Abstract
Natural killer (NK) cells have traditionally been considered nonspecific components of innate immunity, but recent studies have shown features of antigen-specific memory in murine NK cells. However, it has remained unclear whether this phenomenon also exists in primates. Compared to NK cells from uninfected macaques, we found splenic and hepatic NK cells from SHIV-SF162P3- and SIVmac251-infected animals specifically lysed Gag- and Env-pulsed dendritic cells (DCs) in an NKG2-dependent fashion. Moreover, splenic and hepatic NK cells from Ad26-vaccinated macaques efficiently lysed antigen-matched but not antigen-mismatched targets 5 years post-vaccination. These data demonstrate that robust, durable, antigen-specific NK cell memory can be induced in primates following both infection and vaccination, and could be important for vaccines against HIV-1 and other pathogens.
NK cells have traditionally been associated with nonspecific innate killing of virus-infected and neoplastic cells. However, increasing evidence suggests that NK cells also cooperate with adaptive humoral immune responses to mediate antibody-dependent cell-mediated cytotoxicity (ADCC) and modulate CD4+ and CD8+ T cell responses 1–6. Specific to HIV-1 infection, NK cells have been reported to proliferate during primary infection 7 prior to the development of CD8+ T cell responses. In addition, lysis of HIV-1-infected cells by NK cells occurs through a variety of mechanisms including ADCC 8, downmodulation of major histocompatibility complex (MHC) class I molecules 9, and upregulation of NKG2D ligands 10. NK cells can also inhibit CCR5-dependent entry of HIV-1 by secreting β-chemokines CCL3, CCL4, and CCL5 11. In rhesus macaques, NK cells have been shown to lyse SIV-infected cells 12 and SIV-pulsed cells13. Further studies have shown that acute infection of rhesus macaques with SIVmac251 induces rapid NK cell activation and increased cytotoxicity 14, and longitudinal studies suggest that NK cells may be associated with preventing disease progression in SIV-infected macaques 15,16.
To date, antigen-specific NK cell memory has only been described in mice 17–23. Mice lacking T and B cells develop immunologic memory to haptens and viral antigens that was mediated by a transferrable subset of liver-restricted NK cells 18,19,21,23. Certain activating receptors on human and murine NK cells have also been demonstrated to recognize proteins from several viruses and to modulate disease 24–27. However, expression of those surface molecules on NK cells has not been associated with acquisition of antigen-specific NK cell memory responses thus far. Long-lived and transferrable memory responses against murine cytomegalovirus (MCMV) were demonstrated to induce binding of Ly49H on murine NK cells to the virus-encoded protein m157 28, although antigen specificity was not formally tested in that study. Antigen-specific NK cell memory has not been previously demonstrated in any primate species, but a large body of work has long suggested that the NK cell response may not be entirely nonspecific. Increased NK cell antiviral functions in HIV-1-exposed seronegative individuals (HESN) have been associated with protection 29,30 and uninfected infants of HIV-1-positive mothers can mount potent NK cell responses that are associated with blocking transmission in utero 31, both potentially indicating a pre-sensitization to the virus. Similarly, KIR-MHC interactions have been shown to exert significant immunological pressure on HIV-1 replication thereby driving virus evolution and escape 32. During human CMV infection NKG2C+CD57+ NK cells expand in a virus-specific manner 33 and similarly CD27+ NK cells are associated with spontaneous control of HCV 34, but none of these previous studies have definitively shown acquired antigen specificity of NK cells in humans or other primate species. To address this deficit, in this study we assessed potential NK cell memory in both SIV-infected and long-term vaccinated rhesus macaques.
Results
Identification and purification of rhesus macaque NK cells
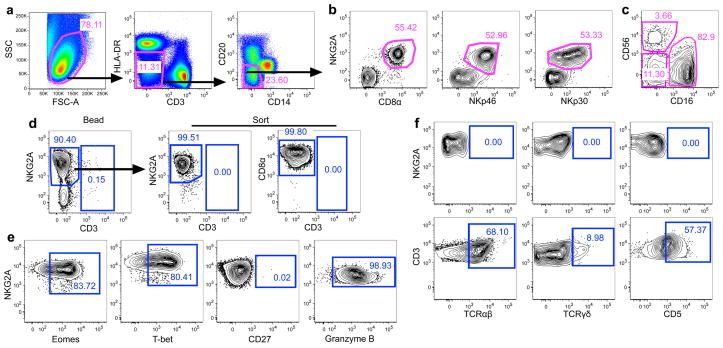
We identified bulk NK cells as CD3−CD14−CD20−HLA-DRdim NKG2A+ mononuclear cells as described previously 15,35,36 (Fig. 1a) and confirmed their identity by demonstrating co-expression of the NK-related markers CD8α, NKp30, and NKp46 (Fig. 1b). Macaque NK cells also had the expected distribution of CD56 and CD16 subpopulations 15,35,36 (Fig. 1c). To evaluate the antigen specificity of NK cells, we developed a flow cytometric assay to measure the ability of purified NK cells to lyse autologous dendritic cells (DCs). Briefly, we purified NK cells from bulk mononuclear cells using magnetic beads and then sorted either CD3−CD14−CD20−NKG2A+ or CD3−CD14−CD20−CD8α NK cells by flow cytometry. This resulted in 99.5–99.8% pure NK cells and with no detectable (0.00%) contaminating T cells (Fig. 1d). NK cells expressed high amounts of intracellular Eomes, T-bet, and Granzyme B, but lacked CD27 expression (Fig. 1e). Due to the highly overlapping phenotypes it is possible purified NK cells could contain minor frequencies of the phylogenetically-related innate lymphoid cell type 1 (ILC1) 37, but since primate ILC1 generally do not express Granzyme B, are CD27+, and have low expression of Eomes 38, contamination is likely to be minimal. To confirm a lack of T cell contamination, we further analyzed purified NK cells by flow cytometry and found no detectable expression of the T cell markers CD5, TCRαβ and TCRγδ (Fig. 1f). Collectively, these data present a comprehensive definition of NK cells in rhesus macaques and our methodologies for highly efficient purification.
