Abstract
We report for the first time the recombinant expression of fully folded bioactive cyclotides inside live yeast cells by using intracellular protein trans-splicing in combination with a highly efficient split-intein. This approach was successfully used to produce the naturally occurring cyclotide MCoTI-I and the engineered bioactive cyclotide MCoCP4. Cyclotide MCoCP4 was shown reduce the toxicity of human α-synuclein in live yeast cells. Cyclotide MCoCP4 was selected by phenotypic screening from cells transformed with a mixture of plasmids encoding MCoCP4 and inactive cyclotide MCoTI-I in a ratio of 1 to 5×104. This demonstrates the potential for using yeast to perform phenotypic screening of genetically-encoded cyclotide-based libraries in eukaryotic cells.
Cyclotides are fascinating micro-proteins (≈30 residues long) present in plants from the Violaceae, Rubiaceae, Cucurbitaceae and more recently Fabaceae families.[1] They display various biological properties such as protease inhibitory, anti-microbial, insecticidal, cytotoxic, anti-HIV and hormone-like activities.[2] They share a unique head-to-tail circular knotted topology of three disulfide bridges, with one disulfide penetrating through a macrocycle formed by the two other disulfides and inter-connecting peptide backbones, forming what is called a cystine knot topology (Fig. 1A). This cyclic cystine knot (CCK) framework gives the cyclotides exceptional rigidity,[3] resistance to thermal and chemical denaturation, and enzymatic stability against degradation.[2] Interestingly, some cyclotides have been shown to be orally bioavailable,[4] and other cyclotides have been shown to cross the cell membrane through macropinocytosis.[5] Recent reports have also shown that engineered cyclotides can be efficiently used to target extracellular [6] and intracellular[7] protein-protein interactions. All of these features make cyclotides ideal tools for drug development to selectively target protein-protein interactions.[8]
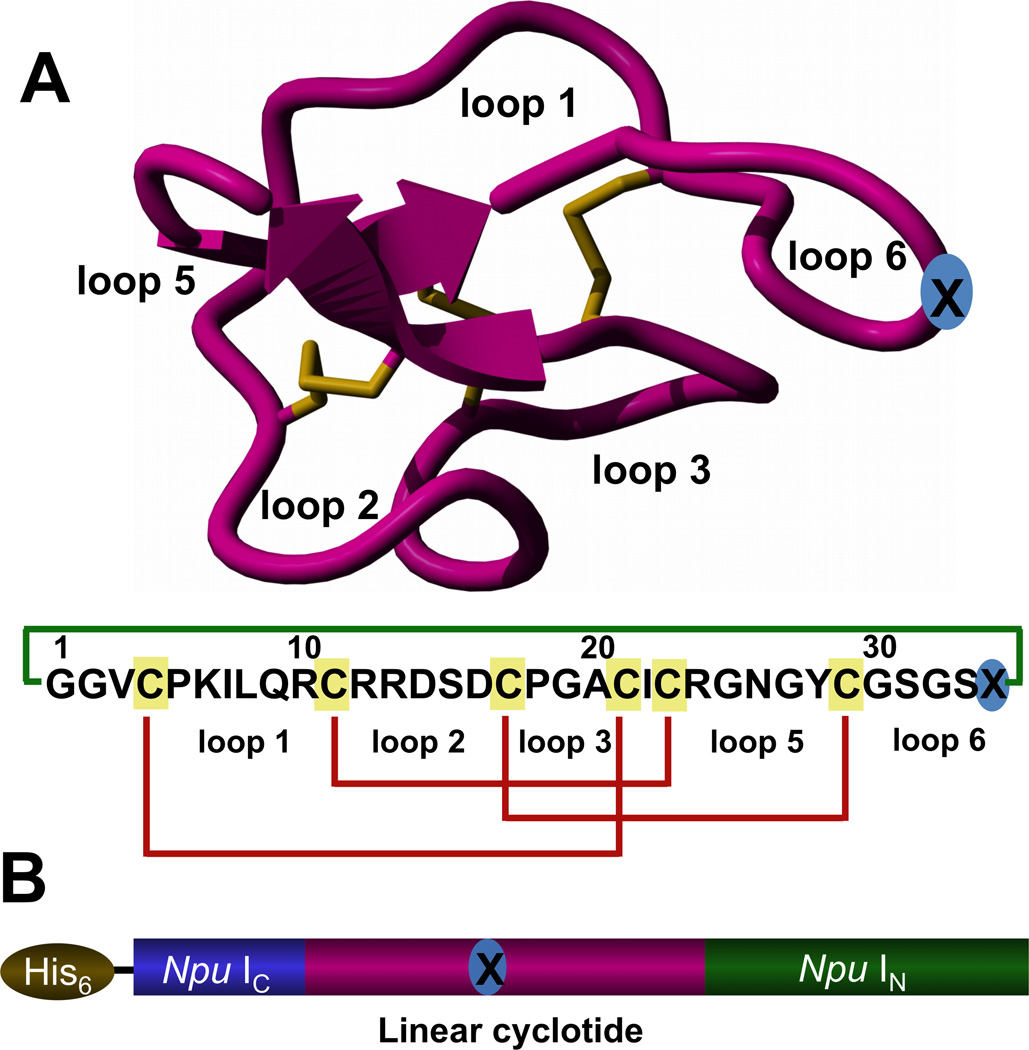
Figure 1.
