the TetActivator transgene at a developmental stage when the regulatory elements driving the TetActivator cassette are active. Transgenic embryos can easily be identified by appearance of AmCyan fluorescence using a stereo fluorescent microscope. NOTE: TetActivator transgene-positive fish may alternatively be identified during adulthood by imaging of anesthetized fish, eg. following amputation of the tail fin and emergence of AmCyan fluorescence in the regenerate.
- Raise selected embryos to adulthood.
- Identify double transgenic adult fish (fish that carry the TetResponder in addition to the TetActivator) by PCR-based genotyping or (preferably) by their ability to induce expression of the TetResponder in the tissue of interest. A protocol for TetResponder induction and detection in the regenerating tail fin is described in section 2.3 and section 4. NOTE: If transient TetResponder expression is compatible with further embryonic or larval development, it is useful to identify double transgenic embryos carrying TetActivator and TetResponder transgenes following DOX treatment during embryonic development as described above (section 2.3, Figure 1H). Double positive embryos will show AmCyan and YPet fluorescence. Raise positive embryos to adulthood.
- Amputation of zebrafish tail fins and induction of transgene expression in the regenerating fin
- Perform partial amputation of the tail fin on anesthetized fish (see section 2.4)12. Treat fish either immediately with DOX or return them to the fish housing system.
- Prepare breeding box (Figure 2A; see Materials table) filled with 1 L of fish system water containing 25 µg/ml DOX (add 500 µl of 2,000x 50 mg/ml stock to 1 L of fish system water). NOTE: Induction of transgene expression immediately after amputation allows for assessment of effects on early events during regeneration, while induction at 3 days post-amputation (3 dpa) should be used to assess transgene expression or function during regenerative growth.
- Transfer up to 10 previously amputated, double transgenic fish to the breeding box and close box with a lid such that fish cannot escape (Figure 2A).
- Place breeding boxes into the dark to reduce stress. We usually place the boxes into a 28.5 °C air incubator to ensure constant darkness and temperature.
- Treat negative control double transgenic fish with EtOH only (250 µl per 1L fish system water). NOTE: Single transgenic or wild-type fish treated with DOX can be used as additional negative control, although we have never observed adverse effects of the DOX doses used here on fin regeneration.
- Anesthetize fish as described in section 2.4 and transfer fish using tweezers or a spoon onto a wetted agarose-coated petri dish. Carefully spread the tail fin using tweezers.
- Screen fish for appearance of YPet within the regenerate using a fluorescent stereo-microscope (Figure 2B). NOTE: Usually, TetResponder-driven fluorescence can be robustly observed following 6 hr of DOX treatment. However, we recommend to characterize the dynamics of TetResponder expression for each line generated.
- Return fish to the fish housing system to terminate TetResponder transgene induction or continue treatments. NOTE: For treatments >24 hr (long-term treatments) it is essential to exchange water and thoroughly clean the breeding boxes daily.
- During long-term treatments, feed fish every 2-3 days by transferring fish into a clean container with fresh fish system water before returning them to breeding boxes after feeding.
4. Characterization of TetResponder Expression in Fin Regenerates
NOTE: We usually verify tissue-specific gene expression in the regenerating tail fin using fluorescent imaging of cryosections at 3 dpa. At this time point the different tissue compartments of a regenerate have formed and can be clearly identified on tissue-sections, and cryosectioning is simpler than at later stages of regeneration. Figure 2C depicts the tissue domains that can be distinguished in the regenerate and lists a few molecular markers for these domains. The following protocol describes the preparation of longitudinal or transverse cryosections for direct imaging or immunostaining.
- Cryosectioning of adult tail fins
- Treat fish whose tail fins were amputated from 2 dpa until 3 dpa with DOX as described in section 3.2
- Anesthetize fish as described in section 2.4
- Use a spoon or tweezers to gently transfer fish onto a lid of a plastic Petri dish.
- Re-amputate the fin regenerate at about 4-5 bony segments proximal to the initial amputation plane using a scalpel and return the fish to fresh fish system water.
- Carefully transfer regenerates to a small petri dish (e.g., 35 mm) containing 4% (w/v) paraformaldehyde (PFA) in 1x Phosphate-buffered saline (PBS; see Materials table) using fine tweezers. Do not touch the regenerate but only the stump part of the tissue.
