Abstract
In the eastern United States, the buckeye butterfly, Junonia coenia, shows seasonal wing colour plasticity where adults emerging in the spring are tan, while those emerging in the autumn are dark red. This variation can be artificially induced in laboratory colonies, thus making J. coenia a useful model system to examine the mechanistic basis of plasticity. To better understand the developmental basis of seasonal plasticity, we used RNA-seq to quantify transcription profiles associated with development of alternative seasonal wing morphs. Depending on the developmental stage, between 547 and 1420 transfrags were significantly differentially expressed between morphs. These extensive differences in gene expression stand in contrast to the much smaller numbers of differentially expressed transcripts identified in previous studies of genetic wing pattern variation in other species and suggest that environmentally induced phenotypic shifts arise from very broad systemic processes. Analyses of candidate endocrine and pigmentation transcripts revealed notable genes upregulated in the red morph, including several ecdysone-associated genes, and cinnabar, an ommochrome pigmentation gene implicated in colour pattern variation in other butterflies. We also found multiple melanin-related transcripts strongly upregulated in the red morph, including tan and yellow-family genes, leading us to speculate that dark red pigmentation in autumn J. coenia may involve nonommochrome pigments. While we identified several endocrine and pigmentation genes as obvious candidates for seasonal colour morph differentiation, we speculate that the majority of observed expression differences were due to thermal stress response. The buckeye transcriptome provides a basis for further developmental studies of phenotypic plasticity.
Keywords: ecdysone, Junonia coenia, ommochrome, phenotypic plasticity, polyphenism, transcriptome, wing patterns
Introduction
Understanding the role of environmental conditions in developmental decision-making processes represents an ongoing challenge in biology. In many species, individuals that are genetically similar, or even identical, can develop into one of several phenotypically distinct morphs in response to specific environmental cues. This phylogenetically widespread mechanism, known as phenotypic plasticity, can allow rapid adaptation to the environment without relying on genetic changes between generations (Nijhout 2003; Moczek 2010; Snell-Rood et al. 2010; Moczek et al. 2011). Plasticity is particularly useful to organisms with short generation times that live in continually changing environments because it allows them to quickly react to changes by adaptively altering gene regulation during development. While much work has been carried out on the biological mechanisms and fitness advantages of plasticity in various study systems, less work has been carried out addressing what manner or magnitude of gene regulatory changes facilitate response to environmental conditions via phenotypic plasticity (Beldade et al. 2011).
The common buckeye butterfly, Junonia coenia, has been extensively studied in terms of how environmental conditions trigger morph-switching across different climatic regions (Smith 1991; Daniels et al. 2012). Buckeye populations in the eastern United States display two primary seasonal wing colour morphs that are determined by the photoperiod and temperature they experience during late larval and early pupal development (Smith 1991). At cool temperatures and short day lengths, such as those experienced in autumn and winter, the majority of individuals will develop a dark red wing colour, whereas at warm temperatures and long day lengths, such as those experienced in spring and summer, the majority of individuals will develop wings that are primarily of a tan colour (Fig. 1a). This seasonal plasticity probably facilitates crypsis and/or thermoregulation (e.g. Brakefield & Larsen 1984), where the red autumn form may more effectively blend in with the senescing foliage of deciduous hardwood forests of the eastern United States (Daniels et al. 2012) or more efficiently absorb heat. Colour morph variation in buckeyes is physiologically controlled by differences in ecdysone levels during a critical period in early pupal development (Rountree & Nijhout 1995) when high levels of ecdysone induce the tan colour morph, while low levels of ecdysone result in the red morph. Differential deposition of ommochrome pigments in wings scales during late pupal development is proposed to be the ultimate cause of colour differences, although the actual identities of all of the key pigments are still not completely certain (Nijhout & Koch 1991; Koch 1993; Nijhout 1997; Daniels & Reed 2012).
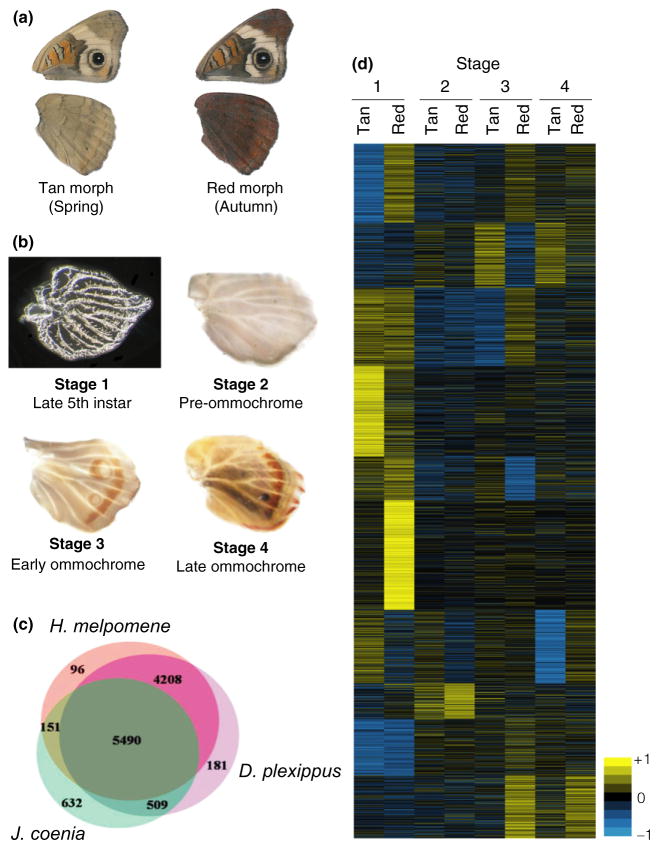
Fig. 1.