A. Tertiary structure of the cyclotide MCoTI-II (PDB code: 1IB9)[31] and primary structures of the cyclotides used in this work, MCoTI-I (X=D) and MCoCP4 (X=SLATWAVG). The CP4-derived peptide was grafted onto loop 6, marked with blue circled X. The backbone cyclized peptide (connecting bond shown in green) is stabilized by the three-disulfide bonds (shown in red). B. Intein precursors used for the expression of cyclotides MCoTI-I (1a, X=D) and MCoCP4 (1b, X=SLATWAVG) in yeast S. cerevisiae using PTS.
Naturally occurring cyclotides are ribosomally produced in plants from precursor proteins[1b] and believed to be processed by specific proteases.[9] More than 200 different cyclotide sequences have been reported in the literature to date,[10] and it has been estimated by genomic analysis that ≈ 50,000 cyclotides may exist.[11] All naturally occurring cyclotides share the same CCK motif despite sequence diversity found in the loops decorating the cysteine-knot. Consequently, cyclotides can be considered as natural combinatorial peptide libraries structurally constrained by the cystine-knot scaffold and head-to-tail cyclization but in which hypermutation of essentially all residues is permitted with the exception of the strictly conserved cysteines that comprise the knot.[12] Cyclotides can be chemically synthesized, thereby permitting the introduction of specific chemical modifications or biophysical probes.[13] More recently, cyclotides have also been biosynthesized in plant-derived cell cultures[14] and prokaryotic expression cells by making use of modified protein splicing units.[15] Cyclotides have been also shown to cross cellular membranes to target intracellular protein-protein interactions.[7] Altogether, these characteristics make cyclotides ideal substrates for in-cell molecular evolution strategies to enable generation and selection of compounds with optimal binding and inhibitory characteristics.
In-cell screening and selection methods of genetically-encoded cyclotide libraries provide several advantages over in vitro techniques: it ensures that hits are non-toxic, can bind the target in the appropriate cellular environment, are not rapidly degraded inside the cell, and possess high selectivity to work in living cells. In addition, this method also enables phenotypic screening for the rapid selection of novel bioactive compounds.