- Place tissue flat onto the petri dish bottom and fix O/N at 4 °C.
- Wash regenerates twice in 1x PBS containing 0.1% Tween (PBT) to remove PFA and incubate regenerates o/n at 4 °C in 0.5 M EDTA-PBS (pH 7.5) to decalcify bone matrix.
- Incubate regenerates at RT in 10% sucrose-PBS, 20% sucrose-PBS, 30% sucrose-PBS and 30% sucrose-PBS:tissue freezing medium at a 1:1 ratio (TFM; see Materials table) for 30 min each. Subsequently, incubate fins at 4 °C for >2 hr in tissue freezing medium.
- Place cryomolds onto a microscope slide and fill them with tissue freezing medium. Avoid introducing air bubbles.
- Transfer regenerates into cryomolds and orient tissue appropriately to obtain longitudinal or transverse sections of fin regenerates (Figure 2D). Ensure that regenerate is straight, which facilitates sectioning.
- Transfer cryomolds together with microscope slide onto a metal rack positioned on a bed of dry ice to start the freezing process (Figure 2D).
- Once the tissue freezing medium has become solid, place cryomold onto bench and let sit at RT for a few minutes such that the tissue freezing medium thaws at the plastic-medium interface.
- Use a pen or similar to push the frozen block out of the cryomold. Store cryoblocks in a box at -80 °C until tissue sectioning.
- Prepare 10-14 µm thick cryosections using a Cryo-microtome and collect sections on adhesion microscope slides (see Materials table). Store slides at -20 °C until needed.
- Characterization of TetResponder expression on fin sections based on YPet fluorescence
- Treat fin sections for 30 min with 100% MeOH chilled to -20 °C to improve adherence to the microscope slide followed by two washes at RT in PBT and 1 wash in PBS, 5 min each.
- Visualize nuclei by incubating sections for 10 min in 4’, 6- diamidino-2-phenylinodole (Dapi) in PBS (1:5,000) followed by two washes 10 min each in PBS.
- Mount sections in 75% Glycerol-PBS or commercially available antifade mounts, and cover with a cover slide.
- Image YPet and AmCyan fluorescence under a confocal or wide-field fluorescent microscope (Figure 1E). NOTE: To facilitate unambiguous identification of cell types expressing YPet, immunofluorescence or immunohistochemistry with antibodies against cell-type specific antigens together with anti-GFP antibodies (which react with YPet, but not AmCyan) can be performed. Alternatively, to facilitate detection of weak YPet expression, RNA in situ hybridization against YPet RNA can be performed either on whole-mount fins or paraffin sections of fins.
Representative Results
To establish a functional TetON system for tissue-specific inducible gene expression, transgenic TetActivator and TetResponder lines need to be generated (Figure 1A). This is accomplished by microinjecting TetActivator (Figure 1B-C) or TetResponder (Figure 1E) constructs into early zebrafish embryos and subsequent germ-line integration. Functional TetActivator constructs can either be generated by cloning of short regulatory sequences (enhancer elements) upstream of the TetActivator cassette (Figure 1B), or by recombining the TetActivator cassette into a BAC containing regulatory elements of the gene of interest (Figure 1C). Germline integration of the TetActivator construct can be assessed by outcrossing individual injected G0 fish and appearance of AmCyan fluorescence in F1 embryos in the expected expression pattern (Figure 1D). Germline integration and functionality of the TetResponder transgene can be assessed by crossing individual G0 injected fish to established TetActivator fish (here ubiquitin:TetA AmCyan), followed by treatment with DOX and appearance of YFP/YPet fluorescence (Figure 1F). Fish showing strong and homogenous YFP/YPet expression should be preferred to establish a stable TetResponder line (Figure 1G). Having established stable TetActivator and TetResponder fish lines, specific TetActivator/TetResponder combinations can be generated by crossing. Carriers of TetActiator and TetResponder transgenes can be identified during embryogenesis following DOX treatment end emergence of YFP/Ypet at the developmental stage the TetActivator is active (Figure 1H). Alternatively, double transgenic fish can be identified following DOX treatment and TetResponder transgene induction in the regenerating tail fin (Figure 2A-B).
