Abstract
PIWI-interacting RNAs (piRNAs) protect the animal germline by silencing transposons. Primary piRNAs, generated from transcripts of genomic transposon ‘junkyards’ (piRNA clusters), are amplified by the “Ping-Pong” pathway, yielding secondary piRNAs. We report that secondary piRNAs, bound to the PIWI protein Ago3, can initiate primary piRNA production from cleaved transposon RNAs. The first ~26 nt of each cleaved RNA becomes a secondary piRNA, but the subsequent ~26 nt becomes the first in a series of phased primary piRNAs that bind Piwi, allowing piRNAs to spread beyond the site of RNA cleavage. The Ping-Pong pathway increases only the abundance of piRNAs, whereas production of phased primary piRNAs from cleaved transposon RNAs adds sequence diversity to the piRNA pool, allowing adaptation to changes in transposon sequence.
In animals, PIWI proteins guided by single-stranded, 23–36 nucleotide (nt) small RNAs, PIWI-interacting RNAs (piRNAs), suppress germline transposon expression. In Drosophila, piRNAs bind the PIWI proteins, Piwi, Aubergine (Aub) and Argonaute3 (Ago3) (1). Fly primary piRNAs derive from long transcripts from piRNA clusters—discrete genomic loci comprising transposon fragments (2). The endonuclease Zucchini (Zuc) is thought to cut cluster transcripts into fragments whose 5′ ends correspond to the 5′ ends of piRNAs, but whose length is longer than piRNAs; these piRNA precursors are loaded into Piwi and Aub and then trimmed from their 3′ ends, yielding mature primary piRNAs (1, 3–5). In the fly oocyte, maternally inherited and primary piRNAs made de novo initiate production of secondary piRNAs, which subsequently self-amplify via reciprocal cycles of Aub- and Ago3-catalyzed cleavage of transposon mRNAs and cluster transcripts, a process known as the Ping-Pong pathway (Fig. S1) (6, 7). The Ping-Pong pathway increases piRNA abundance, but cannot create novel piRNA sequences. Yet piRNA populations are highly diverse, with most individual species of low abundance.
We used genetic mutants to separate primary, maternal, and secondary piRNAs. To assess the mutants’ effects on the germline, we examined piRNAs from the largest piRNA cluster, 42AB (6). aubHN2/QC42; ago3t2/t3 double-mutants lack the Ping-Pong pathway, so they contain only maternal and primary piRNAs (for 42AB, Z10 = 0.6; Z-score ≥ 2.81 corresponds to p-value ≤ 0.005; Fig. 1A). In contrast, zuc mutants contain maternal and secondary, but not primary piRNAs. Loss of Zuc decreased 42AB piRNAs 50-fold (Fig. S2A), but the piRNAs remaining showed significant Ping-Pong amplification (42AB piRNAs, Z10 = 39; all piRNAs, Z10 = 42), consistent with a small pool of maternal piRNAs being amplified into secondary piRNAs.
Fig. 1.
