Abstract
2,6-Diamino-4-hydroxy-5N-methylformamidopyrimidine (MeFapy-dG) arises from N7-methylation of guanine followed by imidazole ring opening. The lesion has been reported to persist in animal tissues. Previous in vitro replication bypass investigations of the MeFapy-dG adduct revealed predominant insertion of C opposite the lesion, dependent on the identity of the DNA polymerase (Pol) and the local sequence context. Here we report crystal structures of ternary Pol•DNA•dNTP complexes between MeFapy-dG-adducted DNA template:primer duplexes and the Y-family polymerases human Pol η and P2 Pol IV (Dpo4) from Sulfolobus solfataricus. The structures of the hPol η and Dpo4 complexes at the insertion and extension stages, respectively, are representative of error-free replication, with MeFapydG in the anti conformation and forming Watson-Crick pairs with dCTP or dC.
Alkylating agents are the earliest class of chemotherapy drugs and are still commonly used to treat different types of cancers. These include monofunctional methylating agents such as Temozolomide, and bifunctional alkylating agents such as nitrogen mustards or chloroethylating agents.1
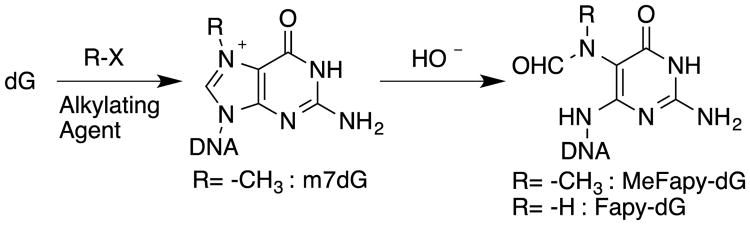
The guanine N-7 position constitutes the most nucleophilic site in DNA.2 Thus, DNA methylation occurs predominantly at that site, resulting in a cationic N7-methyl-deoxyguanosine adduct.3 This product can undergo further hydrolysis, yielding an abasic (AP) site4,5 or the imidazole ring fragmented lesion, N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine (MeFapy-dG) (Figure 1).3,6 Opening of the purine imidazole ring depends on the N-7 substituent and the pH of the medium. Whereas the opening rate under physiological conditions is slow, it becomes accelerated at higher temperature and in alkaline solution.
Figure 1.
Formation of formamidopyrimidine lesions.
MeFapy-dG has been characterized in vivo, in the liver of rats,7, 8 and has also been observed in the urine of healthy humans.9 In general, the MeFapy-dG adduct is considered non-miscoding, owing to the fact that the Watson-Crick face remains unaltered. However, various in vitro experiments using E. coli DNA polymerase I Klenow fragment and T4 polymerase show that MeFapy-dG blocks DNA chain elongation.10,11 Replication of a site-specific MeFapydG lesion in primate cells gave complex mutational spectra with frequencies between 7 – 21 % depending on local the sequence. Common mutations were G→T transversions and deletions.12 An in vitro replication study using the MeFapy-dG lesion with S. solfataricus Dpo4 found miscoding with the incorporation of all four nucleotides with various efficiencies depending on the DNA template sequence around the lesion.13 Oligonucleotides with 5′-T-(MeFapy-dG)-G-3′ resulted in error-free bypass, with insertion of dC opposite the adduct and full-length extension of the primer strand. By comparison, a 5′-T-(MeFapy-dG)-T-3′ template triggered a one base deletion, or misincorporation of dA opposite the MeFapy-dG lesion. The error-free bypass and extension efficiency by Dpo4 was estimated to be 74% for 5′-T-(MeFapy-dG)-G-3′ and 51% for 5′-T-(MeFapy-dG)-T-3′, along with 11% one-base deletion product for the latter template. Recent in vitro replication bypass experiments using human Y-family Pols (hPols) η, κ, ι, and Rev1 showed efficient translesion synthesis (TLS) by hPols η and κ, with error-free insertion of dCTP opposite MeFapy-dG and extension in the above sequence contexts.14 Among these TLS pols, hPol η is the most efficient in the error-free bypass of MeFapy-dG (>70%).