Figure 1.
Purification of macaque NK cells. (a) Representative gating strategy identifying non-lineage cells in bulk mononuclear cells from rhesus macaques. (b) Co-expression of NKG2A and various NK cell markers among CD3−CD14−CD20−HLA-DRdim mononuclear cells identifying NKG2A as the most inclusive NK cell marker in rhesus macaques. (c) Expression of CD56 and CD16 on bulk NK cells identifying putative NK cell subpopulations. Plots are representative of over 30 naïve and SIV-infected rhesus macaques. (d) Representative flow cytometry plots demonstrating NK cell purity after bead-enrichment and subsequent sorting for CD3−CD14−CD20−CD8α+ cells, resulting in 99.5–99.8% purity. (e) Lack of ILC1-related phenotype; NKG2A+CD3− macaque NK cells were analyzed for intracellular Eomes, T-bet, and Granzyme B, and surface CD27. (f) Assessment of T cell-related markers on purified NK cells; NKG2A+CD3− macaque NK cells were analyzed for surface TCRαβ, TCRγδ, and CD5 (upper panels) and compared to bulk T cells (lower panels). Flow plots are representative of four to ten animals per marker.
Antigen-specific NK cell responses in SHIV-SF162P3 infection
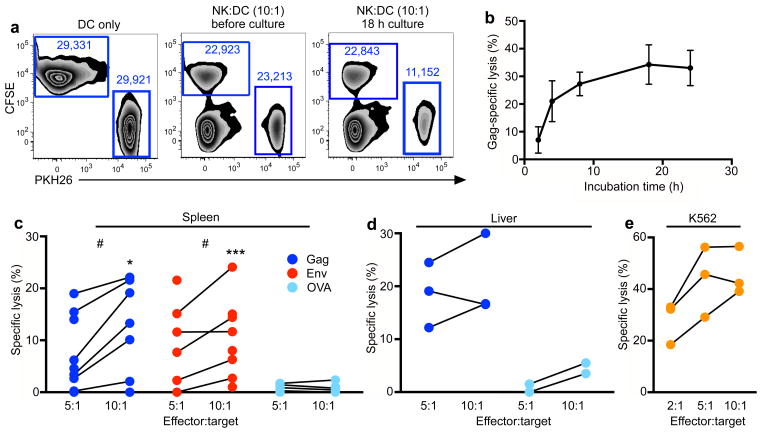
To evaluate the ability of purified NK cells to lyse cells in an antigen-specific manner, we performed co-cultures with autologous DCs pulsed with antigens and labeled with the red dye PKH26. Cultures also contained unpulsed autologous DCs labeled with the green dye CFSE as internal controls (Fig. 2a). Although many NK cell-killing assays perform co-cultures of only six hours, time-course experiments determined maximum killing was achieved with an 18 h co-incubation (Fig. 2b). This could be due, in part, to the use of frozen cells in our assays, differences among species, or differences in the mechanism of ‘innate’ versus ‘antigen-specific’ killing by NK cells. Using this assay, we first tested the ability of purified splenic and hepatic NK cells from a cohort of 8 SHIV-SF162P3-infected macaques to lyse DCs pulsed with SIVmac239 Gag or HIV-1 Env. Splenic NK cells exhibited killing of DCs pulsed with Gag or Env (median specific lysis 13% and 10%, respectively at a 10:1 effector:target (E:T) ratio), but not the irrelevant protein ovalbumin (Ova) (median specific lysis 0.3% at a 10:1 E:T ratio) (Fig. 2c). Responses to the viral proteins versus Ova were significantly different at both 10:1 (Gag, P = 0.015; Env, P = 0.001) and 5:1 (Gag, P = 0.017; Env, P = 0.023) E:T ratios. Hepatic NK cells showed a median specific lysis of 16–18% of Gag-pulsed DCs (Fig. 2d). As an additional positive control, we also demonstrated that bulk NK cells, regardless of their state of antigen experience, were functionally capable of nonspecific lysis of standard NK cell targets, MHC-devoid K562 cells (Fig. 2e). These data confirm that highly purified tissue NK cells from SHIV-infected macaques could recognize and lyse autologous DCs in an antigen-specific manner.