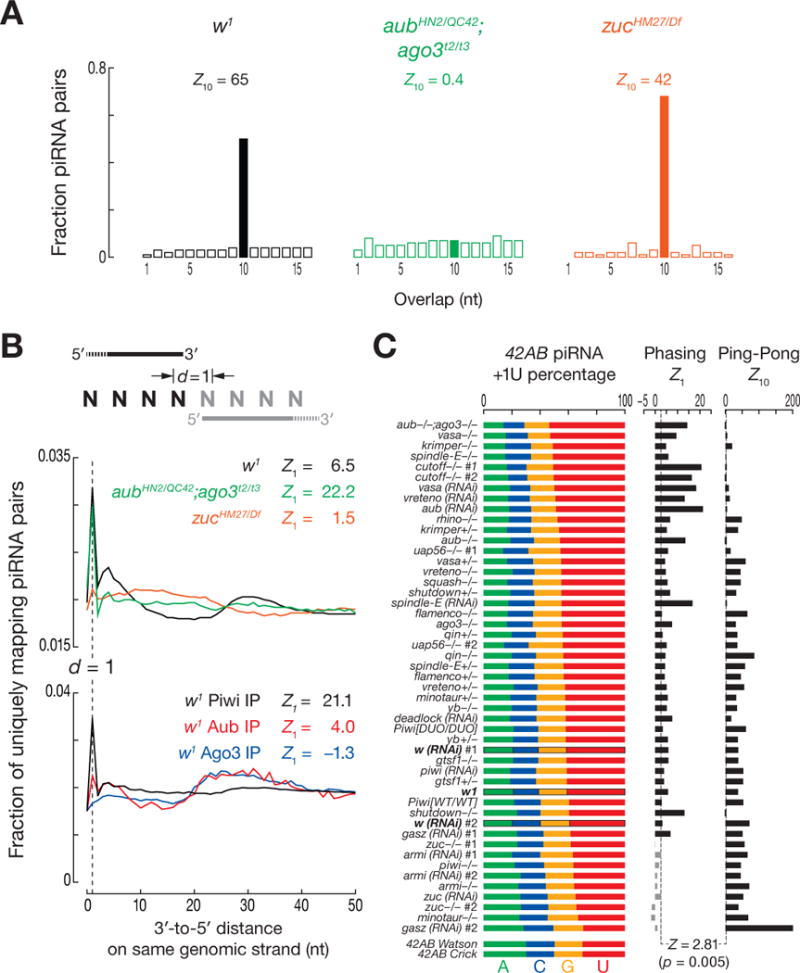
Zuc-dependent phasing of primary piRNAs. (A) Ping-Pong analysis of all piRNAs from w1, aubHN2/QC42; ago3t2/t3 and zucHM27/Df. (B) Distance from the 3′ ends of upstream piRNAs to the 5′ ends of downstream piRNAs on the same genomic strand. (C) Nucleotide composition immediately after the 3′ ends of uniquely mapping 42AB piRNAs. Z10: Ping-Pong; Z1: phasing.
The 5′ ends of piRNAs mapping to the same genomic strand and present in aubHN2/QC42; ago3t2/t3 but not zucHM27/Df, typically lay 25–28 nt apart, the same length as piRNAs themselves (Fig. S2B). Thus, the maternal and primary piRNAs remaining in aubHN2/QC42; ago3t2/t3 double-mutant ovaries were phased, suggesting that a nuclease initiates production of piRNAs from one end of a piRNA precursor, moving 5′-to-3′ to clip off successive piRNAs.
The distance from the 3′ end of each piRNA to the 5′ end of the next downstream piRNA measures piRNA phasing (Fig. 1B). The most common 3′-to-5′ distance was 1 nt: a single cleavage event appears to produce the 3′ end of one piRNA and the 5′ end of the adjacent, downstream piRNA more often than expected by chance (Z1 for w1 = 6.5). Production of phased piRNAs required Zuc but not Ping-Pong: the 1 nt peak was more prominent in aubHN2/QC42; ago3t2/t3 ovaries (Z1 = 22) than in w1, but undetectable in zucHM27/Df (Z1 = 1.5).
piRNA phasing differed among the three Drosophila PIWI proteins (Fig. 1B). By 3′-to-5′ distance, Piwi-bound piRNAs displayed the most significant phasing (Z1 = 21); Aub-bound piRNAs displayed reduced, but still significant phasing (Z1 = 4.0); Ago3-bound piRNAs were not phased (Z1 = −1.3). Thus, Piwi- and Aub-, but not Ago3-bound primary piRNAs are produced by a processive mechanism that requires Zuc.