Seasonal variation and wing development in Junonia coenia. (a) Different light and temperature conditions can induce North Carolina J. coenia to display extremely different seasonal morphs. (b) The four developmental stages sampled in this study: (1) late 5th instar hindwing discs, (2) preommochrome pupal hind-wings, (3) early ommochrome pupal hindwings, and (4) late ommochrome pupal hindwings. (c) Comparisons of transfrag similarity between Heliconius melpomene, Danaus plexippus and J. coenia using OrthoMCL from amino acid sequences. Note that the complete genomes have been sequenced for H. melpomene and D. plexippus, while J. coenia is represented only by wing tissue-specific transcripts. (d) A unified heat map illustrating the differential expression of 3145 loci between four developmental stages and two colour morphs of J. coenia. The diagram shows the relative expression levels from all samples compared against background levels. The relative levels of upregulation or downregulation are represented by a coloured field that ranges from yellow for highly upregulated genes to blue for highly downregulated genes.
Nothing is currently known about the developmental genetic mechanisms that underlie developmental switching between the seasonal buckeye morphs. To better understand the gene regulatory basis of this phenomenon, we undertook a comparative transcriptomic approach, using RNA-seq to allow characterization of transcripts while simultaneously determining their expression levels. Because of the known involvement of ecdysone and ommochromes in wing pattern plasticity, the immediate aim of this study was to assess differential expression of genes related to endocrine and pigment pathways. We specifically sought to assess three hypotheses: (i) morph-specific gene expression differences should be highest later in pupal development, after batteries of downstream response genes (e.g. pigment genes) are activated, (ii) genes implicated in ecdysone response should show differential expression between morphs, and (iii) genes associated with ommochrome pigmentation should be upregulated in the red morph. In addition to testing these predictions, we also sought to more generally assess the number and type of transcripts differentially regulated during seasonal polyphenism to develop an initial working profile of J. coenia’s gene regulatory response to environmental variation.
Materials and methods
Animal rearing
Populations of Junonia coenia from Durham, North Carolina (35°59′19″N, 78°54′26″W), were reared in incubators and fed a standard artificial diet (Yamamoto 1969; Nijhout 1980). Two sets of conditions known to induce alternative colour morphs in North Carolina populations (Smith 1991) were applied: 21 °C with 8 h light per day for short/cool day conditions (red morph) and 27 °C with 16 h light for long/warm day conditions (tan morph).
Tissue sampling
Hindwings from four developmental stages were sampled for both seasonal colour morphs. Hindwings were used for this work because they show much more extensive colour plasticity compared to forewings. The four stages sampled include imaginal discs from late stage fifth instar larvae, prepigment pupae, early ommochrome development and late ommochrome development (Fig. 1b). Two biological replicates were prepared for each developmental stage and colour morph combination, for a total of 16 samples. Each sample represented a pool of hindwings taken from multiple male and female individuals. To preserve mRNA quality, wings were rapidly dissected into RNA-later (Life Technologies, Grand Island, NY, USA), incubated for c. 24 h at 4 °C to allow the solution to permeate tissues and then stored at −80 °C until mRNA extraction.
RNA-seq
Total RNA was extracted from wing tissues using the RNeasy Mini Kit spin column protocol (QIAGEN, Valencia, CA, USA). TURBOTM DNase treatment (Life Technologies) was then used to degrade contaminating DNA, and mRNA was purified from the total RNA using the Oligotex mRNA Mini Kit (QIAGEN). The Ovation® RNA-seq system (NuGEN, San Carlos, CA, USA) was used to prepare double-stranded cDNA from mRNA, and this cDNA product was then purified with QIAquick PCR purification columns (QIAGEN). The purified double-stranded cDNA was sheared by a Covaris Ultrasonicator to 300 bp. The EncoreTM NGS Multiplex System I (NuGEN) was used for final library preparation. Multiplexed libraries were sequenced as 75-bp paired-end Illumina reads or 100-bp paired-end Illumina reads. Sequencing lanes were randomized between samples.
Data analysis
Each RNA-seq data set was processed with Khmer (Brown et al. 2012) to even out the coverage, to remove redundant reads, and to remove errors in reads, thus reducing the data sets to 30–54% of their read counts. Reads from replicates of each developmental stage in each colour morph were pooled into eight individual sets (Table S1, Supporting information). The filtered reads were assembled into developmental stage transcriptomes with OASES v.1.2.03 (Schulz et al. 2012) using a range of hash lengths from 29 to 61 bp. For transcripts with more than 50 transcription fragments (transfrags) per assembly, the longest 50 per transcript were kept. The eight assemblies were consolidated with CD-HIT-EST of CD-HIT package v.4.5.7 (Li & Godzik 2006) into a reference assembly. Only transfrags longer than 1 kb were used in subsequent analyses. This semi-arbitrary length cut-off was selected to improve annotation efficiency while minimizing potential problems associated with isoform variation.
Reads were mapped to the reference transcriptome using BOWTIE v.0.12.7 (Langmead et al. 2009), and transcript expression levels were obtained using EXPRESS v.1.0.0 (Roberts & Pachter 2013). Using Bioconductor package EDGER v.2.4.6 (Robinson et al. 2010) and biological replicates for each developmental stage, differentially expressed transcripts were identified. Functional annotation and gene ontology assignment of transfrags were carried out using Blast2GO (Conesa et al. 2005). The transcriptome was translated to find the longest open reading frame for each transcript. ORTHOMCL 1.4 (Li et al. 2003) was used to identify orthologous protein clusters with Heliconius melpomene (The Heliconius Genome Consortium 2012) and Danaus plexippus (Zhan et al. 2011) transcriptomes.
We tested the sets of upregulated and downregulated genes of each stage-specific comparison among the morphs for gene ontology enrichment using the Fisher’s Exact Test function of Blast2GO. Each of the eight sets of upregulated or downregulated genes was tested for GO enrichment with the annotations of all 16 251 transfrags in the transcriptome as the reference. A P-value of 0.00625 (i.e. multiple testing corrected value for P = 0.05) was used as the threshold to call significant expression differences between morphs. The same analysis was repeated for the upregulated and downregulated gene sets of comparisons between the developmental time points of each the two morphs. A P-value of 0.00417 (i.e. multiple testing corrected value for P = 0.05) was used as the threshold for these GO enrichment analyses.