To analyze the structural basis for the mostly error-free bypass of the MeFapy-dG adduct by hPol η we determined the crystal structure of a hPol η complex trapped at the insertion stage, with MeFapy-dG opposite the non-hydrolyzable dCMPNPP analog, in which an N-atom bridges the α and β P-atoms, and in the presence of Mg2+ (Table 1). Further crystal structures concern the hPol κ homolog Dpo4 in complex with a MeFapy-dG containing template-primer DNA duplex in the presence of Ca2+ and trapped in two different phases of bypass. In the first complex, representative of the insertion stage and a -1 frameshift, MeFapy-dG is unopposed by a residue from the primer and, instead, the incoming dATP pairs with the T that is 5′-adjacent to the adduct on the template. In the second complex, representative of the extension stage and error-free bypass, MeFapy-dG pairs with dC at the -1 position, followed by the incoming dATP pairing with the downstream T of the template.
Table 1. DNA sequences used in the crystallizations.
| Complex | DNA sequence (X = MeFapy-dG) | Incoming nucleotide |
|---|---|---|
| hPol η (Insertion) | 3′- TCG CAG TAX TAC -5′ | |
| 5′- AGC GTC AT -3′ | dCMPNPP | |
| SsDpo4 (Insertion) | 3′- CCC CCT TCC TAA GXT ACT -5′ | |
| 5′- GGG GGA AGG ATT C -3′ | dATP | |
| SsDpo4(Extension) | 3′- CCC CCT TCC TAA TXT ACT -5′ | |
| 5′- GGG GGA AGG ATT AC -3′ | dATP |
The structure of the hPol η ternary complex with MeFapy-dG paired opposite incoming dCMPNPP was determined at a resolution of 2.48 Å (Table 2, Figure 2). The DNA duplex consists of a 12mer template containing MeFapy-dG and an 8mer primer (Table 1). In the structure, all primer nucleotides were visible in the electron density maps along with 11 of 12 template nucleotides (Table 2). An example of the quality of the final electron density is shown in Figure S1. At the active site, the MeFapy-dG:dCMPNPP pair displays the expected Watson-Crick geometry, with the formamide moiety adopting an orientation that is more or less perpendicular to the plane of the six-membered ring (Figure 2A). Thus, the active site configuration in the structure of the hPol η MeFapy-dG complex is similar to that in the crystal structure of the complex between hPol η and native DNA with a G:dCMPNPP pair lodged at the active site.15 Superimposition of the active sites of these two structures indicates that amino acids from the hPol η finger domain (i.e. Gln-38 and Arg-61) adopt similar orientations relative to the nascent base pair (Figure S2).
Table 2. Selected crystal data, diffraction data collection, and refinement parameter statistics.
| Complex | hPol η (Insertion) | SsDpo4 (Insertion) | SsDpo4 (Extension) |
|---|---|---|---|
| Data Collection | |||
|
| |||
| Space group | P61 | P21212 | P21212 |
| Resolution [Å]a | 50.0-2.65 (2.70-2.65) | 50.0-3.10 (3.15-3.10) | 30.0-2.90 (2.95-2.90) |
| Unit cell a,b,c [Å] | 98.97, 98.97, 81.62 | 95.45, 102.72, 53.43 | 94.38, 103.97, 52.56 |
| Completeness [%] | 98.9 (100) | 98.4 (84.7) | 99.8 (100) |
| I/σ(I) | 14.5 (1.9) | 21.2 (2.9) | 18.7 (1.8) |
| R-merge [%] | 14.7 (98.5) | 11.2 (68.0) | 14.3 (61.7) |
| Redundancy | 5.7 (5.6) | 6.2 (3.1) | 7.1 (7.1) |
|
| |||
| Refinement | |||
|
| |||
| R-work [%] | 15.9 (22.1) | 16.4 (25.9) | 17.1 (26.9) |
| R-free [%]b | 22.9 (34.1) | 25.6 (38.6) | 23.5 (35.3) |
| Avg. B [Å2] | 47.7 | 88.2 | 69.8 |
| R.m.s.d. bonds [Å] | 0.011 | 0.010 | 0.010 |
| R.m.s.d. angles [deg.] | 1.5 | 1.5 | 1.4 |
| PDB IDc | 4RU9 | 4RUA | 4RUC |
Statistics for the highest-resolution shell are shown in parentheses.
