Abstract
Noisy, high-dimensional time series observations can often be described by a set of low-dimensional latent variables. Commonly-used methods to extract these latent variables typically assume instantaneous relationships between the latent and observed variables. In many physical systems, changes in the latent variables manifest as changes in the observed variables after time delays. Techniques that do not account for these delays can recover a larger number of latent variables than are present in the system, thereby making the latent representation more difficult to interpret. In this work, we introduce a novel probabilistic technique, time-delay Gaussian-process factor analysis (TD-GPFA), that performs dimensionality reduction in the presence of a different time delay between each pair of latent and observed variables. We demonstrate how using a Gaussian process to model the evolution of each latent variable allows us to tractably learn these delays over a continuous domain. Additionally, we show how TD-GPFA combines temporal smoothing and dimensionality reduction into a common probabilistic framework. We present an Expectation/Conditional Maximization Either (ECME) algorithm to learn the model parameters. Our simulations demonstrate that when time delays are present, TD-GPFA is able to correctly identify these delays and recover the latent space. We then applied TD-GPFA to the activity of tens of neurons recorded simultaneously in the macaque motor cortex during a reaching task. TD-GPFA is able to better describe the neural activity using a more parsimonious latent space than GPFA, which is a method that has been used to interpret motor cortex data, but does not account for time delays. More broadly, TD-GPFA can help to unravel the mechanisms underlying high-dimensional time series data by taking into account physical delays in the system.
Keywords: dimensionality reduction, time series analysis, latent variable models, time delay estimation, neuroscience
1 Introduction
High-dimensional data are commonly seen in domains ranging from medicine to finance. In many cases these high-dimensional data can be explained by a smaller number of explanatory variables. Dimensionality reduction techniques are a family of techniques that seek to recover these explanatory variables. Such techniques have been successfully applied to a wide variety of problems such as analyzing neural activity (Cunningham and Yu, 2014), modeling video textures (Siddiqi et al., 2007), economic forecasting (Aoki and Havenner, 1997), analyzing human gait (Omlor and Giese, 2011), seismic series analysis (Posadas et al., 1993), as well as face recognition (Turk and Pentland, 1991). For time series data, dimensionality reduction techniques can capture the temporal evolution of the observed variables by a low-dimensional unobserved (or latent) driving process that provides a succinct explanation for the observed data.
The simplest class of dimensionality reduction techniques applied to time series data considers linear, instantaneous relationships between the latent variables and the high-dimensional observations. According to these techniques the ith observed variable yi(t), (i = 1 … q) is a noisy linear combination of the p low-dimensional latent variables xj(t), (j = 1 … p, p < q).
| (1) |
where ci,j ∈ ℝ, di ∈ ℝ and εi(t) ∈ ℝ is additive noise. Many techniques based on latent dynamical systems (Roweis and Ghahramani, 1999) as well as Gaussian-process factor analysis (GPFA) (Yu et al., 2009) fall into this category. This implicit assumption of instantaneous relationships between the latent and observed variables is also made while applying static dimensionality reduction techniques to time series data. Examples of such techniques include principal component analysis (PCA) (Pearson, 1901), probabilistic principal component analysis (PPCA) (Roweis and Ghahramani, 1999; Tipping and Bishop, 1999) and factor analysis (FA) (Everitt, 1984).