Results
Transcriptome assembly of the wing development time-course
Because the genome of Junonia coenia is not currently sequenced, we performed a de novo transcriptome assembly using our Illumina sequence data. Multiplexed libraries yielded 41.5 million 75-bp paired-end Illumina reads and 246 million 100-bp paired-end Illumina reads. We assembled a reference transcriptome of 77.55 Mb with 121 076 transfrags and an N50 of 687 bp. Remaining analyses were focused on the 16 251 assembled transcripts >1 kb (26.65 Mb of the transcriptome), as the longer sequences were more likely to be annotatable. The longest transfrag was 22 289 bp (Table 1). DNA sequences and expression metadata are available at NCBI GEO with Accession no. GSE54819. Assembled transcript sequences are available for download at Dryad (doi:10.5061/dryad.5n5h6), and download and BLAST searches at butterflygenome.org.
Table 1.
Statistics for de novo assembly of Junonia coenia wing transcriptome
| Assembly size (Mb) | N50 (bp) | Contigs >N50 | Longest contig (bp) | Contigs >300 bp | Contigs >500 bp | Contigs >1000 bp |
|---|---|---|---|---|---|---|
| 77.55 | 687 | 31 057 | 22 289 | 121 076 | 53 707 | 16 251 |
About two-thirds (10 787 of 16 251) of the assembled J. coenia transfrags clustered with at least one other butterfly transcript from H. melpomene or D. plexippus (Fig. 1c). 3517 one-to-one orthologue clusters were obtained between all three species. Forty-nine per cent of the ORTHOMCL clusters contained transcripts from all three species (5490 of 11 267). Thirty-seven per cent of the total clusters contained both H. melpomene and D. plexippus, but not J. coenia, transfrags (4208 of 11 267), probably representing genes not expressed during wing development. Approximately 81% of clusters with J. coenia transfrags also included both sequenced butterfly species (5490 of 6782), 10% of the total clusters which contain J. coenia transfrags clustered with one of the sequenced butterfly species but not the other (660 of 6782), while about nine per cent only clustered with other J. coenia transfrags (632 of 6782) (Fig. 1c).
Gene ontology groups
Gene Ontology (GO) analysis of differentially expressed transcripts indicated that there were more ontology groups significantly differentially expressed between the colour forms at the fifth instar larval stage than at any other time point, with 33 GO terms over- or under-expressed (P < 0.00625) compared to six GO terms at early pupal, seven GO terms at early ommochrome, and 27 GO terms at the late ommochrome and melanin pigment development stages (full list of GO terms P < 0.00625 in Table S2). Several GO terms with very significant enrichments of interest between the colour morphs were identified. Genes associated with structural constituents of ribosomes (P = 1.52E-05) and oxygen transport (P = 1.49E-04) were significantly upregulated in the tan morph when compared to the red morph at the fifth instar larval stage of development. Genes associated with peptidase activity (P = 6.06E-10) were very significantly overexpressed in the red morph at the fifth instar larval stage of development.
One hundred and sixty-three GO terms were significantly upregulated or downregulated (P < 0.00417), between the fifth instar larval stage and the earliest pupal development stage compared with only 26 GO terms between early pupal and early ommochrome stage, and 37 GO terms between early ommochrome and the late ommochrome and melanin pigment development stages. A full list of enriched GO terms is presented in Table S2 (Supporting information). Of particular biological interest, genes involved in the function of translation were significantly overrepresented in the downregulated genes (P = 1.86E-17) during early pupal development of the tan morph when compared to the larval stage of this morph. Between these stages, cellular macromolecule biosynthetic process genes were also very significantly overrepresented in the downregulated genes (P = 1.53E-15), as were genes associated with ribosome functions (P = 1.48E-13), and genes involved with cellular macromolecule metabolic process (P = 2.09E-13). Genes associated with structural constituent of cuticle were extremely significantly overrepresented in the upregulated genes (P = 3.58E-18 and 4.41E-19, respectively), between the larval development stage and the earliest pupal stage of the tan and red morphs.
Differential expression analyses
There were 3145 significantly differentially expressed transfrags between the different stages and colour morphs (Fig. 1d and Table S3, Supporting information). 1420 transfrags were differentially expressed when comparing the tan and red colour morphs at the fifth instar stage, more than at any other time point sampled. Five hundred and forty-seven transfrags were differentially expressed between the colour morphs during early pupal development, 1231 transfrags were differentially expressed during early ommochrome development, and 854 transfrags were differentially expressed during late ommochrome and early melanin development.