Based on 5% of the reflections.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://wwpdb.org/).
Figure 2.
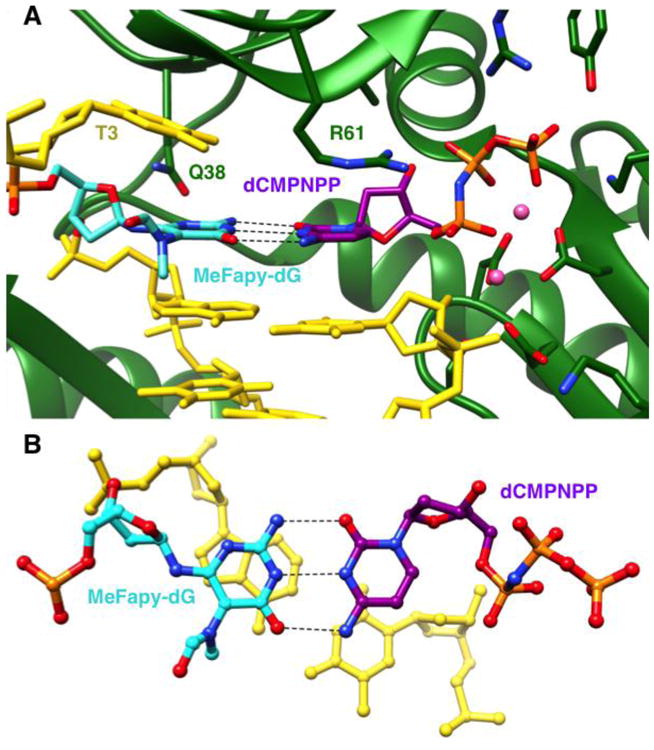
Active site configuration in the ternary hPol η insertion-step complex with dCMPNPP opposite MeFapy-dG. (A) View into the DNA major groove, and (B) rotated by ∼90° around the horizontal axis and looking perpendicularly onto the nucleobase plane of the incoming dCMPNPP. Carbon atoms of MeFapy-dG and incoming dCMPNPP are colored in cyan and purple, respectively, and Mg2+ ions are pink spheres. Gln-38 forms a H-bond to O4′ of MeFapy-dG, but N3 in the minor groove is too far removed (3.9 Å) from the Gln-38 amide oxygen for H-bond formation.
Two structures were determined for Dpo4 in complex with DNA duplexes containing MeFapy-dG– modified template strands (Table 1). Both crystals diffracted to ca. 3 Å and belong to space group P21212 with a single complex per asymmetric unit (Table 2). The first structure features template sequence 5′-T[MeFapy-dG]G-3′ and represents a so-called type II complex, with the adduct unopposed by a primer base and resulting in a-1 frameshift (Figures 3, S3). As is characteristic for a type II complex,16 Dpo4 simultaneously accommodates two template nucleotides inside the catalytic pocket, namely MeFapy-dG and the 5′-adjacent T. Pairing between this T and the incoming dATP leaves a 6 Å gap between the α-phosphate of the nucleotide triphosphate and the 3′-hydroxyl group at the primer terminus (Figure 3). The ability of Dpo4 to accommodate two template bases in its active site forms the basis for correct bypass of cyclic pyrimidine dimers (CPDs) by this Pol, with the type II configuration and a misaligned template strand being consequences of the spacious active site.16
Figure 3.