Figure 2.
Antigen-specific lysis of autologous dendritic cells in chronically SHIV-SF162P3-infected macaques by NK cells. (a) Flow cytometric visualization of NK-DC co-culture; representative of over 50 NK-DC co-culture assays visualizing DCs only, immediately after addition of NK cells, and lysis after co-culture. Collected numbers of events are indicated and are used to calculate lysis. (b) Time course experiment demonstrating maximized killing at 18 h co-incubation. Bars represent mean ± SEM of 4 independent experiments. (c) Specific lysis of Gag- or Env-pulsed dendritic cells from SHIV-SF162P3-infected macaques by splenic NK cells at 10:1 and 5:1 E:T ratios. (d) Specific lysis of Gag- or Env-pulsed dendritic cells from SHIV-SF162P3-infected macaques by hepatic NK cells at 10:1 and 5:1 E:T ratios. (e) Specific lysis of PKH26-labeled K562 cells by splenic NK cells from SHIV-SF162P3-infected macaques. NK cell-resistant RAJI cells labeled with CFSE were included in all wells as internal controls. Statistical comparisons between parallel E:T comparing antigen (Gag or Env) with Ova, *, P < 0.05; **, P < 0.01; ***, P < 0.001; Mann-Whitney U test. Statistical comparisons between 10:1 and 5:1 E:T ratios of same animals, #, P < 0.05, Wilcoxon-Matched pairs test.
Antigen-specific NK cell responses in SIVmac251 infection
We next evaluated NK cells from a cohort of 8 rhesus macaques chronically infected with SIVmac251 and 6 naive uninfected macaques. Splenic NK cells from infected animals were highly reactive to Gag-pulsed DCs at a 10:1 ratio with a median specific lysis of 40% as compared with 0.1% in uninfected age-matched controls (Fig. 3a) (P = 0.018). In contrast, NK cells from SIV-infected animals were not reactive to unpulsed DCs (Fig. 3b). These data demonstrate robust anti-Gag NK cell responses in the spleen of SIV-infected animals but not in uninfected animals, confirming true antigen-specificity. Only marginal Gag-specific NK cell responses were observed in peripheral blood, although these responses were still slightly greater than those observed in uninfected animals (P = 0.05) (Fig. 3c). These findings suggest that antigen-specific NK cells are either located primarily in tissues or alternatively may reflect sequestration in sites of higher antigen concentration. As a confirmatory assay, CD107a was also specifically upregulated on splenic NK cells cultured with Gag-pulsed DCs (Fig. 3d), and CD107a expression correlated with NK cell lysis (P = 0.02; Fig. 3e). These data corroborate the antigen-specific NK cell killing assays and also suggest that CD107a may be a relevant surrogate marker for antigen-specific NK cell cytotoxicity. Taken together, these experiments demonstrate antigen-specific NK cell activity in both SHIV- and SIV-infected rhesus macaques. The greater NK cell killing in the animals infected with SIVmac251 as compared with SHIV-SF162P3 may reflect higher viral loads in these animals or virus-specific differences. Indeed, viral loads modestly correlated with both percentage specific lysis and CD107a expression (Fig. 3f), suggesting an antigen-driven expansion of SIV Gag-specific NK cells.
Figure 3.
Antigen-specific NK cell responses in SIVmac251-infected macaques. (a) Specific lysis of Gag-pulsed dendritic cells from SIV-infected and SIV-uninfected rhesus macaques in NK:DC co-cultures from spleen at 10:1 and 5:1 E:T ratios. (b) Specific lysis of mock-pulsed dendritic cells in splenic NK:DC co-cultures at 10:1 E:T ratios. (c) Specific lysis of Gag-pulsed dendritic cells in NK:DC co-cultures from peripheral blood at indicated E:T ratios. (d) CD107a expression in splenic and peripheral NK cells in response to Gag-pulsed dendritic cells from SIV-infected rhesus macaques in NK:DC (10:1) co-cultures. (e) Relationship of data from (b) and (d) (n = 7). (f) Correlation of SIV-specific NK cell responses with plasma viral load (n = 9; n = 7). . Cross-sectional statistical comparisons are between SIV-infected and SIV-uninfected samples at the 10:1 E:T. *, P < 0.05; Mann-Whitney U test. Spearman’s test was used for all correlations, P < 0.05.