piRNAs associated with Piwi and Aub, but not Ago3, typically begin with uridine (6). Phased piRNAs beginning with U could be produced by a processive nuclease complex measuring out ~26 nt, then cleaving at the nearest U. Alternatively, they could be made by the same nuclease measuring out ~26 nt, but cleaving at all nucleotides with similar efficiency; subsequent binding of Piwi and Aub would select for piRNAs starting with U. The first model predicts that the nucleotide immediately following the 3′ end of a piRNA—in genomic sequence but not mature piRNAs—is more likely to be U than expected. The second model predicts that when one piRNA follows another in phase, the second piRNA is more likely to begin with U because of the preference of Aub and Piwi; the genomic nucleotide following a piRNA would not have any sequence bias, because selection for a 5′ U follows piRNA precursor cleavage. To distinguish between the models, we measured the composition of the nucleotide after the 3′ ends of piRNAs (“+1U percentage”). This nucleotide was typically uridine in both w1 and aubHN2/QC42; ago3t2/t3, but not in zucHM27/Df (Fig. S2D), indicating that phased piRNAs are likely produced by direct cleavage 5′ to U, before pre-piRNAs are loaded into PIWI proteins. Since purified Zuc shows no nucleotide preference (4, 5), we propose that other factors direct Zuc to cleave before U.
We analyzed piRNAs derived from 42AB for 21 different mutant or germline RNAi strains. Phasing was detected in all strains except those with an impaired primary piRNA pathway (Fig. 1C) (2, 8–13). Compared to wild-type, phasing was more readily detected in mutants defective in Ping-Pong, likely because loss of secondary piRNAs reduced background signal. We also detected Zuc-dependent phasing for somatic piRNAs, which are always primary (Fig. S2E). We conclude that phasing is an inherent feature of primary, but not secondary, piRNA production.
To test whether the production of phased piRNAs depends on maternal piRNAs, we used a strain bearing a ~7 kbp transgene, P{GSV6}, inserted into 42AB. P{GSV6} carries both gfp and w+mC and produces both sense and antisense piRNAs (Figs. S3A and S3B). Both transgene piRNA abundance and Ping-Pong were greater when P{GSV6}42A18 was inherited maternally (Figs. 2A and S3A) (14–16), but primary piRNA phasing was unaltered by the parental source of the transgene (Z1 maternal = 13; Z1 paternal = 13). As an additional test of the idea that phased piRNAs are primary, not maternal, we sequenced piRNAs from vasaD5/PH165 ovaries that had inherited the P{GSV6} transgene maternally or paternally (Fig. 2A); Vasa is required for Ping-Pong amplification. Regardless of which parent contributed the transgene, P{GSV6}-derived piRNAs displayed significant phasing (paternal, Z1 = 12; maternal, Z1 = 9.0; wild-type, Z1 = 13), consistent with the idea that phasing is a primary piRNA signature that requires neither maternal piRNAs nor Ping-Pong amplification.
Fig. 2.
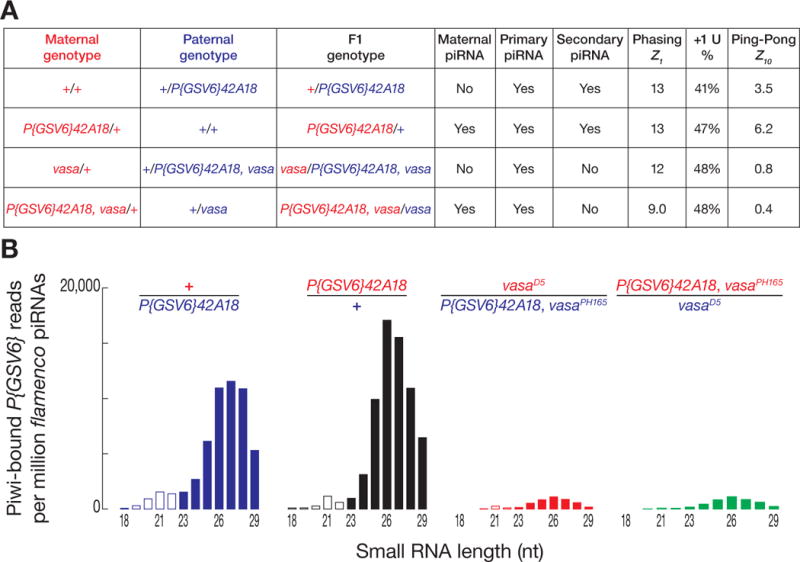
Contribution of maternal and secondary piRNAs to phasing. (A) Ping-Pong, phasing, and +1U percentage were analyzed for P{GSV6}42A18-derived piRNAs inherited paternally or maternally, with or without Vasa. (B) Length distribution of Piwi-bound, uniquely mapping P{GSV6}42A18 piRNAs. Reads were normalized to flamenco-derived, uniquely mapping piRNAs.