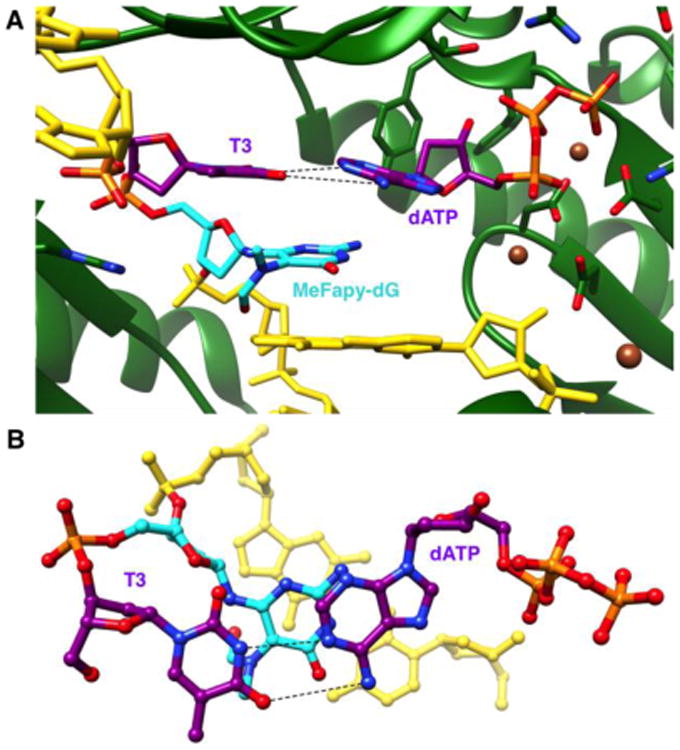
Active site configuration in the ternary Dpo4 insertion-step complex with unpaired MeFapy-dG and incoming dATP opposite template T. (A) View into the DNA major groove, and (B) rotated by ∼90° around the horizontal axis and looking perpendicularly onto the nucleobase plane of the incoming dATP. Color codes match those in Fig. 2 except that carbon atoms of template T5′-adjacent to the adduct are purple. Ca2+ ions are brown spheres.
The second structure is of a post-insertion ternary complex, involving the template sequence 5′-T[MeFapy-dG]T-3′ and a 14mer primer of which the 3′ terminal cytosine is intended to pair with template MeFapy-dG (Figures 4, S4). The incoming dATP pairs with the thymidine base to the 5′-side of MeFapydG. Both base pairs, namely MeFapy-G:dC and dT:dATP are found in the standard Watson-Crick H-bonding configuration. As in the case of the hPol η complex (Figure 2), formamide is rotated out of the plane of the six-membered ring in the two Dpo4 complexes (Figures 3, 4). However, unlike in the hPol η complex, the formamide C=O is directed toward the residue 3′-adjacent to the adduct.
Figure 4.