Ad26 vectors induce potent memory NK cell responses
We then evaluated whether NK cells were capable of long-term antigen-specific memory following vaccination in the absence of ongoing antigen stimulation. Nine rhesus macaques were vaccinated twice 6 months apart with 109 viral particles of either replication-incompetent Ad26 vectors39 expressing HIV-1 Env (n = 4), or a DNA prime, Ad26 boost regimen expressing SIVmac239 Gag (n= 5). These animals developed durable Env- and Gag-specific T cell responses to their respective vaccine antigens as expected (Fig. 4a & 4b) and were not further boosted or challenged. Five years after the final vaccination, we necropsied both groups of animals and assessed for potential antigen-specific memory NK cell responses in peripheral blood, liver and spleen. As added controls we evaluated antigen-specific responses to both the vaccine antigen and the non-vaccine antigen (i.e., mismatched = evaluation of anti-Env responses in Gag-vaccinated animals). At both 10:1 and 5:1 E:T ratios, splenic and hepatic NK cells lysed antigen-matched targets efficiently but exhibited minimal to no reactivity against antigen-mismatched targets (Fig. 4c & 4d) (P = 0.02 for hepatic NK cells, P = 0.04 for splenic NK cells). In particular, at the 10:1 E:T ratio, splenic NK cells from vaccinated animals showed a median of 27% specific lysis of antigen-matched targets but only 2% specific lysis of antigen-mismatched targets. Consistent with the previous experiment (Fig. 3), responses in peripheral blood were marginal (Fig. 4e). We also observed CD107a responses in response to the matched but not the mismatched antigen (Fig. 4f & 4g), and these data correlated with the killing assays (P = 0.04; Fig. 4h). The observation of antigen-specific NK cell responses 5 years following vaccination indicate the durability of these memory NK cell responses. Memory NK cell responses were comparable in liver and spleen, which contrasts with some evaluations of murine memory NK cell responses that have demonstrated restriction of memory NK cell activity to the liver 19,21,23.
Figure 4.
Antigen-specific NK cell memory in vaccinated macaques. Gag (a) and Env (b) specific T cell responses in Ad26-vaccinated macaques were measured longitudinally by IFN-γ ELISPOT as previously described 39. Specific lysis of antigen-pulsed DCs with ‘matched’ antigens (i.e., Gag-pulsed DCs in Gag-vaccinated animals, Env-pulsed DCs in Env-vaccinated animals) and ‘mismatched’ antigens (i.e, Gag-pulsed DCs in Env-vaccinated animals, Env-pulsed DCs in Gag-vaccinated animals) by purified (c) splenic and (d) hepatic NK cells at 10:1 and 5:1 E:T ratios. (f) Specific lysis of Gag-pulsed DCs by purified NK cells from PBMC of Gag-vaccinated rhesus macaques. CD107a expression in (f) splenic and (g) hepatic NK cells in response to antigen-pulsed DCs with ‘matched’ antigens and ‘mismatched’ antigens. Assays were performed at 10:1 NK:DC ratios and are background subtracted using unpulsed DC co-culture controls. (h) Correlation of Gag-specific lysis and CD107a upregulation in ‘matched’ and ‘mismatched’ animals (n = 28). (i) Lack of CD8+ T cell responses at low E:T ratios to exclude possibility of contaminating CD8+ T cell-mediated killing. CD8+ T cells were bead-purified from Ad26-Gag vaccinated macaques as described in the Methods and co-cultured with Gag-pulsed DCs. Statistical comparisons between parallel E:T comparing matched antigen with mismatched antigen, *, P < 0.05; Mann-Whitney U test. Statistical comparisons between 10:1 and 5:1 E:T ratios of same animals, #, P < 0.05, Wilcoxon-Matched pairs test. Spearman’s test was used for all correlations, P < 0.05.
Sorting of NK cells eliminated all contaminating CD8+ T cells (Fig. 1d). To further exclude the remote possibility that contaminating T cells may still have contributed to the observed specific lysis, we performed parallel cell killing experiments with CD8+ T cells from the vaccinated animals at 1:1, 0.1:1, and 0.01:1 E:T ratios. These low ratios were chosen not to evaluate CD8+ T cell-mediated lysis but to simulate T cell contamination, and demonstrated little to no killing, even at the 1:1 ratio (Fig. 4i). In re-sort analyses, we demonstrate 0.00% T cell contamination (Fig. 1d), and even if we consider the potential theoretical error rate of the flow cytometer of 1 in 10,000 cells, the 1:1 E:T ratio represents 5 logs greater than the maximal possible levels of T cell contamination in any of the NK killing assays. These considerations collectively eliminate the possibility that the observed antigen-specific NK killing could be due to contaminating CD8+ T cells.