Expression of ecdysone-associated genes
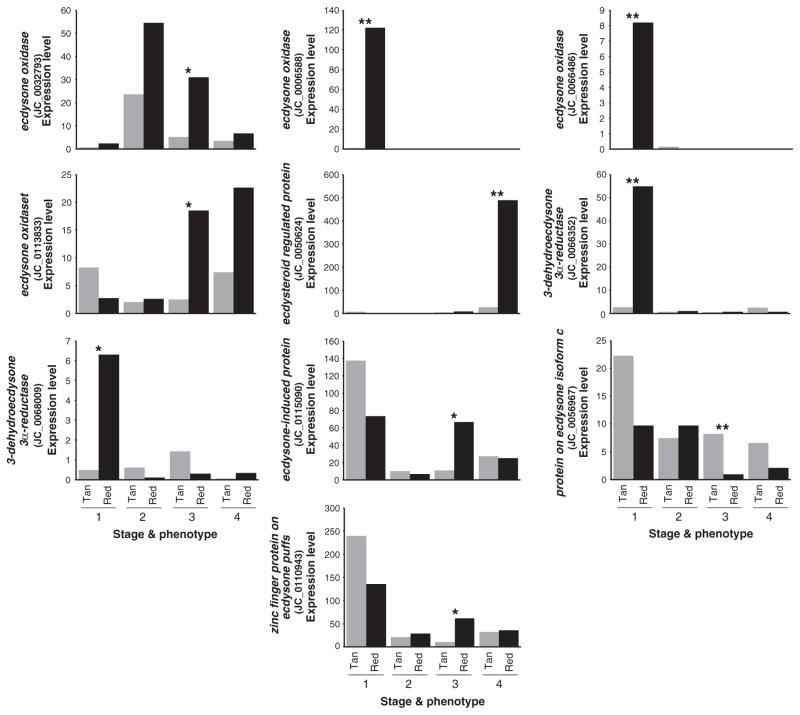
Of 11 ecdysone-associated transfrags identified by BLAST similarity, 10 showed significant differential expression between seasonal morphs (Fig. 2), and all but one of these showed significant upregulation specifically in the red morph. Transfrags encoding ecdysone-altering enzymes, including ecdysone oxidase and 3-dehydroecdy-sone 3α-reductase, showed the most dramatic patterns of differential expression at both early and late stages of development.
Fig. 2.
Stage- and phenotype-specific expression levels (FPKM) of selected ecdysone-associated transfrags that show significant differences between induced seasonal morphs. Stages are as shown in Fig. 1b. (*P < 0.05; **P < 0.005).
Expression of pigmentation-associated genes
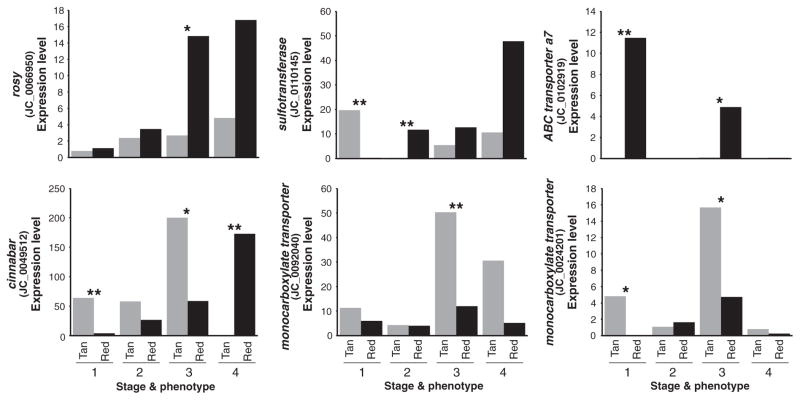
Based on BLAST similarity, we identified 19 transfrags potentially associated with ommochrome pigmentation, 14 of which were differentially expressed between morphs. Interestingly, 11 of these were differentially expressed in fifth instar wing discs, including significant red morph-specific upregulation of transfrags encoding ATP-binding cassette (ABC) transporters (JC_0023776, JC_0033677, JC_0036375, JC_0036375, JC_0092468, JC_0024281, JC_0028972) and monocarboxylate transporters (JC_0115080, JC_0024201, JC_0013098) (Table S2, Supporting information). Six transfrags showed differential expression during pupal development, including three transporters, rosy, a sulfotransferase (potentially involved in synthesis of sulphur-bearing ommatin-D) and cinnabar (kynurenine 3-monooxygenase) (Fig. 3). Interestingly, cinnabar was expressed at significantly higher levels in the tan morph during earlier developmental time points, but at significantly lower levels in the tan morph during late ommochrome/melanin development stage when compared to the red morph, thus suggesting a heterochronic shift in peak transcription between morphs.
Fig. 3.
Stage- and phenotype-specific expression levels (FPKM) of selected ommochrome-associated transfrags that show significant differences between induced seasonal morphs. Stages are as shown in Fig. 1b. (*P < 0.05; **P < 0.005).
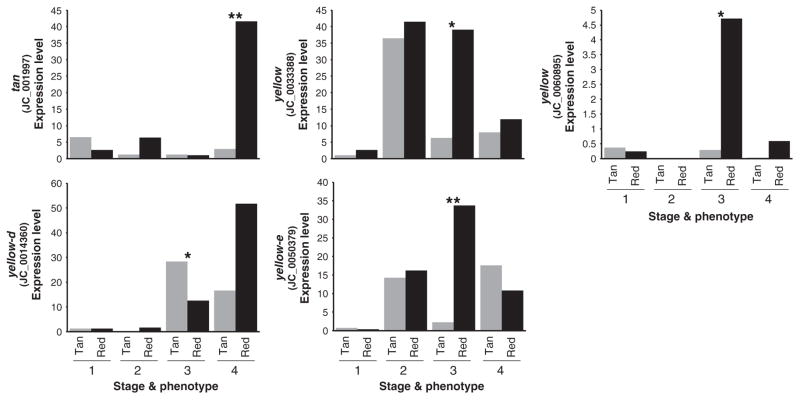
Of 13 transfrags identified as melanin gene transcripts based on BLAST similarity, six showed significant expression differences between colour morphs (Fig. 4). In the red morph, there was significant upregulation of tan at the final stage of pupal development, and yellow and yellow-e during early ommochrome development. yellow-h3 and black also showed striking patterns of specific upregulation in the red morph; however, their expression levels were relatively low and the differential expression was not statistically significant (Table S2, Supporting information).
Fig. 4.
Stage- and phenotype-specific expression levels (FPKM) of melanin-associated transfrags that show significant differences between induced seasonal morphs. Only transfrags showing differences during pupal development are shown. Stages are as shown in Fig. 1b. (*P < 0.05; **P < 0.005).