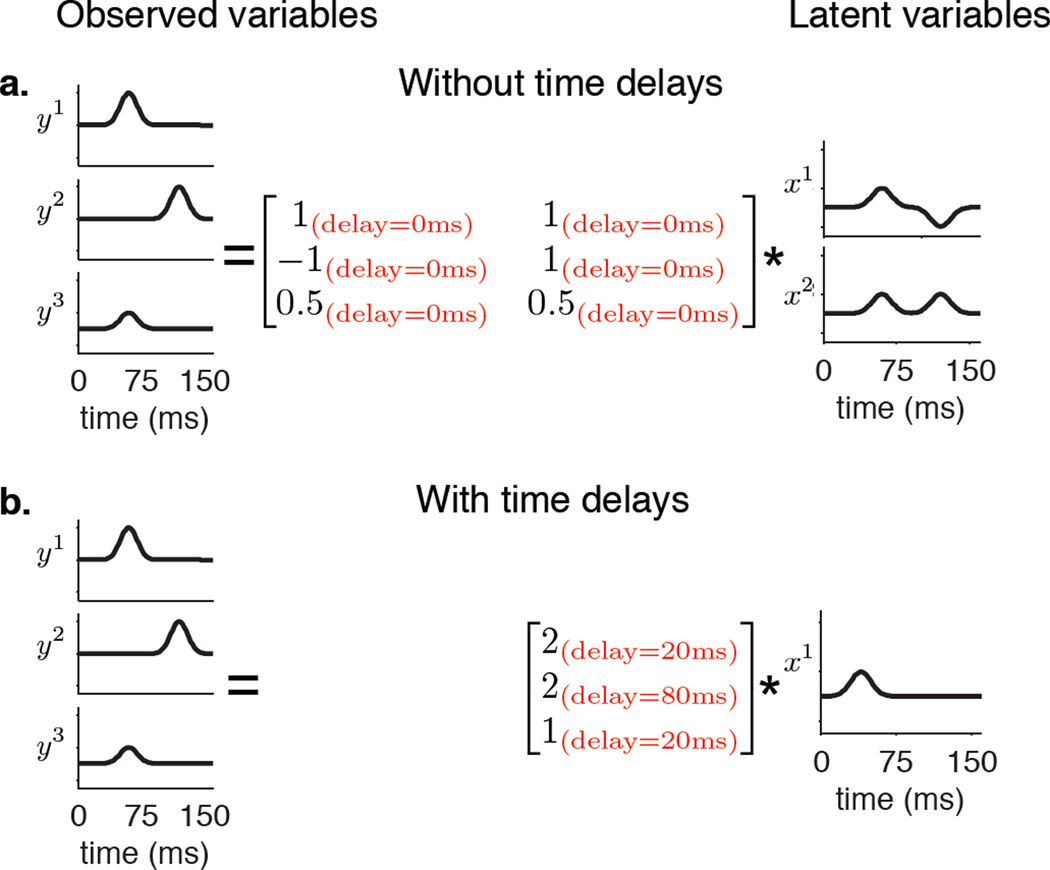
Many real world problems involve physical time delays whereby a change in a latent variable may not manifest as an instantaneous change in an observed variable. These time delays may be different for each latent-observed variable pair. For example, such time delays appear when a few sound sources are picked up by microphones placed at various locations in a room. Each sound source has a different delay to each microphone that depends on the distance between them (Morup et al., 2007; Omlor and Giese, 2011). Other examples where such time delays appear include fMRI imaging, where these delays could arise from hemodynamic delays (Morup et al., 2008) and human motion analysis, where the same control signal may drive muscles after different delays (Barliya et al., 2009). Dimensionality reduction algorithms that only consider instantaneous latent-observed variable relationships can explain these delays only by the introduction of additional latent variables. To see this, consider a conceptual illustration (Figure 1) that shows 3 observed variables, each of which exhibits a single hill of activity but at different times. A model that ignores delays presents a more complicated picture of the underlying latent space, needing 2 latent variables to explain the observations (Figure 1a). In contrast, a model that accounts for these time delays only needs a single latent variable to describe the observations (Figure 1b). Because the goal of dimensionality reduction is to obtain the most parsimonious explanation of the observations, a latent space that is lower dimensional is more desirable.
Figure 1.
Conceptual illustration showing how dimensionality reduction algorithms that consider time delays can present a more compact view of the latent space. a. Three observed variables y1, y2 and y3 that each show a single hill of activity, but at different times. If only instantaneous latent-observation relationships are considered, 2 latent variables are needed to explain these observed variables. b. If non-zero time delays between latent and observed variables are considered, only one latent variable with a single hill of activity is needed to explain the same observed variables. The matrices show the delays as well as the weights that linearly combine the latent variables to reconstruct the observed variables.
We can extend the observation model in Equation (1) to explicitly account for these delays:
| (2) |
where ci,j, di, εi(t) ∈ ℝ. In Equation (2) each observed variable is a linear combination of latent variables, with ci,j as the weights, such that there is a constant time delay Di,j between observed variable yi(t) and latent variable xj(t). εi(t) is the observation noise. Equation (2) is known in the signal processing literature as the anechoic mixing model (Omlor and Giese, 2011). In this work we present time-delay Gaussian-process factor analysis (TD-GPFA), a novel technique that uses the observation model defined in Equation (2) to capture time delays between latent and observed variables, while also accounting for temporal dynamics in the latent space.