NK cell memory responses are NKG2-dependent
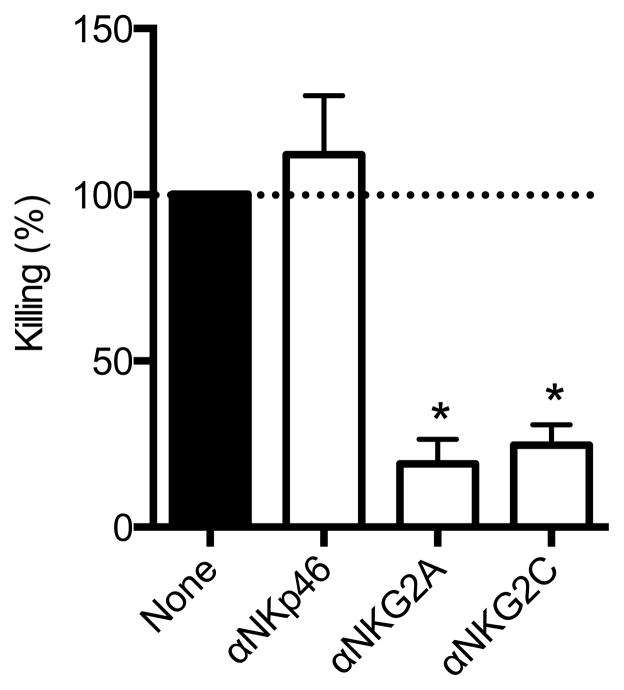
To determine the relative contribution of various NK cell subsets to the observed memory NK cell responses, we repeated the experiments in Fig. 2 in the presence or absence of blocking antibodies to various NK cell receptors. Blocking NKp46 showed no effect on NK cell-mediated killing, but blocking NKG2A and NKG2C markedly reduced NK cell killing of Gag-pulsed DCs by 80% and 75%, respectively (P < 0.05; Fig. 5). These data suggest that molecules of the NKG2 family may play a role in target cell recognition or as a co-receptor, analogous to what has been shown for NKG2D in murine memory NK responses to haptens 21.
Figure 5.
Antigen-specific NK cell killing is dependent on NKG2a/c. Specific lysis of antigen-pulsed DCs from animals shown in Fig. 2. Antibodies were added prior to co-culture and percent (%) killing is shown as a fraction of control wells with no blocking (none; normalized to 100%). Summarizes data from five independent experiments. Mean ± SEM are shown. *, P < 0.05; Wilcoxon Matched Pairs test.
Discussion
In these studies, we demonstrate clear evidence of antigen-specific memory NK cell responses in a primate species. Specifically we show anti-Gag and anti-Env NK cell responses in both SIVmac251- and SHIV162P3-infected rhesus macaques demonstrating NK cell memory generated against active replication of lentiviruses. Furthermore, we found that antigen-specific NK cells were inducible after Ad26 vaccination and the responses were durable 5 years post-vaccination without challenge.
Prior to these studies, antigen-specific NK cell memory has only been described in mice, by showing21 transferrable hapten-specific contact hypersensitivity (CHS) in mice lacking T and B cells was dependent on a subpopulation of liver NK cells. These findings of NK cell-mediated CHS were subsequently confirmed 17–19,22, and NK cell memory has now been demonstrated against VSV, HIV-1, and influenza vaccine antigens in mice and found to contribute to efficient protection against vaccinia and influenza challenges 19,23. Pre-sensitized memory-like NK cells have also been described against MCMV and can expand upon virus exposure and to protect upon adoptive transfer 28.
Our data extend these prior studies by showing antigen-specific NK cell memory in primates, following both infection and vaccination. Interestingly, since SIVmac251 and SHIV-SF162P3 are not natural pathogens in macaques, it seems unlikely that memory responses to these viruses occur through typical NK cell receptor interactions, such as the effect mediated by Ly49H-m157 in mice infected with MCMV 28. The most specific associations with control of HIV-1 by NK cells have been KIR-mediated including epidemiologic demonstration that expression of KIR3DS1 on NK cells, in combination with its ligand HLA-Bw4-80I, are associated with slower disease progression 40, and in vitro evidence that KIR3DS1+ NK cells can inhibit HIV-1 replication in vitro 41. However, KIR-mediated effects require interaction with cognate ligands that are often disparately expressed in individuals. Perhaps more likely is antigen-specific imprinting and recall by a subset of NK memory cells which remain incompletely defined in primates, similar to observations in mice 21. Our data also suggest that this process may be dependent on NKG2 molecules. Although the full mechanism is unclear, our data indicates that both the inhibitory NKG2A and the activating NKG2C are required, but not the unrelated NKp46. NKG2A is known to bind MHC-I, and absence of this inhibitory interaction results in cell killing as demonstrated by lysis of MHC-devoid K562 cells. Our data find that NKG2 molecules may also be necessary for antigen-specific killing, perhaps suggesting a distinction specific to memory NK cells. Interestingly, NKG2C expression on human NK cells is associated with subset expansion in response to HCMV, but not other herpesviruses 33. NKG2C also delineates a sub-population of ‘memory-like’ NK cells that use antibody binding for specificity and display enhanced ADCC, but become hyporesponsive to cytokine stimulation 42–44. These alterations in NK cell function reflect widespread epigenetic modifications with changes in DNA methylation patterns resembling those of cytotoxic T cells 43,44. NK cells and NKG2 expression have also been linked to elite control of viremia and reduced acquisition of HIV-1 32,45,46.