We routinely use this system to achieve inducible, tissue-specific gene expression in the regenerating zebrafish tail fin. We verify tissue-specific TetResponder transgene expression by fluorescence imaging of cryosections of 3 dpa fin regenerates, since the different tissue compartments of a regenerate can be clearly identified on tissue-sections at this time-point (Figure 2C). Longitudinal or transverse fin regenerate sections are obtained by cryosectioning after cryoprotecting tissue and embedding fin regenerate appropriately (Figure 2D). Following sectioning, TetResponder transgene expression (Ypet/YFP fluorescence) can be imaged using fluorescence or confocal microscopy (Figure 2E). Note that the her4.3 promoter-driven TetActivator induces TetResponder expression in the proximal medial blastema, while the distal blastema (arrowhead) and the epidermis (arrow) are devoid of expression.
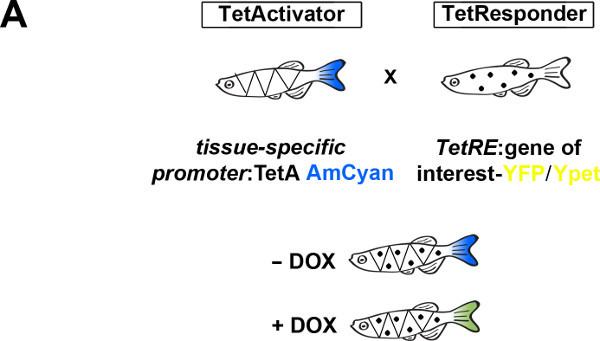
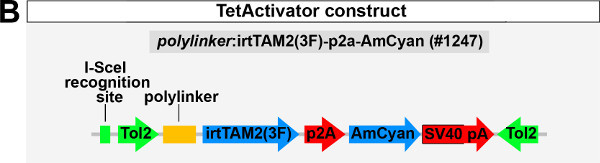
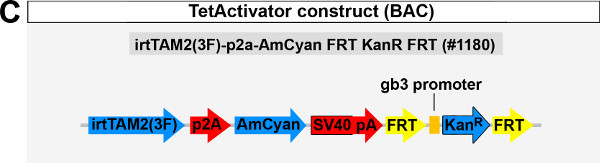
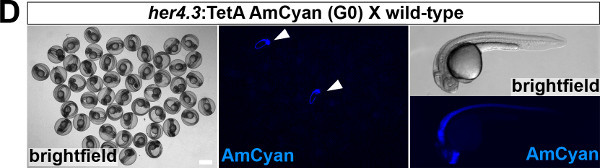
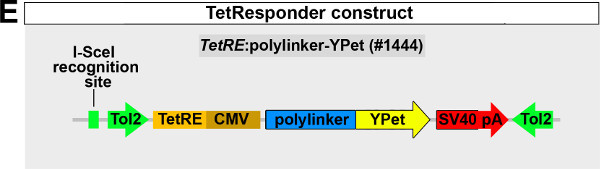
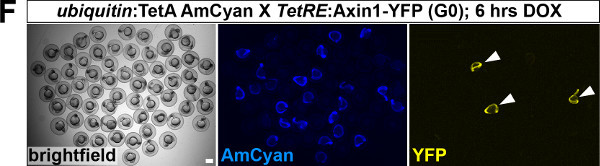
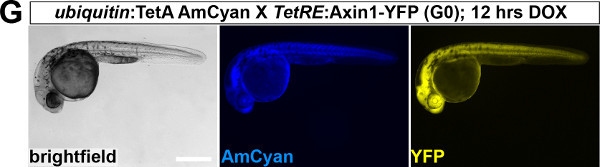
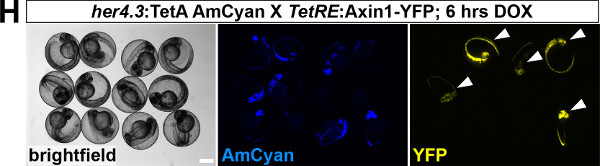 Figure 1: Generation of transgenic TetActivator and TetResponder lines.(A) Cartoon showing strategy for tissue-specific inducible gene expression using the TetON system. (B) Transgenic TetActivator construct (Weidinger lab plasmid database no. 1247). A polylinker 5’ of the TetActivator irtTAM2(3F) facilitates cloning of regulatory sequences of interest. The AmCyan coding sequence is separated from the TetActivator via a p2a ‘self-cleaving’ peptide. The cassette also contains a SV40 polyadenylation signal, is flanked by Tol2 transposon inverted repeats and contains a single I-SceI meganuclease recognition site. (C) TetActivator cassette for BAC recombineering (Weidinger lab plasmid database no. 1180). The cassette includes the Kanamycin gene for selection, which is driven by a gb3 prokaryotic promoter. FRT sites flanking the Kanamycin resistance gene allow for its Flp-recombinase mediated removal following successful integration into the BAC. (D) Identification of G0 founder fish transmitting the her4.3:TetA AmCyan construct through their germ-line. Carriers are identified via emergence of AmCyan fluorescence in some F1 embryos (arrowheads). (E) Transgenic TetResponder construct (Weidinger lab plasmid database no. 1444). This constructs consists of a polylinker and YPet CDS under control of optimized Tet response elements (TetRE-tight, Clontech) and terminated by the SV40 polyadenylation signal. It is flanked by Tol2 transposon inverted repeats and contains a single I-SceI meganuclease recognition site. (F) Identification of G0 founder fish transmitting a functional TetResponder TetRE:Axin1-YFP construct