Discussion
Large numbers of differentially expressed transcripts
In addition to assembling the first transcriptome resource for Junonia coenia, we generated RNA-seq developmental time-courses for two distinct wing morphs to address the question of how developmental gene transcription can be influenced by environmental conditions. In part to make a preliminary assessment of the quality of our assembled transcripts, we used the trans-frag sequences to make comparisons of gene conservation with the postman butterfly H. melpomene (The Heliconius Genome Consortium 2012) and the monarch butterfly D. plexippus (Zhan et al. 2011)—two butterfly species with completely sequenced genomes. Using comparative sequence analyses, we found that about half of the gene clusters are shared by all three species. We speculate that most of the 39% of transfrags shared solely between H. melpomene and D. plexippus, and not J. coenia, are likely to represent genes not expressed during J. coenia wing development but that are present in the genome sequences. About 81% J. coenia transfrags were also represented in both other butterfly species, indicating a high level of conservation of the majority of genes between all three species.
Compared to a previous transcriptomic study that identified only 51 genes associated with colour pattern polymorphism in Heliconius wings (Hines et al. 2012), the number of transcripts differentially expressed between J. coenia seasonal morphs was remarkably high. This trend was strongest when comparing the red and tan morphs at the fifth instar developmental stage, which showed 1420 differentially expressed transfrags (as opposed to 547, 1231, and 854 transfrags at subsequent sampled stages). This finding that gene expression varied most strongly at the earliest sampled time point challenges our initial hypothesis that gene expression differences should be most pronounced at later stages of development when downstream effector genes are making pigments and determining scale morphology. We speculate that many of these transcriptional differences could be the result of nonpigmentation-related effects related to general thermal stress. The time for a pupa to develop under our defined cold temperature condition was about 2 weeks, as opposed to 1 week under our defined warm temperature condition. It is likely that this doubling of developmental time influenced wing cellular and metabolic processes in ways extending beyond the expected changes in wing pigment synthesis. In support of the thermal stress, hypothesis was our observation that transcripts representing genes in the oxygen transport GO group were differentially regulated between temperature conditions—a finding consistent with a transcriptional response to cold-induced hypoxia (Al-Fageeh & Smales 2006).
On another level, the unusual complexity of our data set may be due to the fact that wing pigmentation in J. coenia may be more complex than in Heliconius wings. The Heliconius butterfly wings for which transcriptomic data exist have simple patterns consisting of only three dominant pigments (yellow, red and black). In contrast, the wing scales of J. coenia form a very complex mosaic of a range of many different scale colours. Therefore, in J. coenia, different individual scales in close proximity will likely have several different pigments, or relative quantities of pigments, and thus could result in a more complex gene expression profile during development.
Finally, the method we used may have contributed somewhat to the relatively high number of differentially expressed transcripts observed. RNA-seq provides increased sensitivity to detect expression level changes over microarrays, which makes it particularly useful for accurate expression analyses (Mortazavi et al. 2008). At the same time, RNA-seq allows detection of transcripts across the entire genome, whereas microarray studies are limited to only a subset of genes. This increased sampling in itself should lead to uncovering a potentially higher number of differentially expressed genes; however, this effect would probably not be sufficient to wholly explain the magnitude of difference between the previous Heliconius work and our current results.
Expression of ecdysone-associated genes: evidence for morph-specific ecdysone inactivation
Differences in ecdysone levels during early pupal development (i.e. lower in the red morph, higher in the tan morph) are directly responsible for seasonal wing pattern variation in J. coenia (Rountree & Nijhout 1995). Because (i) ecdysone titres themselves are determined by activity of the prothoracic gland, independent of wing development, and (ii) ecdysone levels alone are sufficient to determine wing pattern morph, we hypothesized that differential expression of genes involved with synthesis of ecdysone or reception of ecdysone signals was unlikely to be associated with plasticity, and instead we would see differential expression of genes associated with downstream ecdysone response. Indeed, to a small degree, our findings supported this idea, in that we observed differential expression of transfrags identified as genes such as ecdysone-induced protein and zinc finger protein on ecdysone puffs (Fig. 2) as opposed to synthesis genes such as Halloween-family P450s or receptors such as ultraspiracle or ecdysone receptor.
One of the more interesting findings to come from our study was a surprise, however—namely the strong morph-specific expression of ecdysone-inactivating enzymes. Specifically, multiple transfrags representing ecdysone oxidase and 3-dehydroecdysone 3α-reductase were strongly upregulated during early wing development of the red morph (Fig. 2). These two enzymes catalyse an irreversible two-step inactivation of ecdysone to 3-epiec-dysteroid (Rees 1995). Therefore, high levels of these two enzymes could potentially result in reduced levels of ecdysone during the critical period in the red morph and potentially suggest a novel mechanism for localized modulation of ecdysone levels in response to environmental conditions. Future work should examine the potential roles of these genes in controlling seasonal phenotypes.
Ommochrome genes: transporters and enzymes associated with red morph
Because previous studies have identified ommochrome and ommochrome-related pigments in J. coenia wings (Nijhout 1997; Daniels & Reed 2012), ommochrome-associated transcripts are obvious candidates as causative agents for seasonal colour plasticity. Work in several nymphalid species has shed some light on which ommochromes genes may underlie wing pigment development and polymorphism. Reed & Nagy (2005) showed that the ommochrome genes vermilion, cinnabar and white were expressed in developing wings of the nymphalid Vanessa cardui. Later work in Heliconius (Reed et al. 2008; Ferguson & Jiggins 2009; Hines et al. 2012) demonstrated a clear spatial correlation between red wing pattern elements and cinnabar expression, thus implicating regulation of this gene in colour pattern development. Importantly, the most recent Heliconius microarray work suggests that butterflies have an expanded repertoire of uncharacterized ommochrome genes compared to Drosophila (Hines et al. 2012), including several novel transporter-encoding genes. As J. coenia wings are known to bear ommochrome pigments, and J. coenia is a nymphalid such as Vanessa and Heliconius, we hypothesized that similar ommochrome pathway genes may be upregulated during development of the red seasonal morph, leading to increased quantities of red ommochrome pigments.