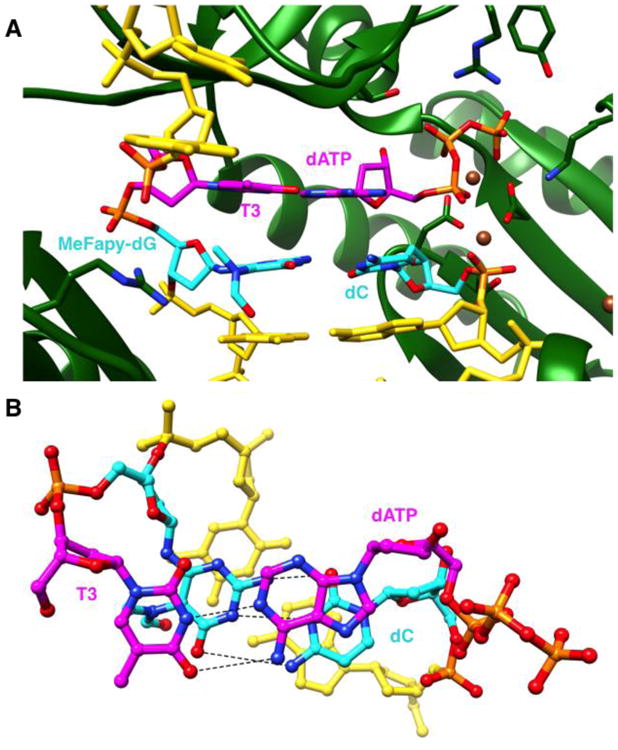
Active site configuration in the ternary Dpo4 extension-step complex with primer dC opposite the MeFapy-dG, followed by dATP opposite template dT. (A) View into the DNA major groove, and (B) rotated by ∼90° around the horizontal axis and looking perpendicularly onto the nucleobase plane of the incoming dATP. Carbon atoms of the MeFapy-G:dC and dT:dATP pairs are colored in cyan and magenta, respectively
In vitro or in vivo formations of methylated or nonmethylated formamidopyrimidines (MeFapy-dG or Fapy-dG, respectively) in DNA have been studied extensively.17 Although the origins of these lesions are different (Fapy-dG is the result of oxidative damage), they are structurally very similar and only weakly mutagenic. Earlier studies established that Pols exhibit a strong preference for dCTP incorporation opposite the Fapy-dG lesion.18-21 Recent work by Gehrke et al. showed that the replication of the carbocyclic sugar analog Fapy lesion (cFaPydG) by high-fidelity polymerase I from Geobacillus stearothermophilus (Bst Pol I) results in error-free bypass alongside a smaller amount of error-prone bypass.22 A previous NMR structure of a cFaPydG:dC containing DNA duplex was also consistent with cFaPydG pairing with dC in a standard Watson-Crick fashion.23
N7-Methyl-2′-deoxyguanosine (m7dG), the initial lesion formed by methylating agents (Figure 1), has also been the subject of several studies and classified as non-promutagenic.24,25 Previous crystallographic studies using the chemically stable 2′-fluoro-m7dG analog in a DNA sequence, either in complex with E. coli DNA glycosylase AlkA26 or human DNA polymerase β27 revealed that this polymerase bypasses m7dG accurately and that the lesion forms a canonical Watson-Crick base pair with incoming dCTP.
In summary, the three X-ray crystal structures of TLS Pols trapped either at the insertion or extension stages of MeFapy-dG bypass synthesis provide detailed insight into the basis of the mostly error-free replication of the adduct by hPol η and Dpo4, a hPol κ homolog. The structure of the hPol η•MeFapy-dG•dCMPNPP ternary complex is consistent with kinetic data of error free bypass and extension by hPol η. The major groove is wide open at the active site of hPol η, allowing this Pol to potentially also bypass more bulky lesions, such as BenzylFapy-dG (Figure S5).
In the hPol η complex the formamide C=O points toward the 5′-adjacent T, whereas it is directed toward the 3′-adjacent G in the Dpo4 structure(s), perhaps as a result of the proximity between formamide and the 5′-template T in the latter case (Figures 2, 4, S6). Thus, partial unstacking of the template T 5′-adjacent to the lesion at the active site of hPol η creates somewhat more room, allowing a virtually upright orientation of formamide. By comparison, the equivalent T and the six-membered ring of MeFapy-dG are tightly stacked at the Dpo4 active site and an orientation of formamide similar to that in the hPol η complex would create a tight spacing between C=O and the T C5-methyl substituent (Figure S6).
Supplementary Material
Acknowledgments
Supported by NIH grants P01 CA160032, P30 ES00267, and P30 CA068485.
Footnotes
Associated Content Supporting Information. Crystallographic experimental procedures, crystal data, electron density maps, and structural superimpositions. “This material is available free of charge via the Internet at http://pubs.acs.org.”