through their germline. Carriers are identified by crossing with an established TetActivator line (here ubiquitin:TetA AmCyan) and induction of YFP fluorescence following DOX treatment for 6 hr (arrowheads). (G) Close up ofembryos shown in (F) after a total of 12 hr of DOX treatment. Note fairly ubiquitous induction of YFP fluorescence. (H) Identification of double transgenic embryos carrying TetActivator (her4.3:TetA AmCyan) and TetResponder (TetRE:Axin1-YFP) transgenes (arrowheads). Embryos were treated with DOX for 6 hr. (A-H) Scale bar: 500 µm Please click here to view a larger version of this figure.
Figure 1: Generation of transgenic TetActivator and TetResponder lines.(A) Cartoon showing strategy for tissue-specific inducible gene expression using the TetON system. (B) Transgenic TetActivator construct (Weidinger lab plasmid database no. 1247). A polylinker 5’ of the TetActivator irtTAM2(3F) facilitates cloning of regulatory sequences of interest. The AmCyan coding sequence is separated from the TetActivator via a p2a ‘self-cleaving’ peptide. The cassette also contains a SV40 polyadenylation signal, is flanked by Tol2 transposon inverted repeats and contains a single I-SceI meganuclease recognition site. (C) TetActivator cassette for BAC recombineering (Weidinger lab plasmid database no. 1180). The cassette includes the Kanamycin gene for selection, which is driven by a gb3 prokaryotic promoter. FRT sites flanking the Kanamycin resistance gene allow for its Flp-recombinase mediated removal following successful integration into the BAC. (D) Identification of G0 founder fish transmitting the her4.3:TetA AmCyan construct through their germ-line. Carriers are identified via emergence of AmCyan fluorescence in some F1 embryos (arrowheads). (E) Transgenic TetResponder construct (Weidinger lab plasmid database no. 1444). This constructs consists of a polylinker and YPet CDS under control of optimized Tet response elements (TetRE-tight, Clontech) and terminated by the SV40 polyadenylation signal. It is flanked by Tol2 transposon inverted repeats and contains a single I-SceI meganuclease recognition site. (F) Identification of G0 founder fish transmitting a functional TetResponder TetRE:Axin1-YFP construct through their germline. Carriers are identified by crossing with an established TetActivator line (here ubiquitin:TetA AmCyan) and induction of YFP fluorescence following DOX treatment for 6 hr (arrowheads). (G) Close up ofembryos shown in (F) after a total of 12 hr of DOX treatment. Note fairly ubiquitous induction of YFP fluorescence. (H) Identification of double transgenic embryos carrying TetActivator (her4.3:TetA AmCyan) and TetResponder (TetRE:Axin1-YFP) transgenes (arrowheads). Embryos were treated with DOX for 6 hr. (A-H) Scale bar: 500 µm Please click here to view a larger version of this figure.