In accord with our prediction, in the red morph, we observed a significant upregulation of multiple transcripts encoding ABC and monocarboxylate transporters phylogenetically related to the ommochrome genes white and karmoisin, respectively—primarily during very early wing development (Table S2, Supporting information, Fig. 3). Upregulation of these transporters may allow butterflies to more efficiently transport ommochrome precursors into scale cells or pigment granules, thus facilitating synthesis of red pigments. Interestingly, however, we also observed that two monocarboxylate transporters were upregulated in the tan form during the ommochrome stage of development (Fig. 3), possibly suggesting a role in the synthesis of a tan morph-specific pigments.
We observed that cinnabar was expressed at significantly higher levels in the red morph during late pupal development when pigments are actively being synthesized. The product of cinnabar, kynurenine-3-monooxy-genase, catalyses the conversion of kynurenine to 3-hydroxykynurenine, which is a key intermediate in the production of red and orange ommochrome pigments including xanthommatin, ommatin-D and the recently characterized J. coenia wing pigment xanthurenic acid (Daniels & Reed 2012). Therefore, it is reasonable to speculate that upregulation of cinnabar facilitates increased ommochrome synthesis in the red morph. One unexpected finding, however, was that cinnabar was significantly upregulated in the tan morph early in pupal development before pigment synthesis occurs. This difference in temporal expression mirrors the temporal shift in ecdysone titres responsible for inducing the polyphenism, so it interesting to consider whether this heterochronic shift in cinnabar transcription may be regulated by ecdysone. More broadly, our finding that cinnabar is the only ommochrome enzyme transcript showing a strong association with colour variation is similar to what has been observed in Heliconius (Reed et al. 2008; Ferguson & Jiggins 2009; Hines et al. 2012), thus suggesting that the cinnabar gene may be predisposed towards a repeated role in butterfly wing pattern evolution. We also noted a complex pattern of differential expression of a sulfotransferase transcript (Fig. 3). This enzyme could play a role in the synthesis of sulphur-containing ommochromes such as ommatin-D—a pigment proposed to occur in nymphalids (Linzen 1974). Finally, we observed red morph-specific upregulation of the xanthine dehydrogenase gene rosy during ommochrome development (Fig. 3). While rosy is typically associated with pteridine pigmentation (Reaume et al. 1991), Drosophila mutants for this gene show significant changes in tryptophan (including ommochrome) metabolism, although the role of rosy in this process is still not well understood (Kamleh et al. 2008).
Melanin genes: yellow-family genes and tan are associated with red morph
Work in Drosophila has functionally implicated a number of genes in the evolution and development of melanin pigmentation (e.g. True et al. 2005; Prud’homme et al. 2006; Jeong et al. 2008; Rebeiz et al. 2009; Wittkopp et al. 2009; Kronforst et al. 2012). Interestingly, we found that the majority of homologues to these Drosophila melanin genes showed elevated levels of expression during development of the red morph, and many of these trends were significant. The yellow family of genes in particular displayed a strong pattern of association with the red morph. Three transfrags representing yellow and yellow-e showed significant upregulation in red morphs. In addition to the yellow genes, the melanin enzyme gene tan was highly upregulated during late pigment development in red wings.
It was surprising to find that so many genes associated with black melanin pigmentation in other Lepidoptera were so strongly upregulated in red wings in our study. In Heliconius wing development yellow and tan are specifically associated only with melanic colour pattern elements, not red ommochrome elements (Ferguson et al. 2011a, b; Hines et al. 2012). In addition, in Papilio xuthus epidermal development tan shows strong specific spatial association with only black colour patterns (Futahashi et al. 2010; Futahashi et al. 2012). The only yellow-family gene previously associated with red ommochrome pigmentation is yellow-d, which was observed to be upregulated in red colour pattern in H. erato (Hines et al. 2012). While we did see strong upregulation of yellow-d during late ommochrome development in the J. coenia red morph, the difference did not reach statistical significance. Surprisingly, we observed that yellow-d was significantly upregulated in the tan morph during early ommochrome development (Fig. 3).
It was fascinating to find this association between melanin gene expression and a red phenotype, especially given that our ommochrome gene expression data are consistent with previous work that identified ommochromes as the primary red pigments in J. coenia (Nijhout 1997). One possible explanation is that the red coloration of the autumn morph is produced by a combination of ommochromes and red pheomelanins. Although pheomelanin pigments have not been rigorously characterized from J. coenia wings, there are three lines of preliminary evidence supporting their presence. First of all, labelled tyrosine—the precursor of melanin —is incorporated into scales of red regions at slightly elevated levels (Nijhout & Koch 1991). Second, acid methanol extraction of pigments from whole J. coenia wings does not completely remove red coloration from wings, as it does for red ommochrome wing patterns in Heliconius and Vanessa butterflies (R. D. Reed, personal observation), therefore suggesting that some of the red coloration in J. coenia is not due solely to ommochrome pigmentation. Third, cysteinyldopa melanin, a pheomelanin precursor, was characterized from developing J. coenia wings (Wakamatsu & Ito 2002). Although more work needs to be performed, our data hint at a complex relationship between the regulation of yellow-family genes and nonblack pigment synthesis.
Conclusion: The transcriptional profile of seasonal polyphenism
We used an unbiased whole transcriptome approach to study phenotypic plasticity in the wings of the buckeye butterfly J. coenia. Our work revealed a large set of transcripts that show significantly different expression levels during the development of alternative seasonal colour pattern morphs. This magnitude of transcription difference between wing morphs indicates that environmentally induced phenotypic plasticity is associated with very deep and broad shifts in developmental gene expression. This stands in marked contrast to previous work on genetic colour pattern variation, which shows an order of magnitude fewer differences in transcriptional changes. Many of the differentially expressed transcripts we identified were of unknown function, and hopefully future analyses benefitting from improved annotation and a fully sequenced genome will help determine whether they may play a functional role in phenotypic plasticity. Our focused analysis on endocrine and pigmentation genes revealed a number of candidates that may be specifically involved in differentiation of wing colour morphs. Among the interesting candidates were ecdysone-inactivating enzymes and genes related to ommochrome pigment synthesis. Surprisingly, we also found that numerous melanin genes were associated with the red colour morph, including tan and several members of the yellow gene family. This finding leads us to speculate that the red coloration in autumn J. coenia may be attributable to both ommochromes and red pheomelanins.