The use of an adequate microorganism that allows the production of large genetically-encoded libraries is key for the phenotypic screening of these type of libraries. The baker’s yeast Saccharomyces cerevisiae has been used for decades as a versatile and robust model system for eukaryotic cellular biology.[16] For example, many proteins important in human biology, including cell cycle proteins, signaling proteins, and protein-processing enzymes, were first discovered by studying their homologs in yeast.[17] In addition, several human pathologies derived from protein misfolding have been successfully modeled in simple eukaryotic organisms such as yeast Saccharomyces cerevisiae.[18]
To test the feasibility of expressing folded cyclotides in S. cerevisiae, we used the cyclotide MCoTI-I (Fig. 1A). This cyclotide is a potent trypsin inhibitor (Ki ≈ 20 pM)[19] that is naturally found in the dormant seeds of Momordica cochinchinensis, a plant member of the Cucurbitaceae family.[20] Trypsin inhibitor cyclotides are interesting candidates for drug design because they show very low toxicities to mammalian cells and can be used as natural scaffolds to generate novel biological activities.[6–7, 13b, 21]
To express cyclotide MCoTI-I inside living yeast cells we made use of protein trans-splicing (PTS) to facilitate the intracellular backbone cyclization (Fig. 2). This process has been previously used to express small cyclic peptides[22] and more recently cyclotides[15a] in bacterial expression systems but never used before in a eukaryotic expression system to express large disulfide-containing cyclic proteins such as cyclotides. PTS-mediated backbone cyclization can be accomplished by rearranging the order of the intein fragments, i.e. by fusing the IN and IC fragments to the C- and N-termini of the linear polypeptide precursor to be cyclized (Fig. 1B). To boost the intracellular expression of folded cyclotide MCoTI-I in yeast we used the Nostoc puntiforme PCC73102 (Npu) DnaE split-intein. This DnaE intein has the highest reported rate of protein trans-splicing (τ1/2 ≈ 60 s) and has a high splicing yield.[23]
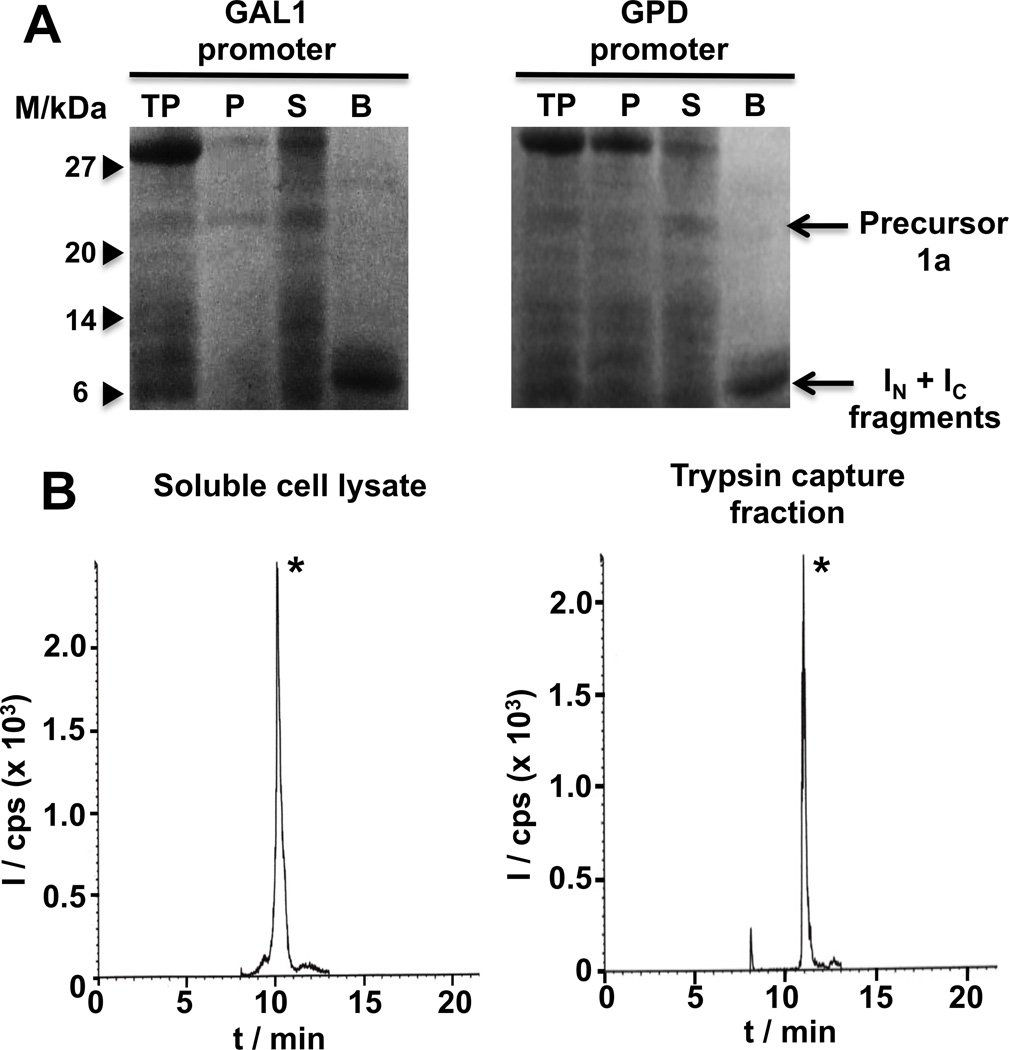
Figure 2.