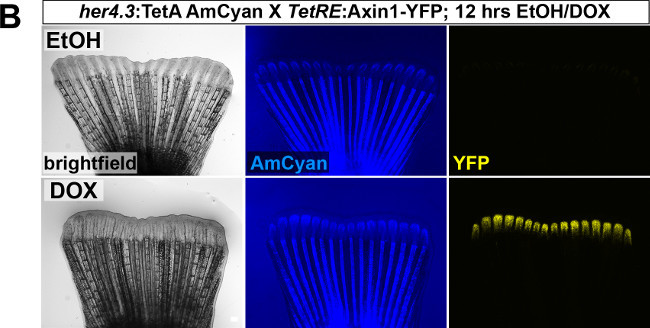
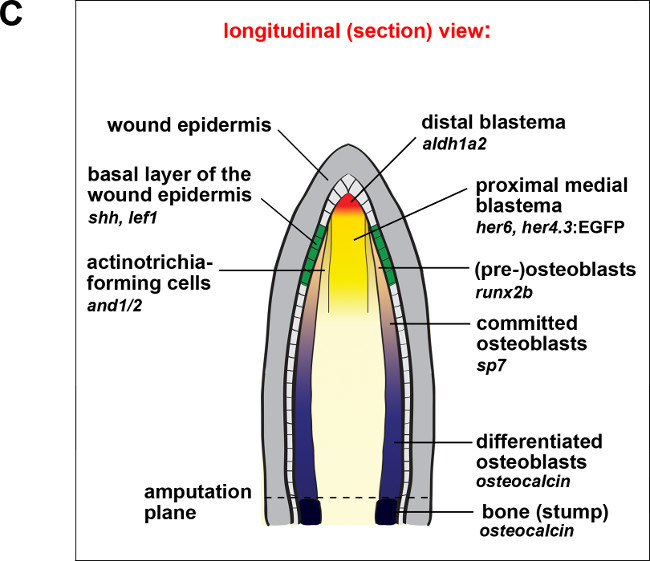

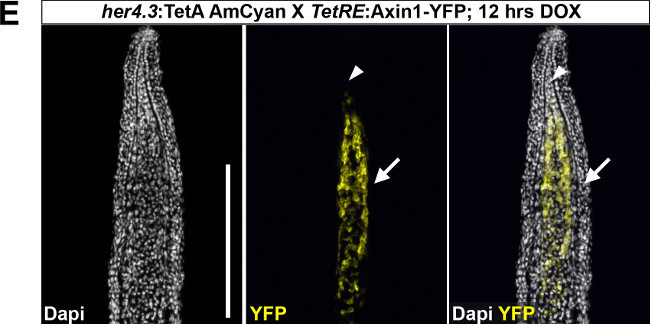 Figure 2: Use of the TetON system for tissue-specific gene expression in the regenerating zebrafish tail fin.(A) Breeding boxes used for DOX treatment of adult zebrafish. (B) Induction of TetRE:Axin1-YFP TetResponder transgene by the her4.3:TetA AmCyan TetActivator in the regenerating tail fin following 12 hr of DOX treatment. Note the absence of YFP fluorescence in the EtOH-treated control fish. (C) Cartoon depicting some of the tissue compartments found within a fin regenerate at 3 dpa in a longitudinal section view. (D) Sample preparation for cryosectioning. Fin regenerates are placed in tissue-freezing medium (TFM)-filled cryomolds, oriented appropriately to obtain either longitudinal or transverse sections of the regenerate, and transferred to a metal rack sitting on top of dry-ice until TFM has solidified. Sectioning plane is indicated in red. Abbreviations: dist.: distal, prox.: proximal, vent.: ventral, dors.: dorsal. (E) Confocal image of YFP fluorescence in a longitudinal section of a fin ray regenerate derived from fins shown in (B). Note that the her4.3:TetA AmCyan TetActivator line drives YFP induction specifically in the proximal-medial blastema. YFP fluorescence is not detected in a variety of compartments, including the wound epidermis (arrow), and the distal-most domain of the mesenchyme (arrowhead) (B-C) Scale bar: 500 µm; (E) Scale bar: 100 µm. Please click here to view a larger version of this figure.