Without Vasa, piRNA phasing (Z1) and the percentage of uridine at the genomic nucleotide immediately after the 3′ ends of the piRNAs (+1U percentage) was unchanged, but the abundance of Piwi-bound piRNAs was less than one-tenth of wild-type (Figs. 2B and S3C). Piwi is likely loaded only with primary piRNAs (Fig. S3D) (16–18). Why then should Vasa, a central component of the secondary piRNA pathway, affect the abundance of Piwi-bound piRNAs? One explanation is that production of Piwi-loaded, phased primary piRNAs requires precursor cleavage directed by secondary piRNAs. To test this idea, we sequenced the “degradome”—RNAs >200 nt and bearing 5′ monophosphates—to detect the RNAs cleaved by secondary piRNAs bound to Aub or Ago3. In w1 control ovaries, we readily identified long transposon RNAs whose 5′ ends were generated by Aub or Ago3 (Fig. S4A) (19); such degradome reads were absent from aubHN2/QC42; ago3t2/t3 mutants (Z10 = 0.8). Moreover, degradome reads corresponding to Piwi-catalyzed cleavage were indistinguishable from background (Z10 = 0.4), consistent with Piwi silencing via transcriptional repression, rather than RNA cleavage (18). Thus 3′ cleavage products of Aub- or Ago3-catalyzed slicing are subsequently used to produce phased primary piRNAs.
We determined the fraction of piRNA 5′ ends at each position ±150 nt from an Aub or Ago3 cleavage site (Fig. S4B). In both w1 and zuc mutant ovaries, piRNA 5′ ends were more likely to map to the cleavage site than expected by chance (w1, Z0 = 27; zucHM27/Df, Z0 = 34; Fig. S4C). These piRNAs correspond to secondary piRNAs produced by the Ping-Pong cycle. We also detected phased piRNAs in w1 as a peak ~26 nt after the Aub or Ago3 cleavage sites that corresponds to the 5′ ends of primary piRNAs immediately following the 3′ end of secondary piRNAs, and as a ~53 nt peak representing the 5′ end of another primary piRNA immediately following the 3′ end of the first primary piRNA. zucHM27/Df ovaries lacked both peaks.
We separated degradome reads based on the likelihood (p-value ≤ 0.005, χ2 test) that they were produced by Aub versus Ago3 (Fig. S4D) (19), then analyzed the distance between the 5′ ends of Piwi-bound piRNAs and the sites of Aub- or Ago3-catalyzed cleavage. The 5′ ends of Piwi-bound piRNAs coincided with the Zuc-dependent ~26 and ~53 nt peaks for both Aub- and Ago3-cleaved RNAs (Fig. 3). A small but significant fraction of Aub-, but not Ago3-bound piRNAs also began ~26 and ~53 nt after the Ago3-cleaved sites.
Fig. 3.