The full role of NK cells in either modulation of HIV-SIV disease or prevention against acquisition also remains uncertain. NK cells are known to lyse both HIV- and SIV-infected cells, but to what extent this occurs in vivo has been difficult to determine 7,11,14,41. Increased NK cell function and sensitization in HIV-1-exposed seronegative individuals HESN suggests these cells could contribute, at least in part, to blocking transmission 29,30, although whether antigen-specificity is generated in this context is unknown. Contradictory evidence alternatively suggests NK cell may have little to no role in limiting lentivirus transmission or elimination of infected cells 36,47, but these studies also did not address antigen specificity. Although significant empirical evidence exists it has been difficult to define a specific role for NK cells in HIV or SIV disease, partly due to the fact that currently, strategies to fully and accurately deplete NK cells in SIV-infected macaque models are lacking 15,48. Intriguingly SIV-specific NK cell activity positively correlated with plasma viral load indicating the presence of antigen can likely influence at least the magnitude of antigen-specific NK cell responses. The longevity of memory NK cell responses following Ad26 vaccination in the absence of virus replication also suggests ongoing antigenic stimulation is not necessarily required to maintain long-term responses. Future studies will be necessary to determine the kinetics and full anatomic distribution of antigen-specific NK cells following lentivirus infection and vaccination, as well as the relative contributions to preventing virus acquisition and disease modulation.
Overall, our findings show antigen-specific NK cell responses in primates following both infection and vaccination. Although antigen-specific NK cells are likely being modulated by ongoing virus replication in infected animals, the finding that Ad26 vaccination can induce true memory NK cells that are maintained 5 years after vaccination is particularly intriguing. The longevity, functionality, and specificity of these responses suggest their potential utility in the development of vaccines against both HIV-1 and other pathogens. Further research to evaluate the protective efficacy of these responses in primates and to define the detailed molecular mechanisms of target cell engagement are warranted. Regardless, our new data and the extensive work previously done in mice suggest that NK cell memory could be conserved among mammalian species and underscores the incomplete understanding and heterogeneity of innate versus adaptive immune responses.
ONLINE METHODS
Animals and SIV infections
Thirty-one adult male and female Indian rhesus macaques were analyzed in this study, including 6 naïve, 8 chronically infected with SHIV-SF162P3 (median duration of infection 802 days), 8 chronically infected with SIVmac251 (median duration of infection 371 days), and 9 vaccinated IM twice with 109 viral particles Ad26 vectors expressing HIV-1 Env inserts 39 or primed with DNA vaccines and boosted with Ad26 vectors expressing SIVmac239 Gag. All animals were free of simian retrovirus type D and simian T-lymphotrophic virus type 1 and were housed at New England Primate Research Center, Southborough, MA. All animal studies were performed in accordance with the American Association for Accreditation of Laboratory Animal Care standards and in compliance with protocols approved by the Institutional Animal Care and Use Committee of Harvard Medical School.
Tissue collection and processing
Macaques were euthanized and spleen and liver were collected and mechanically disrupted. EDTA-treated venous blood was also collected at indicated time points. Mononuclear cells were isolated from blood and single-cell spleen suspensions by density gradient centrifugation over lymphocyte separation media (MP Biomedicals, Solon, OH). Hepatic mononuclear cells were isolated from the interface of a 30/60% discontinuous Percoll gradient. A hypotonic ammonium chloride solution was used to lyse contaminating red blood cells. Cells were either immediately cultured or cryopreserved in 90% fetal bovine serum/10% DMSO and stored in liquid nitrogen vapor.
Antigen-specific NK cell killing assay
Hepatic and splenic NK cells were isolated from bulk mononuclear cells using custom negative magnetic bead selection protocols, routinely yielding >90% NKG2A+CD3− NK cell purity with <1% T cell contamination as described. 49 For some experiments, NK cells were further purified by positive sorting for CD3−CD14−CD20−CD8α+ on a FACS Aria (Purity mode; 2,500 events/second) resulting in 99.5 – 99.8% NK cell purity. Input numbers of between 2.0 × 107 and 1.0 × 108 mononuclear cells, depending on the experiment, yielded between 5.0 × 105 and 5.0 × 106 sort-purified NK cells. Dead cells were excluded by Aqua dye and only live cells were sorted with > 95% viability. Autologous DCs were similarly purified by bead-based separation using antibodies against macaque CD3, CD20, CD8α, and NKG2A. DC purity resulted in > 90% HLA-DR+NKG2A−CD3− cells, with 0% contamination by CD3+ or NKG2A+ cells. DCs were then pulsed with intact Gag or Env antigens (2 μg/ml) and were labeled with the red dye PKH26 (Sigma-Aldrich) for identification in our flow cytometric assay. Non-pulsed DCs serving as intra-well controls were labeled with the green dye CFSE (Sigma-Aldrich). Purified NK cells were co-cultured with DCs at multiple effector-to-target (E:T) ratios (equal mixture of pulsed target DCs and non-pulsed control DCs) for 18 h, and specific lysis of DCs was calculated as (% sample lysis with NK effectors - % basal lysis without NK effectors) / (100 - % basal lysis without NK effectors) (Fig. 1g). In alternative experiments, anti-CD107a was added for the duration of culture and measured after 18 h as described previously 35. In blocking experiments, 2 μg/ml of anti-NKG2A (clone Z199, Beckman-Coulter), anti-NKG2C (clone REA110, Bioss), or anti-NKp46 (clone BAB281, Beckman-Coulter) were added 1 h prior to co-culture.