References
- 1.Beranek DT. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 2.Reiner B, Zamenhof S. J Biol Chem. 1957;228:475–486. [PubMed] [Google Scholar]
- 3.Gates KS, Nooner T, Dutta S. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 4.Boiteux S, Guillet M. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Loeb LA, Preston DB. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 6.Tudek B. J Biochem Mol Biol. 2003;36:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- 7.Beranek DT, Weis CC, Evans FE, Chetsanga CJ, Kadlubar FF. Biochem Biophys Res Commun. 1983;110:625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- 8.Kadlubar FF, Beranek DT, Weis CC, Evans FE, Cox R, Irving CC. Carcinogenesis. 1984;5:587–592. doi: 10.1093/carcin/5.5.587. [DOI] [PubMed] [Google Scholar]
- 9.Barak R, Vincze A, Bel P, Dutta SP, Chedda GB. Chem-Biol Interact. 1993;86:29–40. doi: 10.1016/0009-2797(93)90109-c. [DOI] [PubMed] [Google Scholar]
- 10.Boiteux S, Laval J. Biochem Biophys Res Commun. 1983;110:552–558. doi: 10.1016/0006-291x(83)91185-3. [DOI] [PubMed] [Google Scholar]
- 11.O'Connor TR, Boiteux S, Laval J. Nucleic Acids Res. 1988;16:5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earley LF, Minko IG, Christov PP, Rizzo CJ, Lloyd RS. Chem Res Toxicol. 2013;26:1108–1114. doi: 10.1021/tx4001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christov PP, Angel KC, Guengerich FP, Rizzo CJ. Chem Res Toxicol. 2009;22:1086–1095. doi: 10.1021/tx900047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christov PP, Yamanaka K, Choi JY, Takata K, Wood RD, Guengerich FP, Lloyd RS, Rizzo CJ. Chem Res Toxicol. 2012;25:1652–1661. doi: 10.1021/tx300113e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patra A, Nagy LD, Zhang Q, Su Y, Muller L, Guengerich FP, Egli M. J Biol Chem. 2014;289:16867–16882. doi: 10.1074/jbc.M114.551820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling H, Boudsocq F, Woodgate R, Yang W. Mol Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 17.Dizdaroglu M, Kirkal G, Jaruga P. Free Radical Biol Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Wiederholt CJ, Greenberg MM. J Am Chem Soc. 2002;124:7278–7279. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 19.Patro JN, Wiederholt CJ, Jiang YL, Delaney JC, Essigmann JM, Greenberg MM. Biochemistry. 2007;46:10202–10212. doi: 10.1021/bi700628c. [DOI] [PubMed] [Google Scholar]
- 20.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ober M, Müller H, Pieck C, Gierlich J, Carell T. J Am Chem Soc. 2005;127:18143–18149. doi: 10.1021/ja0549188. [DOI] [PubMed] [Google Scholar]
- 22.Gehrke TH, Lischke U, Gasteiger KL, Schneider S, Arnold S, Muller HC, Stephenson DS, Zipse H, Carell T. Nat Chem Biol. 2013;9:455–461. doi: 10.1038/nchembio.1254. [DOI] [PubMed] [Google Scholar]
- 23.Lukin M, Zaliznyak T, Attaluri S, Johnson F, de los Santos C. Chem Res Toxicol. 2012;25:2423–2431. doi: 10.1021/tx300290b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gates KS, Nooner T, Dutta S. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 25.Boysen G, Pachkowski BF, Nakamura J, Swenberg JA. Mutat Res. 2009;678:76–94. doi: 10.1016/j.mrgentox.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. J Am Chem Soc. 2008;130:11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koag MC, Kou Y, Ouzon-Shubeita H, Lee S. Nucleic Acids Res. 2014;42:8755–8766. doi: 10.1093/nar/gku554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.