Figure 2: Use of the TetON system for tissue-specific gene expression in the regenerating zebrafish tail fin.(A) Breeding boxes used for DOX treatment of adult zebrafish. (B) Induction of TetRE:Axin1-YFP TetResponder transgene by the her4.3:TetA AmCyan TetActivator in the regenerating tail fin following 12 hr of DOX treatment. Note the absence of YFP fluorescence in the EtOH-treated control fish. (C) Cartoon depicting some of the tissue compartments found within a fin regenerate at 3 dpa in a longitudinal section view. (D) Sample preparation for cryosectioning. Fin regenerates are placed in tissue-freezing medium (TFM)-filled cryomolds, oriented appropriately to obtain either longitudinal or transverse sections of the regenerate, and transferred to a metal rack sitting on top of dry-ice until TFM has solidified. Sectioning plane is indicated in red. Abbreviations: dist.: distal, prox.: proximal, vent.: ventral, dors.: dorsal. (E) Confocal image of YFP fluorescence in a longitudinal section of a fin ray regenerate derived from fins shown in (B). Note that the her4.3:TetA AmCyan TetActivator line drives YFP induction specifically in the proximal-medial blastema. YFP fluorescence is not detected in a variety of compartments, including the wound epidermis (arrow), and the distal-most domain of the mesenchyme (arrowhead) (B-C) Scale bar: 500 µm; (E) Scale bar: 100 µm. Please click here to view a larger version of this figure.
| TetActivator line | Reference | Regulatory elements used | Primarily expressed in |
| 7xTCF-Xla.Siam:irtTAM2(3F)-p2a-AmCyan | Wehner &Weidinger, unpublished | 7xTCF-Xla.Siam Wnt reporter 1 | Wnt-responsive tissues |
| myl7: irtTAM2(3F)-p2a-mCherry | Haase & Weidinger, unpublished | myl7 (cmlc2) 2 | mature cardiomyocytes |
| her4.3: irtTAM2(3F)-p2a-AmCyanulm6 | 3 | her4.3 4 | central nervous system |
| keratin4: irtTAM2(3F)-p2a-AmCyanulm5 | 3 | keratin4 5 | epidermis, in the adult fin excluding the basal layer |
| keratin18: irtTAM2(3F)-p2a-AmCyanulm4 | 3 | keratin18 6 | epidermis, in the adult fin exclusively in the basal layer |
| sp7: irtTAM2(3F)-p2a-AmCyanulm3 | 3 | sp7/osx 7 | (committed) osteoblasts |
| ubiquitin:irtTAM2(3F)-p2a-AmCyanulm2 | 3 | ubiquitin 8 | (fairly) ubiquitous |
Table 1: Available transgenic TetActivator lines. This table lists transgenic TetActivator lines for tissue-specific expression of the TetActivator in both embryonic and adult fish, which are available from the Weidinger lab upon request.
Discussion
The adult zebrafish has an amazing capacity to successfully regenerate many internal organs and appendages. A thorough understanding of the molecular and cellular mechanisms involved requires tissue-specific analysis of gene functions and signaling pathways. Towards this, the TetON system provides an efficient tool for spatiotemporally controlled gene expression in embryonic and adult zebrafish. The TetON system constructs and methodology described in this manuscript have been successfully used in a recent study of our laboratory3. The following additional issues should be considered when establishing the TetON system:
TetActivator transgene design considerations
We have previously shown that a single-inducible TetActivator consisting of the M2 mutant variant of the reverse Tet repressor domain fused with the Herpes simplex virus VP16 transactivation domain-derivative 3F [irtTAM2(3F), in short TetA-M2] confers efficient induction, but can show low, albeit measurable leakiness, that is background inducing activity in the absence of Doxycycline (DOX)5. Therefore, we have described dually inducible systems using fusions with a glucocorticoid receptor (TetA-GBD) or a domain of the Ecdysone receptor (TetA-EcR), which eliminate leakiness5. Thus, if the system will be used to express toxic or very potent proteins (e.g., oncogenes) the use of a dually inducible variant should be considered. Alternatively, during establishment of TetActivator and TetResponder lines a range of lines producing different expression levels can be selected and weaker inducers chosen in case leakiness is an issue with strongly inducing lines. TetActivator constructs containing the TetA-GBD or TetA-EcR variant are available from the Weidinger lab upon request.