CD8+ T cell isolation
Macaque CD8+ T cells were isolated from blood using Miltenyi nonhuman primate CD8+ T cell isolation kits according to the manufacturer’s suggested protocol (Miltenyi Biotec). Purity was verified by flow cytometry to be greater than 95% CD8+CD3+ T cells.
Nonspecific NK cell killing assay
To serve as an additional positive control and to ensure our purified NK cells were reactive to known NK cell stimuli, we modified our killing assay to use MHC-devoid K562 cells as susceptible NK cell targets, labeling them with PKH26. NK cell-resistant RAJI cells served as intra-well controls and were labeled with the green dye CFSE. Purified NK cells were co-cultured with these cell lines at multiple E:T ratios for 18 hours and specific lysis of K562 cells was calculated as (% sample lysis with NK effectors - % basal lysis without NK effectors) / (100 - % basal lysis without NK effectors).
Flow cytometry
Isolated mononuclear cells or Q-Prep (Beckman-Coulter) on whole blood was used for surface staining. Caltag Fix & Perm (Invitrogen) was used for all intracellular staining. Antibodies used are shown in Supplementary Table 1. Flow cytometry acquisitions were performed on an LSR II (BD Biosciences, La Jolla, CA) and FlowJo software (version 9.6.4, Tree Star Inc., Ashland, OR) was used for all analyses.
Statistical analyses
All statistical and graphic analyses were performed using GraphPad Prism 6.0 software (GraphPad Software Inc., La Jolla, CA). Nonparametric Mann-Whitney U tests were used where indicated, and a P value of < 0.05 (by a 2-tailed test) was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants AI060354, AI078526, AI096040, AI09585 (D.H.B), AI069259, AI111595 and AI112521 (U.H.v.A), AI067031 (M.A.), AI118468 (RKR), as well as American Foundation for AIDS Research grant 108547-53-RGRL, Harvard Center for AIDS Research grant AI060354 (both to R.K.R), and by the Ragon Institute of MGH, MIT, and Harvard (S.J., M.A., D.H.B, U.H.v.A.).
Footnotes
Author contributions
R.K.R., M.A., U.H.v.A., and D.H.B designed the studies. H.L., S.J., E.B., H.L., J.L.S., V.V., C.M., and L.E conducted the assays. R.K.R. and D.H.B. wrote the paper with the assistance of all the co-authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Alpert MD, et al. ADCC develops over time during persistent infection with live-attenuated SIV and is associated with complete protection against SIV(mac)251 challenge. PLoS Pathog. 2012;8:e1002890. doi: 10.1371/journal.ppat.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks ND, Kinsey N, Clements J, Hildreth JE. Sustained antibody-dependent cell-mediated cytotoxicity (ADCC) in SIV-infected macaques correlates with delayed progression to AIDS. AIDS Res Hum Retroviruses. 2002;18:1197–1205. doi: 10.1089/08892220260387940. [DOI] [PubMed] [Google Scholar]
- 3.Johansson SE, et al. NK cell function and antibodies mediating ADCC in HIV-1-infected viremic and controller patients. Viral immunology. 2011;24:359–368. doi: 10.1089/vim.2011.0025. [DOI] [PubMed] [Google Scholar]
- 4.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2012;481:394–398. doi: 10.1038/nature10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soderquest K, et al. Cutting edge: CD8+ T cell priming in the absence of NK cells leads to enhanced memory responses. J Immunol. 2011;186:3304–3308. doi: 10.4049/jimmunol.1004122. [DOI] [PubMed] [Google Scholar]
- 6.Lang PA, et al. Natural killer cell activation enhances immune pathology and promotes chronic infection by limiting CD8+ T-cell immunity. Proc Natl Acad Sci U S A. 2012;109:1210–1215. doi: 10.1073/pnas.1118834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alter G, et al. Evolution of innate and adaptive effector cell functions during acute HIV-1 infection. J Infect Dis. 2007;195:1452–1460. doi: 10.1086/513878. [DOI] [PubMed] [Google Scholar]
- 8.Bandyopadhyay S, et al. Natural killer cell-mediated lysis of T cell lines chronically infected with HIV-1. Clinical and experimental immunology. 1990;79:430–435. doi: 10.1111/j.1365-2249.1990.tb08107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonaparte MI, Barker E. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood. 2004;104:2087–2094. doi: 10.1182/blood-2004-02-0696. [DOI] [PubMed] [Google Scholar]
- 10.Ward J, et al. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood. 2007;110:1207–1214. doi: 10.1182/blood-2006-06-028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehniger TA, et al. Natural killer cells from HIV-1+ patients produce C-C chemokines and inhibit HIV-1 infection. J Immunol. 1998;161:6433–6438. [PubMed] [Google Scholar]
- 12.Shieh TM, et al. Functional analyses of natural killer cells in macaques infected with neurovirulent simian immunodeficiency virus. J Neurovirol. 2001;7:11–24. doi: 10.1080/135502801300069593. [DOI] [PubMed] [Google Scholar]
- 13.Vowels BR, Gershwin ME, Gardner MB, McGraw TP. Natural killer cell activity of rhesus macaques against retrovirus-pulsed CD4+ target cells. AIDS Res Hum Retroviruses. 1990;6:905–918. doi: 10.1089/aid.1990.6.905. [DOI] [PubMed] [Google Scholar]