A more general question that arises from this work concerns the nature of the physiological ‘switch’ that underlies phenotypic plasticity. Many of the gene expression differences we observed seemed to be related to thermal response. Low temperatures are indeed stressful for buckeyes, and development time in butterflies reared at 21 °C is essentially double of those reared at 27 °C. Together, our observations raise the question to what extent seasonal morph differences are directly triggered by temperature cues, vs. to what extent they are a downstream by-product of stress and/ or delayed development that indirectly induces heterochronic shifts in hormone titres or gene expression. There is some circumstantial evidence for direct regulation of colour patterns by ecdysone in J. coenia, in that Koch et al. (2003) observed coexpression of ecdysone receptor and the transcription factor Distal-less in plastic eyespot patterns. This is interesting because changes in Distal-less expression are correlated with eyespot plasticity in the butterfly Bicyclus anyana (Brakefield et al. 1996). To address this question more thoroughly, we are now working to better define the relative contributions of light, temperature and critical period in buck-eye morph determination. We are also conducting comparative work between populations with different reaction norms to understand how seasonal plasticity varies between populations adapted for life at different latitudes and elevations. The insights and transcriptome resources generated through the work described here will form an important foundation for this and other future work in the buckeye system.
Supporting information
Additional supporting information may be found in the online version of this article.
Supplementary Material
Assembly statistics by stage and morph before being condensed by CD-HIT.
List of GO terms that are significantly overrepresented in the upregulated and downregulated genes lists (P < 0.05/number of gene sets used for multiple comparisons) between color forms and developmental stages of J. coenia.
List of differentially expressed genes from the J. coenia transcriptome, with P-values, FPKM expression levels for all four developmental stages, the log2 expression fold changes, and associated GO terms. Read counts were normalized to the number of kilobases of transcript length and for number of reads sequenced per sample. FPKM values are the average of two replicates for each developmental stage and morph.
Acknowledgments
This research was supported by the United States National Science Foundation grant DEB 1011619. We would like to thank Fred Nijhout for access to his colony of North Carolina buck-eye butterflies. We also thank Riccardo Papa, Arnaud Martin, Kailen Mooney, John Avise and members of the Mortazavi laboratory for feedback and discussion spanning all aspects of this project, and three anonymous reviewers for their helpful comments and suggestions.
Footnotes
E.V.D. and R.D.R. designed the study. E.V.D. performed the experiments. R.M. performed assemblies and computational analyses with guidance from E.V.D., A.M. and R.D.R. E.V.D. and R.D.R. wrote the manuscript with input from R.M. and A.M. All the authors read and approved the final version of the manuscript.
Data accessibility
DNA sequences, assemblies and associated metadata: NCBI GEO Accession no. GSE54819. Junonia coenia transcriptome download available at Dryad (doi:10.5061/ dryad.5n5h6), and download and BLAST interface at butterflygenome.org.
References
- Al-Fageeh MB, Smales CM. Control and regulation of the cellular responses to cold shock: the responses in yeast and mammalian systems. Journal of Biochemistry. 2006;397:247–259. doi: 10.1042/BJ20060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beldade P, Mateus ARA, Keller RA. Evolution and molecular mechanisms of adaptive developmental plasticity. Molecular Ecology. 2011;7:1347–1363. doi: 10.1111/j.1365-294X.2011.05016.x. [DOI] [PubMed] [Google Scholar]
- Brakefield PM, Larsen TB. The evolutionary significance of dry and wet season forms in some tropical butterflies. Biological Journal of the Linnean Society. 1984;22:1–12. [Google Scholar]
- Brakefield PM, Gates J, Keys D, et al. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 1996;384:236–242. doi: 10.1038/384236a0. [DOI] [PubMed] [Google Scholar]
- Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH. A reference-free algorithm for computational normalization of shotgun sequencing data. 2012. arXiv:1203.4802v2. [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Daniels EV, Reed RD. Xanthurenic acid is a pigment in Junonia coenia butterfly wings. Biochemical Systematics and Ecology. 2012;44:161–163. [Google Scholar]
- Daniels EV, Mooney KA, Reed RD. Seasonal wing colour plasticity varies dramatically between buckeye butterfly populations in different climatic zones. Ecological Entomology. 2012;37:155–159. [Google Scholar]
- Ferguson LC, Jiggins CD. Shared and divergent expression domains on mimetic Heliconius wings. Evolution & Development. 2009;11:498–512. doi: 10.1111/j.1525-142X.2009.00358.x. [DOI] [PubMed] [Google Scholar]
- Ferguson LC, Maroja L, Jiggins CD. Convergent, modular expression of ebony and tan in the mimetic wing patterns of Heliconius butterflies. Development Genes and Evolution. 2011a;221:297–308. doi: 10.1007/s00427-011-0380-6. [DOI] [PubMed] [Google Scholar]
- Ferguson LC, Green J, Surridge A, Jiggins CD. Evolution of the insect yellow gene family. Molecular Biology and Evolution. 2011b;28:257–272. doi: 10.1093/molbev/msq192. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Banno Y, Fujiwara H. Caterpillar color patterns are determined by a two-phase melanin gene prepatterning process: new evidence from tan and laccase2. Evolution & Development. 2010;12:157–167. doi: 10.1111/j.1525-142X.2010.00401.x. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Shirataki H, Narita T, Mita K, Fujiwara H. Comprehensive microarray-based analysis for stage-specific larval camouflage pattern-associated genes in the swallowtail butterfly, Papilio xuthus. BMC Biology. 2012;10:46. doi: 10.1186/1741-7007-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines HM, Papa R, Ruiz M, et al. Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genomics. 2012;13:288. doi: 10.1186/1471-2164-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True JR, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Kamleh MA, Hobani Y, Dow JAT, Watson DG. Metabolomic profiling of Drosophila using liquid chromatography Fourier transform mass spectrometry. FEBS Letters. 2008;582:2916–2922. doi: 10.1016/j.febslet.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Koch PB. Production of [14C]-labeled 3-hydroxy-L-ky-nurenine in a butterfly, Heliconius charitonia L. (Heliconidae), and precursor studies in butterfly wing ommatins. Pigment Cell Research. 1993;6:85–90. doi: 10.1111/j.1600-0749.1993.tb00586.x. [DOI] [PubMed] [Google Scholar]