In-cell expression of MCoTI-I based cyclotides in S. cerevisiae cells using Npu DnaE intein-mediated PTS. A. SDS-PAGE analysis of the recombinant expression of cyclotide precursors 1a using a high-copy µ episomal plasmid under the control of the GAL1 (inducible, left) or GPD (constitutive, right) promoters (M stands for protein markers, TP for total cell lysate, P and S for insoluble and soluble cell lysate fractions, and B for fraction captured by Ni-NTA-agarose beads). B. Analytical HPLC-MS/MS traces of the soluble cell extract (left) or trypsin pull-down (right) fraction from S. cerevisiae cells expressing precursor 1a under the control of a GAL1 inducible promoter. The peak marked with an asterisk corresponds to folded cyclotide MCoTI-I. MS/MS analysis was performed using the ion (m/z = 871.6) for both Q1 and Q3 (I and cps stand for intensity and ion counts per second, respectively).
Accordingly, we designed the split-intein construct 1a (Fig. 1B), where the MCoTI-I linear precursor was fused directly to the N- and C-termini of the Npu DnaE IC and IN polypeptides, respectively. To facilitate backbone cyclization we used the native Cys residue located at the N-terminal position of loop 6 (Fig. 1). A His-tag was also added at the N-terminus of the construct to facilitate identification of the precursor and intein-containing byproducts of the cellular cyclization process.
Expression in yeast S. cerevisiae of cyclotide MCoTI-I using PTS was tested by employing two different high-copy 2µ episomal expression plasmids, pYES2/NT (under the control of a GAL1 inducible promoter) and p426GPD (under the control of a GPD constitutive promoter). Expression plasmids encoding the split-intein precursor 1a derived from plasmids pYC2/NT and p426GPD were transformed into S. cerevisiae strains INVSc1 and W303-1a, respectively, by electroporation. Expression of the MCoTI-precursor split-intein was accomplished for 48 h at 30° C in media containing either 2% galactose (inducible GAL1 promoter) or 2% glucose (constitutive GPD promoter). Under these conditions the precursor was expressed at relatively high levels in both cases, ≈ 10 mg/L (GAL1 promoter) and ≈ 7 mg/L (GPD promoter). In both cases the precursor was completely cleaved (Fig. 2A), indicating the intrinsic high reactivity of the split-intein construct to undergo protein trans-splicing. Next, we quantified the amount of natively folded MCoTI-I generated in-cell by LC-MS analysis using pure MCoTI-I as standard. Correctly folded MCoTI-cyclotides are able to bind trypsin with high affinity (Ki ≈ 20–30 pM). Therefore, this step can be used for affinity purification and to test the biological activity of the recombinant cyclotides. By either using the whole cell lysate or the fraction purified with trypsin-immobilized sepharose beads the LC-MS analysis revealed in both fractions the presence of a major peak that had the expected mass of the natively folded MCoTI-I cyclotide (Figs. 2B and S1). Quantification of the amount of cyclotide gave similar yields in both fractions ≈ 50 µg/L (GAL1 promoter) and ≈ 60 µg/L (GPD promoter), which correspond approximately to an intracellular concentration of approximately 450 nM and 660 nM, respectively. These results indicate that in cell produced cyclotide MCoTI-I is biologically active and therefore adopts a native cyclotide fold.