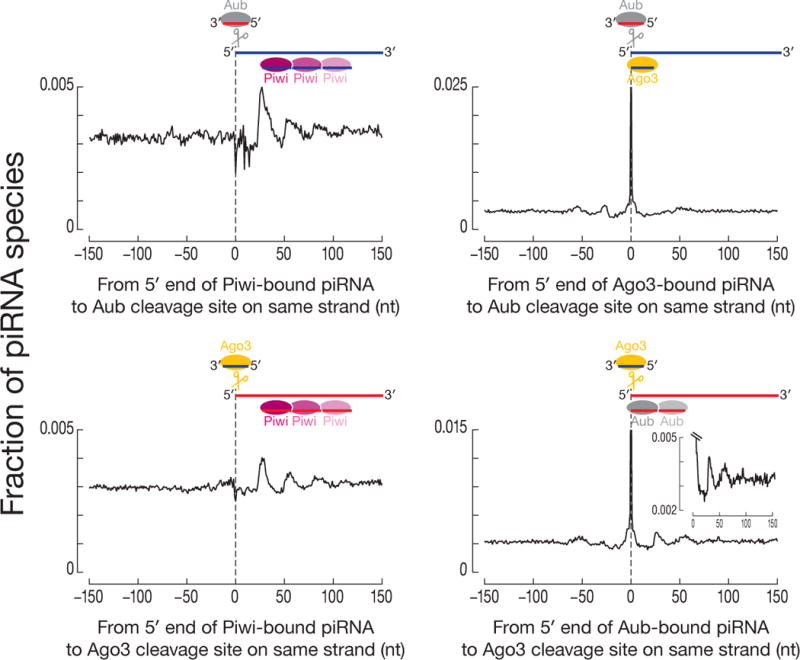
Phasing of Piwi-bound piRNAs after sites of Aub- or Ago3-cleavage. Distance was measured from the 5′ ends of Piwi-, Ago3-, and Aub-bound piRNAs to sites of Aub- or Ago3-cleavage on the same genomic strand.
Small RNA and degradome sequencing data from zuc mutant ovaries unambiguously identified sites cleaved by Aub or Ago3. Using this data, the 5′ ends of Piwi-bound piRNAs in w1 ovaries were typically ~26 and ~53 nt downstream from where Aub or Ago3 cleaved (Fig. S4E), a relationship not detected using degradome data from aubHN2/QC42; ago3t2/t3 ovaries, which lack secondary piRNAs (Fig. S4F). Next, we measured the distance from the 5′ ends of Aub- and Ago3-bound piRNAs to the 5′ ends of Piwi-bound piRNAs on the same genomic strand (Fig. S5A). Again, the 5′ ends of Piwi-bound piRNAs were typically 26 nt downstream from the 5′ ends of Ago3-piRNAs and 27–29 nt downstream of the 5′ ends of Aub-piRNAs. Similarly, the 5′ ends of Aub-bound piRNAs lay ~26 nt downstream from the 5′ ends of Ago3-bound piRNAs. In contrast, the 5′ ends of Ago3-bound piRNAs were no more likely to be ~26 nt downstream from the 5′ ends of Aub-bound piRNAs than would be expected by chance. Thus, RNAs cut by Ago3 produce phased, Aub-bound piRNAs, but RNAs cut by Aub do not make phased, Ago3-bound piRNAs.
The distance between the 5′ ends of Piwi-bound piRNAs and the 5′ ends of Ago3- or Aub-bound piRNAs on the opposite genomic strand (i.e., Ping-Pong analysis) again suggests that the 3′ cleavage product generated by Ago3 or Aub is initially processed into a secondary piRNA, and thereafter is used for the production of phased primary piRNAs loaded into Piwi (Fig. S5B). Piwi does not directly participate in Ping-Pong, and the 5′ ends of Piwi-bound piRNAs did not map 10 nt from the 5′ ends of Aub- or Ago3-bound piRNAs. Instead, Piwi-bound piRNAs lay 15–19 nt after the 5′ ends of Aub- or Ago3-bound piRNAs. Such phased piRNAs have been detected previously, but were attributed to Ping-Pong amplification (20).
In the absence of Ago3 or Vasa, 42AB-derived, Piwi-bound piRNAs decreased to ~10% of the w1 level (Fig. 4A). Loss of Aub had a more modest effect: 42AB-derived, Piwi-bound piRNAs were ~47% of w1. Thus, Ago3 initiates the production of most phased, Piwi-bound primary piRNAs. These data help explain why transposon silencing requires heterotypic Aub:Ago3 Ping-Pong amplification (17): homotypic Aub:Aub Ping-Pong cannot generate enough Piwi-bound, antisense, primary piRNAs.