Transgenesis methods for generation of transgenic zebrafish lines
Generation of stable transgenic zebrafish lines is accomplished by microinjection of DNA constructs into the early embryo and subsequent germ-line insertion of the injected DNA. Two methods have been routinely used to achieve efficient germ-line transformation in zebrafish: i) enhancement of plasmid integration efficiency using I-SceI meganuclease14, and ii) Tol2 transposon-mediated transgenesis15. The first relies on co-injection of plasmid DNA together with I-SceI meganuclease protein, which results in linearization of the plasmid and thereby is thought to increase the efficiency of plasmid integration into the host genome16. The latter requires co-injection of tol2 transposase RNA together with DNA containing Tol2 transposable elements for transposase-mediated integration into the genome. The Tol2-mediated transgenesis strategy is generally thought to be more efficient and can be used to integrate large DNA constructs, e.g., bacterial artificial chromosomes (BACs) or P1-derived artificial chromosomes (PACs), however frequently results in multiple single-copy insertions at several genomic loci, which can make establishment of a stable transgenic line showing uniform expression levels and Mendelian inheritance of the transgene time-consuming. In contrast, the I-SceI meganuclease technique is usually less efficient and not recommended for BAC transgenesis, but yields single-copy or tandem array transgene integration at a single genomic locus. We have used both transgenesis methods to successfully create functional TetActivator or TetResponder lines.
TetResponder transgene expression analysis
To detect tissue-specific TetResponder transgene expression in adult fins we usually use fluorescent imaging of cryosections. In our experience, fluorescence of fluorescent proteins expressed by the transgene is preserved throughout the fixation and cryosectioning procedure and thus can be directly imaged. Additional methods for detection of TetResponder expression, which have been described elsewhere, are: i) immunostaining on sections, preferably against the fluorescent protein tag, here YPet, using an anti-GFP antibody, ii) fluorescent or chromogenic ISH on sections, or iii) whole-mount ISH followed by sectioning.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors thank Christa Haase, Doris Weber and Brigitte Korte for technical assistance. Work in the Weidinger lab is supported by grants of the Deutsche Forschungsgemeinschaft WE 4223/3-1, WE 4223/4-1 and by the Deutsche Gesellschaft für Kardiologie via an Oskar-Lapp-Stipendium and a Klaus-Georg-und-Sigrid-Hengstberger-Forschungsstipendium.
References
- Fang Y, et al. Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(33):13416–13421. doi: 10.1073/pnas.1309810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryon RC, Johnson SL. Clonal analysis of kit ligand a functional expression reveals lineage-specific competence to promote melanocyte rescue in the mutant regenerating caudal fin. PloS one. 2014;9(7):e102317. doi: 10.1371/journal.pone.0102317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner D, et al. Wnt/beta-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep. 2014;6(3):467–481. doi: 10.1016/j.celrep.2013.12.036. [DOI] [PubMed] [Google Scholar]
- Huang CJ, et al. Conditional expression of a myocardium-specific transgene in zebrafish transgenic lines. Dev Dyn. 2005;233(4):1294–1303. doi: 10.1002/dvdy.20485. [DOI] [PubMed] [Google Scholar]
- Knopf F, et al. Dually inducible TetON systems for tissue-specific conditional gene expression in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(46):19933–19938. doi: 10.1073/pnas.1007799107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45(10):625–629. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- Suster ML, Abe G, Schouw A, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc. 2011;6(12):1998–2021. doi: 10.1038/nprot.2011.416. [DOI] [PubMed] [Google Scholar]
- Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development. 2011;138(19):4327–4332. doi: 10.1242/dev.068080. [DOI] [PubMed] [Google Scholar]
- Yuan S, Sun Z. Microinjection of mRNA and morpholino antisense oligonucleotides in zebrafish embryos. J Vis Exp. 2009. [DOI] [PMC free article] [PubMed]
- Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one. 2011;6(4):e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews M, Varga ZM. Anesthesia and euthanasia in zebrafish. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2012;53(2):192–204. doi: 10.1093/ilar.53.2.192. [DOI] [PubMed] [Google Scholar]
- Hyde DR, Godwin AR, Thummel R. In vivo electroporation of morpholinos into the regenerating adult zebrafish tail fin. J Vis Exp. 2012. [DOI] [PMC free article] [PubMed]
- Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques. 2007;43(5):610. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- Thermes V, et al. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118(1-2):91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- Suster ML, Kikuta H, Urasaki A, Asakawa K, Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol. 2009. pp. 56141–56163. [DOI] [PubMed]
- Grabher C, Wittbrodt J. Meganuclease and transposon mediated transgenesis in medaka. Genome Biol. 2007;8(1S10) doi: 10.1186/gb-2007-8-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]