Encouraged by these results, we decided to express a bioactive cyclotide that could be used to perform phenotypic screening in yeast. S. cerevisiae has been shown to be a good model for many human diseases involving protein misfolding such as Parkinson’s disease.[18, 24] The human protein α-synuclein (α-syn) is a small lipid-binding protein that is prone to misfolding and aggregation that has been liked to Parkinson’s disease by genetic evidence and its abundance in the Parkinson’s disease-associated intracellular aggregates known as Lewy bodies.[25] Overexpression of human α-syn in yeast S. cerevisiae leads to endoplasmatic reticulum stress, disruption of endoplasmic reticulum-Golgi vesicle trafficking, accumulation of lipid droplets, mitochondrial dysfunction, and ultimately cell death.[25–26] This cellular pathology mirrors many aspects of the dysfunction seen in neurons and glia cells of patients with Parkinson’s disease.[27] In addition, genetic screening using a yeast synucleopathy model has produced inhibitors of α-syn cytotoxicy that resulted also effective in neuronal models.[24, 28]
To engineer the cyclotide MCoTI-I to inhibit α-syn-induced cytotoxicity in yeast S. cerevisiae, we used the sequence of cyclic peptide CP4 (cyclo-CLATWAVG), which was recently shown to reduce α-syn-induced cytotoxicity in a yeast synucleopathy model.[24] A CP4-derived linear peptide, in which the Cys residue was replaced by Ser, was grafted onto the cyclotide scaffold using loop 6 to provide cyclotide MCoCP4 (Fig.1). We have shown that loop 6 has the higher backbone mobility of the MCoTI-I cyclotide[3] and accordingly this loop has been shown to tolerate well the grafting of bioactive peptides.[6a, 7] Replacement of the Cys residue was done to facilitate the folding of MCoCP4. Cyclotide MCoCP4 was expressed using a high-copy 2µ plasmid under the control of a GPD constitutive promoter as described above for cyclotide MCoTI-I. Under these conditions, the MCoCP4 split-intein precursor 1b (Fig. 1B) was expressed with a yield similar to that of the MCoTI-I precursor (≈ 9 mg/L). As before, the precursor was completely processed in vivo (Fig. 3A). LC-MS analysis of the soluble cell lysate revealed the presence of a major peak with the expected mass for the natively folded MCoCP4 cyclotide (Figs. 3B and S3). Quantitative LC-MS analysis of the cell lysate provided an expression yield of 45 µg/L, which corresponds to an intracellular concentration of ≈ 390 nM. To confirm the identity of the cyclotide MCoCP4 expressed in yeast, this cyclotide was chemically produced using a GSH-induced cyclization-folding one-pot reaction as previously described.[6a, 7] The cyclization-folding reaction was very efficient and complete in less than 24 h (Fig. S6). Pure synthetic cyclotide MCoCP4 was shown to co-elute by HPLC-MS with yeast-produced MCoCP4 and adopt a native cyclotide fold by NMR (Table S2 and Figs. S7 and S9).
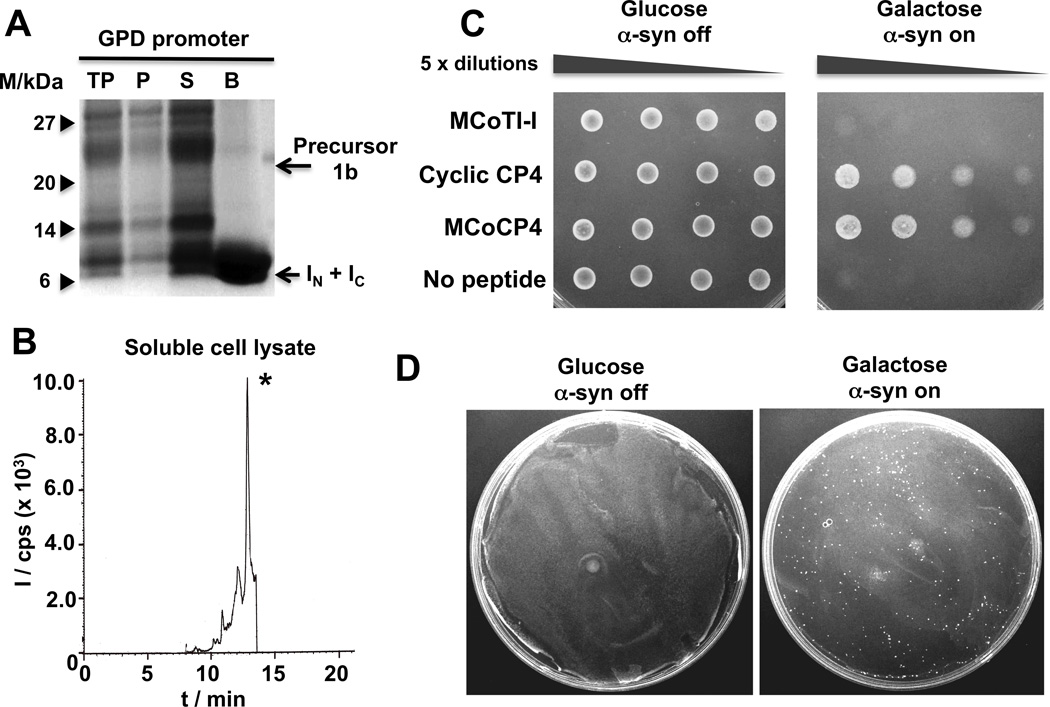
Figure 3.