Fig. 4.
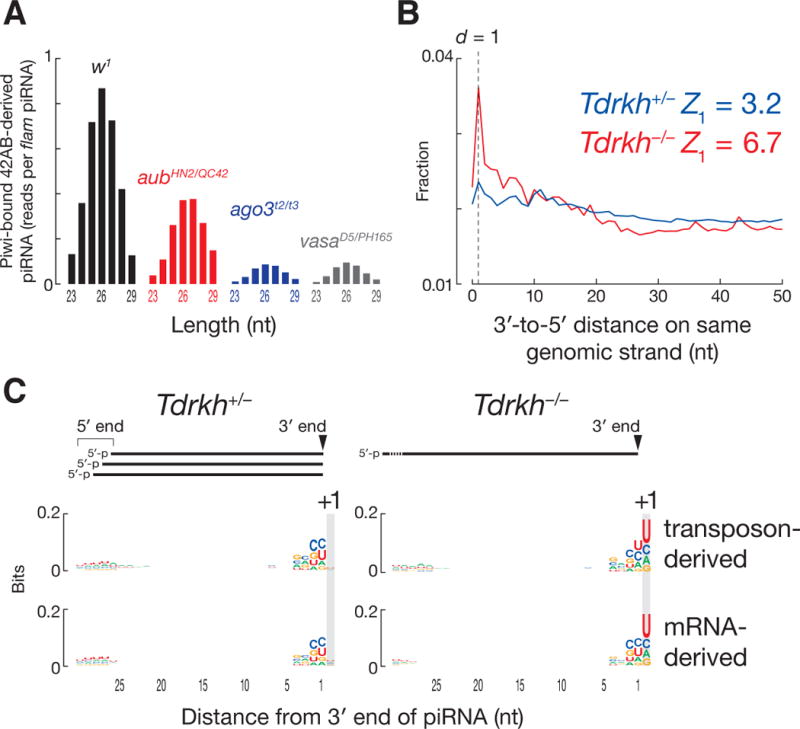
Mouse piRNAs are phased. (A) Length distribution of Piwi-bound, uniquely mapping fly piRNAs derived from 42AB in w1, aubHN2/QC42, ago3t2/t3 and vasaD5/PH165. Reads were normalized to flamenco-derived, uniquely mapping piRNAs. (B) Distance from the 3′ ends of upstream piRNAs to the 5′ ends of downstream piRNAs on the same genomic strand for uniquely mapping piRNAs in Tdrkh+/− and Tdrkh−/− mouse testes at 11 dpp. (C) Genomic nucleotide composition 29 nt before and 1 nt after the 3′ ends of uniquely mapping mouse piRNAs.
Experiments in silkmoth cells and mice implicate the Tudor protein Papi in 3′ piRNA trimming (21, 22). To test the role of 3′ trimming in the biogenesis of phased primary piRNAs, we sequenced small RNAs from papi mutant fly ovaries (Fig. S6A). The median length of piRNAs from nearly all transposon families increased 0.35 nt (p-value < 2.2 × 10−16, Wilcoxon signed-rank test; Figs. S6B–S6D) and germline and somatic piRNA phasing became more pronounced (Figs. S6E and S6F). We propose that 3′ trimming of Piwi-bound piRNAs allows the use of uridines >26 nt after the 5′ end of a pre-piRNA as cleavage sites to make piRNAs.