- 14.Giavedoni LD, Velasquillo MC, Parodi LM, Hubbard GB, Hodara VL. Cytokine expression, natural killer cell activation, and phenotypic changes in lymphoid cells from rhesus macaques during acute infection with pathogenic simian immunodeficiency virus. J Virol. 2000;74:1648–1657. doi: 10.1128/jvi.74.4.1648-1657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi Y, et al. In vivo administration of a JAK3 inhibitor during acute SIV infection leads to significant increases in viral load during chronic infection. PLoS Pathog. 2014;10:e1003929. doi: 10.1371/journal.ppat.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bostik P, et al. Decreased NK cell frequency and function is associated with increased risk of KIR3DL allele polymorphism in simian immunodeficiency virus-infected rhesus macaques with high viral loads. J Immunol. 2009;182:3638–3649. doi: 10.4049/jimmunol.0803580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouzaire P, et al. Natural killer cells and T cells induce different types of skin reactions during recall responses to haptens. Eur J Immunol. 2012;42:80–88. doi: 10.1002/eji.201141820. [DOI] [PubMed] [Google Scholar]
- 18.Majewska-Szczepanik M, Paust S, von Andrian UH, Askenase PW, Szczepanik M. Natural killer cell-mediated contact sensitivity develops rapidly and depends on interferon-alpha, interferon-gamma and interleukin-12. Immunology. 2013;140:98–110. doi: 10.1111/imm.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paust S, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paust S, von Andrian UH. Natural killer cell memory. Nat Immunol. 2011;12:500–508. doi: 10.1038/ni.2032. [DOI] [PubMed] [Google Scholar]
- 21.O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 22.Peng H, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillard GO, et al. Thy1+ NK [corrected] cells from vaccinia virus-primed mice confer protection against vaccinia virus challenge in the absence of adaptive lymphocytes. PLoS Pathog. 2011;7:e1002141. doi: 10.1371/journal.ppat.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gazit R, et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- 25.Mandelboim O, et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 26.Smith HR, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang M, et al. CD94 is essential for NK cell-mediated resistance to a lethal viral disease. Immunity. 2011;34:579–589. doi: 10.1016/j.immuni.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravet S, et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109:4296–4305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 30.Scott-Algara D, et al. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J Immunol. 2003;171:5663–5667. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- 31.Tiemessen CT, et al. Cutting Edge: Unusual NK cell responses to HIV-1 peptides are associated with protection against maternal-infant transmission of HIV-1. J Immunol. 2009;182:5914–5918. doi: 10.4049/jimmunol.0900419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alter G, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Verges S, et al. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A. 2011;108:14725–14732. doi: 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenhardt M, et al. CD27(+)CD56Bright natural killer cells may be involved in spontaneous clearance of acute hepatitis C in HIV-positive patients. AIDS. 2014 doi: 10.1097/QAD.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 35.Reeves RK, et al. CD16- natural killer cells: enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood. 2010;115:4439–4446. doi: 10.1182/blood-2010-01-265595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang L, et al. NK cell responses to simian immunodeficiency virus vaginal exposure in naive and vaccinated rhesus macaques. J Immunol. 2014;193:277–284. doi: 10.4049/jimmunol.1400417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 39.Barouch DH, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flores-Villanueva PO, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci U S A. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alter G, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang T, Scott JM, Hwang I, Kim S. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J Immunol. 2013;190:1402–1406. doi: 10.4049/jimmunol.1203034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. 2015;42:431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlums H, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 2015;42:443–456. doi: 10.1016/j.immuni.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas R, et al. NKG2C deletion is a risk factor of HIV infection. AIDS Res Hum Retroviruses. 2012;28:844–851. doi: 10.1089/aid.2011.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marras F, et al. Natural killer cells in HIV controller patients express an activated effector phenotype and do not up-regulate NKp44 on IL-2 stimulation. Proc Natl Acad Sci U S A. 2013;110:11970–11975. doi: 10.1073/pnas.1302090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Connell KA, Han Y, Williams TM, Siliciano RF, Blankson JN. Role of natural killer cells in a cohort of elite suppressors: low frequency of the protective KIR3DS1 allele and limited inhibition of human immunodeficiency virus type 1 replication in vitro. J Virol. 2009;83:5028–5034. doi: 10.1128/JVI.02551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi EI, Reimann KA, Letvin NL. In vivo natural killer cell depletion during primary simian immunodeficiency virus infection in rhesus monkeys. Journal of Virology. 2008;82:6758–6761. doi: 10.1128/JVI.02277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreland AJ, et al. Characterization of killer immunoglobulin-like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genomics. 2011;12:295. doi: 10.1186/1471-2164-12-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.