Expression of cyclotide MCoCP4 in S. cerevisiae cells using Npu DnaE intein-mediated PTS. A. SDS-PAGE analysis of the recombinant expression of cyclotide precursor 1b in S. cerevisiae strain W303-1a cells for in-cell production of cyclotide MCoCP4 (lane description as in Fig. 2). B. Analytical HPLC-MS/MS trace of the soluble cell extract fraction from S. cerevisiae cells expressing precursor 1b under the control of a GPD constitutive promoter. The peak marked with an asterisk corresponds to folded cyclotide MCoCP4. MS/MS analysis was performed using the ion (m/z = 1038.8) for both Q1 and Q3 (units are as described in Fig. 2B). C. Spotting assays of normalized, serially diluted cells of the synucleopathy model expressing cyclotides MCoTI-I (negative control) and MCoCP4, and cyclic peptide CP4 (positive control). D. Phenotypic screening using a yeast synucleopathy model. Screening cells were transformed with a mixture of p426GPD plasmids encoding cyclotides MCoCP4 (cytoprotective) and MCoTI-I (inactive) in a ratio of 1 to 5×104. Glucose and galactose plates are shown to illustrate how a single bioactive cyclotide can be isolated from more than 107 transformants on a single plate.
To test the cytoprotective activity of cyclotide MCoCP4 we used a yeast synucleopathy model previously described in the literature.[26, 29] This cellular model employs a GAL1 promoter to tightly control the expression of human α-syn encoded in the genomic DNA of the microorganism. In this way, α-syn can be expressed or totally repressed only when the yeast are grown on medium containing either galactose or glucose, respectively. Accordingly, we expressed the cyclotide MCoCP4 using a compatible µ episomal expression vector under the control of a GPD constitutive promoter using the yeast synucleopathy model described above. The inactive cyclotide MCoTI-I and cyclic peptide CP4 were also used as negative and positive controls, respectively. After transforming the cells with the plasmids encoding the different cyclic peptides, the yeast cells were grown first in medium containing 2% raffinose as carbon source and then serially spotted in media containing either 2% glucose (α-syn off) or 2% galactose (α-syn on) (Fig. 3C). Expression of the corresponding cyclotides under these conditions did not affect significantly their expression level. As expected, no cytotoxicity was observed when the cells were grown in the presence of 2% glucose (Fig. 3C), thereby indicating the total repression of α-syn expression as well as the lack of cytotoxicity of cyclotides MCoTI-I and MCoCP4. When cells were grown in the presence of galactose, which activates α-syn expression, cells coexpressing inactive cyclotide MCoTI-I were unable to grown at all dilutions tried in the assay (Figs. 3C and S8). As anticipated, coexpression of cyclic peptide CP4 was able to rescue the cytotoxic phenotype induced by α-syn therefore confirming the protective activity of this peptide.[24] Remarkably, cells expressing the engineered cyclotide MCoCP4 were also able to suppress the α-syn-induced cytotoxicity under the conditions used in this assay. The cytoprotective effect of MCoCP4 was similar to that of the peptide CP4. Preliminary results indicated that cyclic peptide CP4 can be cyclized by PTS using the Npu DnaE split-intein providing an intracellular concentration of ≈ 5 µM as quantified by LC-MS/MS (Fig. S2).