In testes from wild-type mice, one piRNA 5′ end often lies 30–40 nt downstream from another (Figs. S7A–C), possibly because mouse pre-piRNAs are longer than those in flies and require the 3′ trimming of ~3–10 nt. Analysis of Papi (Tdrkh−/−) mutant testes supports this view. Tdrkh−/− testes accumulate 31–37 nt RNAs instead of 26–30 nt piRNAs, and most of these longer species share their 5′ ends with mature piRNAs from Tdrkh+/− heterozygotes (22). At 11 dpp, 3′-to-5′ distance analysis of piRNAs from Tdrkh−/− testes showed clear evidence for phasing (Fig. 4B). Mouse piRNAs typically begin with uridine, and the 3′ ends of the longer RNAs in Tdrkh−/− testes were generally followed by a uridine in genomic sequence (Fig. 4C). piRNA 5′-to-5′ distance analysis of Tdrkh+/− and Tdrkh−/− showed broad peaks at 35–43 nt—the same length as the pre-piRNAs detected in Tdrkh−/− (Fig. S7D). We conclude mammalian primary piRNAs are phased, but are more extensively trimmed than those in flies.
Our findings suggest a model for primary piRNA biogenesis (Fig. S8) in which each RNA cleaved by Ago3 or Aub produces not only a secondary Ping-Pong piRNA, but also primary piRNAs from the sequences immediately 3′ to the secondary piRNA. Such a spreading mechanism calls to mind features of siRNA production in Caenorhabditis elegans and Arabidopsis thaliana (23–26), and primed CRISPR adaptation in Escherichia coli (27, 28). Although the detailed mechanisms differ, signal amplification with sequence diversification is clearly a recurrent theme for RNA-guided silencing in animals, plants, and bacteria.
Supplementary Material
Acknowledgments
We thank Alicia Boucher, Cindy Tipping, Gwen Farley, and Ellen Kittler for technical assistance; Ryuya Fukunaga and Erik Sontheimer for discussions; Julius Brennecke for sharing reagents and unpublished data; and members of the Weng and Zamore laboratories for advice and comments on the manuscript. This work was supported in part by National Institutes of Health grants HG007000 to Z.W. and GM62862 and GM65236 to P.D.Z. Sequencing data are available from the NCBI Sequence Read Archive using accession number SRP045930.
References and Notes
- 1.Luteijn MJ, Ketting RF. Nat Rev Genet. 2013;14:523. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 2.Malone CD, et al. Cell. 2009;137:522. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voigt F, et al. RNA. 2012;18:2128. doi: 10.1261/rna.034967.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishimasu H, et al. Nature. 2012;491:284. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 5.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. Nature. 2012;491:279. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennecke J, et al. Cell. 2007;128:1089. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 7.Gunawardane LS, et al. Science. 2007;315:1587. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 8.Vagin VV, et al. Science. 2006;313:320. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 9.Pane A, Wehr K, Schupbach T. Dev Cell. 2007;12:851. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. EMBO J. 2010;29:3301. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vagin VV, et al. RNA. 2013;19:1064. doi: 10.1261/rna.039669.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czech B, Preall JB, McGinn J, Hannon GJ. Mol Cell. 2013;50:749. doi: 10.1016/j.molcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handler D, et al. Mol Cell. 2013;50:762. doi: 10.1016/j.molcel.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennecke J, et al. Science. 2008;322:1387. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vanssay A, et al. Nature. 2012;490:112. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- 16.Le Thomas A, et al. Genes Dev. 2014;28:1667. doi: 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, et al. Mol Cell. 2011;44:572. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sienski G, Donertas D, Brennecke J. Cell. 2012;151:964. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, et al. Mol Cell. 2014;56:708. doi: 10.1016/j.molcel.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau NC, et al. Genome Res. 2009;19:1776. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda S, et al. RNA. 2013;19:1405. doi: 10.1261/rna.040428.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxe JP, Chen M, Zhao H, Lin H. EMBO J. 2013;32:1869. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Z, Allen E, Wilken A, Carrington JC. Proc Natl Acad Sci U S A. 2005;102:12984. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshikawa M, Peragine A, Park MY, Poethig RS. Genes Dev. 2005;19:2164. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagijn MP, et al. Science. 2012;337:574. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HC, et al. Cell. 2012;150:78. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko KA, et al. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