- Koch PB, Merk R, Reinhardt R, Weber P. Localization of ecdysone receptor protein during colour pattern formation in wings of the butterfly Precis coenia (Lepidoptera: Nymphalidae) and co-expression with Distalless protein. Development Genes and Evolution. 2003;212:571–584. doi: 10.1007/s00427-002-0277-5. [DOI] [PubMed] [Google Scholar]
- Kronforst MR, Barsh GS, Kopp A, et al. Unraveling the thread of nature’s tapestry: the genetics of diversity and convergence in animal pigmentation. Pigment Cell and Melanoma Research. 2012;25:411–433. doi: 10.1111/j.1755-148X.2012.01014.x. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. ORTHOMCL: identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzen B. The tryptophanommochrome pathway in insects. Advances in Insect Physiology. 1974;10:117–246. [Google Scholar]
- Moczek AP. Phenotypic plasticity and diversity in insects. Philosophical Transactions of the Royal Society B – Biological Sciences. 2010;365:593–603. doi: 10.1098/rstb.2009.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP, Sultan S, Foster S, et al. The role of developmental plasticity in evolutionary innovation. Proceedings of the Royal Society B: Biological Sciences. 2011;278:2705–2713. doi: 10.1098/rspb.2011.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Pattern formation on lepidopteran wings: determination of an eye-spot. Developmental Biology. 1980;80:267–274. doi: 10.1016/0012-1606(80)90403-0. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Ommochrome pigmentation of the linea and rosa seasonal forms of Precis coenia (Lepidoptera: Nymphalidae) Archives of Insect Biochemistry and Physiology. 1997;36:215–222. [Google Scholar]
- Nijhout HF. Development and evolution of adaptive polyphenisms. Evolution & Development. 2003;5:9–18. doi: 10.1046/j.1525-142x.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Koch PB. The distribution of radiolabeled pigment precursors in the wing patterns of nymphalid butterflies. Journal of Research on the Lepidoptera. 1991;30:1–13. [Google Scholar]
- Prud’homme B, Gompel N, Rokas A, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Reaume AG, Knecht DA, Chovnick A. The rosy Locus in Drosophila melanogaster: xanthine dehydrogenase and eye pigments. Genetics. 1991;129:1099–1109. doi: 10.1093/genetics/129.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, Nagy LM. Evolutionary redeployment of a bio-synthetic module: expression of eye pigment genes vermil-lion, cinnabar, and white in butterfly wing development. Evolution and Development. 2005;7:301–311. doi: 10.1111/j.1525-142X.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- Reed RD, McMillan WO, Nagy LM. Gene expression underlying adaptive variation in Heliconius wing patterns: non-modular regulation of overlapping cinnabar and vermilion prepatterns. Proceedings of the Royal Society B: Biological Sciences. 2008;275:37–45. doi: 10.1098/rspb.2007.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HH. Ecdysteroid biosynthesis and inactivation in relation to function. European Journal of Entomology. 1995;92:9–39. [Google Scholar]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nature Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree DB, Nijhout HF. Hormonal control of a seasonal polyphenism in Precis coenia (Lepidoptera: Nymphalidae) Journal of Insect Physiology. 1995;41:987–992. [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KC. The effects of temperature and daylength on the rosa polyphenism in the buckeye butterfly, Precis coenia (Lepidoptera: Nymphalidae) Journal of Research on the Lepidoptera. 1991;30:225–236. [Google Scholar]
- Snell-Rood EC, Van Dyken JD, Cruickshank T, Wade MJ, Moc-zek AP. Toward a population genetic framework of developmental evolution: the costs, limits, and consequences of phenotypic plasticity. BioEssays. 2010;32:71–81. doi: 10.1002/bies.200900132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Heliconius Genome Consortium . Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Yeh S, Hovemann BT, et al. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genetics. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu I, Ito S. Advanced chemical methods in melanin determination. Pigment Cell Research. 2002;15:174–183. doi: 10.1034/j.1600-0749.2002.02017.x. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Stewart EE, Arnold LL, et al. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science. 2009;326:540–544. doi: 10.1126/science.1176980. [DOI] [PubMed] [Google Scholar]
- Yamamoto RT. Mass rearing of the tobacco hornworm. II. Larval rearing and pupation. Journal of Economic Entomology. 1969;62:1427–1431. [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM. The Monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Assembly statistics by stage and morph before being condensed by CD-HIT.
List of GO terms that are significantly overrepresented in the upregulated and downregulated genes lists (P < 0.05/number of gene sets used for multiple comparisons) between color forms and developmental stages of J. coenia.
List of differentially expressed genes from the J. coenia transcriptome, with P-values, FPKM expression levels for all four developmental stages, the log2 expression fold changes, and associated GO terms. Read counts were normalized to the number of kilobases of transcript length and for number of reads sequenced per sample. FPKM values are the average of two replicates for each developmental stage and morph.