Finally, we decided to explore the feasibility of using the yeast synucleopathy model for the phenotypic screening of bioactive cyclotides against α-synuclein-induced cytotoxicity. For this purpose we generated a mixture of plasmids encoding MCoCP4 and MCoTI-I in a ratio of 1 to 5×104, respectively. This DNA mixture was transformed by electroporation to provide ≈ 107 transformants as determined by plating on medium containing 2% glucose. To perform the phenotypic screening the whole cell-based mixture was plated on media containing 2% galactose to activate the expression of α-syn and incubated for 3 days. Under these conditions ≈150 colonies were obtained (Fig. 3D). 30 different colonies were picked, the corresponding plasmids isolated and analyzed by DNA sequencing to confirm the identity of the encoded cyclotide. Almost all the colonies analyzed (27 of 30, 90%) provided DNA sequences encoding cyclotide MCoCP4 therefore demonstrating the feasibility of using yeast as eukaryotic platform to perform phenotypic screening of in-cell generated bioactive cyclotides. The appearance of a small amount of false positives probably stemmed from spontaneous genomic suppressor mutations, which are common in most yeast selection schemes.[24]
In summary we have shown that bioactive folded cyclotides can be produced in eukaryotic microorganisms such as yeast S. cerevisiae by PTS using highly efficient split inteins. This approach was successfully used for the production of a novel cyclotide (MCoCP4) that was able to inhibit α-syn-induced cytotoxicity in live yeast cells, therefore allowing us for the first time to perform a phenotypic screen to select cyclotide sequences with biological activity from inactive cyclotides in a yeast synucleopathy model. These exciting results open the possibility to perform in-cell phenotypic screens against other cellular pathologies using cyclotide-based libraries. Using eukaryotic microorganisms for screening purposes should provide a more biologically relevant cellular background for phenotypic screening but also facilitate the production of cyclotide-based libraries containing post-translational modifications such as phosphorylation and/or glycosylation, which are not available in bacterial expression systems. In addition, it should also allow screening cyclotides against proteins containing post-transtational modifications. Moreover, the use of the cyclotide molecular scaffold, which possesses unique pharmacological features such as extreme stability,[30] oral bioavailability[6b] and ability to cross cellular membranes,[5] should provide an extremely valuable screening platform for the selection of novel therapeutic leads.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Research Grants R01-GM090323 (JAC) and R01-GM085006 (AS). We also like to thank Dr. Susan Lindquist (Whitehead Institute for Biomedical Research, MIT) for kindly providing plasmid pRS306-α syn-GFP.
REFERENCES
- 1.a) Poth AG, Colgrave ML, Philip R, Kerenga B, Daly NL, Anderson MA, Craik DJ. ACS chemical biology. 2010;6:345–355. doi: 10.1021/cb100388j. [DOI] [PubMed] [Google Scholar]; b) Poth AG, Colgrave ML, Lyons RE, Daly NL, Craik DJ. Proc Natl Acad Sci U S A. 2011;108:1027–1032. doi: 10.1073/pnas.1103660108. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nguyen GK, Zhang S, Nguyen NT, Nguyen PQ, Chiu MS, Hardjojo A, Tam JP. J Biol Chem. 2011 doi: 10.1074/jbc.M111.229922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Craik DJ, Simonsen S, Daly NL. Curr. Opin. Drug Discov. Devel. 2002;5:251–260. [PubMed] [Google Scholar]; b) Garcia AE, Camarero JA. Curr Mol Pharmacol. 2010;3:153–163. doi: 10.2174/1874467211003030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Angew Chem Int Ed Engl. 2010;49:7030–7034. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Angew Chem Int Ed Engl. 2011;50:6948–6949. [Google Scholar]
- 4.Saether O, Craik DJ, Campbell ID, Sletten K, Juul J, Norman DG. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 5.Contreras J, Elnagar AY, Hamm-Alvarez SF, Camarero JA. J Control Release. 2011;155:134–143. doi: 10.1016/j.jconrel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Aboye TL, Ha H, Majumder S, Christ F, Debyser Z, Shekhtman A, Neamati N, Camarero JA. J Med Chem. 2012;55:10729–10734. doi: 10.1021/jm301468k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wong CT, Rowlands DK, Wong CH, Lo TW, Nguyen GK, Li HY, Tam JP. Angew Chem Int Ed Engl. 2012;51:5620–5624. doi: 10.1002/anie.201200984. [DOI] [PubMed] [Google Scholar]
- 7.Ji Y, Majumder S, Millard M, Borra R, Bi T, Elnagar AY, Neamati N, Shekhtman A, Camarero JA. J Am Chem Soc. 2013;135:11623–11633. doi: 10.1021/ja405108p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark RJ, Daly NL, Craik DJ. The Biochemical journal. 2006;394:85–93. doi: 10.1042/BJ20051691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen GK, Wang S, Qiu Y, Hemu X, Lian Y, Tam JP. Nat Chem Biol. 2014;10:732–738. doi: 10.1038/nchembio.1586. [DOI] [PubMed] [Google Scholar]
- 10.Wang CK, Kaas Q, Chiche L, Craik DJ. Nucleic Acids Res. 2008;36:D206–D210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruber CW, Elliott AG, Ireland DC, Delprete PG, Dessein S, Goransson U, Trabi M, Wang CK, Kinghorn AB, Robbrecht E, Craik DJ. Plant Cell. 2008;20:2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Austin J, Kimura RH, Woo YH, Camarero JA. Amino Acids. 2010;38:1313–1322. doi: 10.1007/s00726-009-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Huang YH, Colgrave ML, Clark RJ, Kotze AC, Craik DJ. J Biol Chem. 2010;285:10797–10805. doi: 10.1074/jbc.M109.089854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Daly NL, Love S, Alewood PF, Craik DJ. Biochemistry. 1999;38:10606–10614. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]; b) Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, Tate EW. Org Biomol Chem. 2008;6:1462–1470. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- 14.Dornenburg H. Biopolymers. 2010;94:602–610. doi: 10.1002/bip.21466. [DOI] [PubMed] [Google Scholar]
- 15.a) Jagadish K, Borra R, Lacey V, Majumder S, Shekhtman A, Wang L, Camarero JA. Angew Chem Int Ed Engl. 2013;52:3126–3131. doi: 10.1002/anie.201209219. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kimura RH, Tran AT, Camarero JA. Angew. Chem. Int. Ed. 2006;45:973–976. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- 16.Botstein D, Fink GR. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foury F. Gene. 1997;195:1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 18.Winderickx J, Delay C, De Vos A, Klinger H, Pellens K, Vanhelmont T, Van Leuven F, Zabrocki P. Biochim Biophys Acta. 2008;1783:1381–1395. doi: 10.1016/j.bbamcr.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Chembiochem. 2007;8:1363–1366. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Trinh Hong T, Pham TT, Le Nguyen D. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 21.Chan LY, Gunasekera S, Henriques ST, Worth NF, Le SJ, Clark RJ, Campbell JH, Craik DJ, Daly NL. Blood. 2011;118:6709–6717. doi: 10.1182/blood-2011-06-359141. [DOI] [PubMed] [Google Scholar]
- 22.Young TS, Young DD, Ahmad I, Louis JM, Benkovic SJ, Schultz PG. Proc Natl Acad Sci USA. 2011;108:11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zettler J, Schutz V, Mootz HD. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Kritzer JA, Hamamichi S, McCaffery JM, Santagata S, Naumann TA, Caldwell KA, Caldwell GA, Lindquist S. Nat Chem Biol. 2009;5:655–663. doi: 10.1038/nchembio.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, Barlowe C, Lindquist S. Proc Natl Acad Sci U S A. 2008;105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee VM, Trojanowski JQ. Neuron. 2006;52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Gitler AD, Chesi A, Geddie ML, Strathearn KE, Hamamichi S, Hill KJ, Caldwell KA, Caldwell GA, Cooper AA, Rochet JC, Lindquist S. Nat Genet. 2009;41:308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Outeiro TF, Lindquist S. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colgrave ML, Craik DJ. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 31.Felizmenio-Quimio ME, Daly NL, Craik DJ. J Biol Chem. 2001;276:22875–22882. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.