While the TD-GPFA framework is applicable in any domain with noisy, high dimensional time-series data where time delays may play a role, in this work we focus on neuroscience as the area of application. The field of neuroscience is witnessing a rapid development of recording technologies that promise to increase the number of simultaneously recorded neurons by orders of magnitude (Ahrens et al., 2013). Dimensionality reduction methods provide an attractive way to study the neural mechanisms underlying various brain functions (Cunningham and Yu, 2014). In this context, the latent process can be thought of as a common input (Kulkarni and Paninski, 2007; Vidne et al., 2010) that drives the system, while the neurons are noisy sensors that reflect the time evolution of this underlying process. However, this common input from a driving area in the brain may follow different pathways to reach each neuron. Physical conduction delays as well as different information processing times along each pathway may introduce different time delays from the driving area to each neuron. Previous work in neuroscience has focused on dimensionality reduction techniques that do not consider time delays between latent variables and neurons. In this work we investigate the effects of incorporating time delays into dimensionality reduction for neural data.
The rest of this paper is structured as follows. In section 2 we briefly discuss related work and provide motivation for the TD-GPFA model. In section 3 we provide a formal mathematical description and describe how the model parameters can be learnt using the Expectation/Conditional Maximization Either (ECME) algorithm (Liu and Rubin, 1994). In section 4 we demonstrate the benefits of TD-GPFA on simulated data and show that TD-GPFA is an attractive tool for the analysis of neural spike train recordings. Finally in section 5 we discuss our findings and mention possible avenues for future work.
2 Background and Related Work
Factor Analysis (FA) (Everitt, 1984) is a commonly used dimensionality reduction technique that has been applied across many domains ranging from psychology (Spearman, 1904) to neuroscience (Sadtler et al., 2014; Santhanam et al., 2009). FA is a linear dimensionality reduction method that incorporates the concept of observation noise, whose variance can be different for each observed variable. An extension of FA, Shifted Factor Analysis (SFA) (Harshman et al., 2003a), allows integer delays between latent and observed variables but requires an exhaustive integer search to identify the values of these delays (Harshman et al., 2003b). One way to avoid this combinatorial search over delays is to work in the frequency domain, since a shift by τ in time domain can be approximated by multiplication by e−ιωτ in the frequency domain, where and ω is the frequency. This property has been exploited to tackle the so called “anechoic blind source separation” problem where the aim is to separate mixtures of sounds which are received at microphones that have a different delay to each source, assuming no echoes (“anechoic”). There has been a sizable body of work in the underdetermined case, where the number of sources is greater than or equal to the number of observed mixtures (Torkkola, 1996; Yeredor, 2003; Be’ery and Yeredor, 2008). The overdetermined case, where the number of sources is strictly less than the number of observed mixtures, is similar in flavor to dimensionality reduction and has received less attention. One approach uses an iterative procedure to perform joint diagonalization of the observations’ cross spectra (Yeredor, 2001). Approaches based on alternating least squares that minimize errors in time and frequency domain have been suggested (Morup et al., 2007, 2008). A framework derived from stochastic time-frequency analysis uses properties of the Wigner-Ville spectrum to tackle the blind source separation problem (Omlor and Giese, 2011).
The aforementioned techniques do not explicitly consider temporal dynamics in the latent space. However, in many situations with time series data we might have some a priori information about the latent process that is driving the observations. For example, while analyzing neural spike train recordings we typically seek to extract latent variables that evolve smoothly in time (Smith and Brown, 2003; Kulkarni and Paninski, 2007; Yu et al., 2009). We could model the latent variables using a linear dynamical system but the naive approach runs into the same problem as SFA, where the need for a combinatorial search over integer delays makes the problem intractable in the time domain.
Our novel technique, time-delay Gaussian-process factor analysis (TD-GPFA), uses an independent Gaussian process to model the temporal dynamics of each latent variable along with a linear-Gaussian observation model. As shown in subsequent sections, this allows us to tractably learn delays over a continuous space while also combining smoothing and dimensionality reduction into a unified probabilistic framework. Our approach can be viewed as an extension of Gaussian-process factor analysis (GPFA) (Yu et al., 2009) that introduces time delays between latent and observed variables.
3 Methods
3.1 TD-GPFA model
Given q observed variables y(t) ∈ ℝq×1 recorded at times t = 1, 2, … T, we seek to identify p latent variables x(t) ∈ ℝp×1 that provide a succinct explanation for these observations, where p < q.
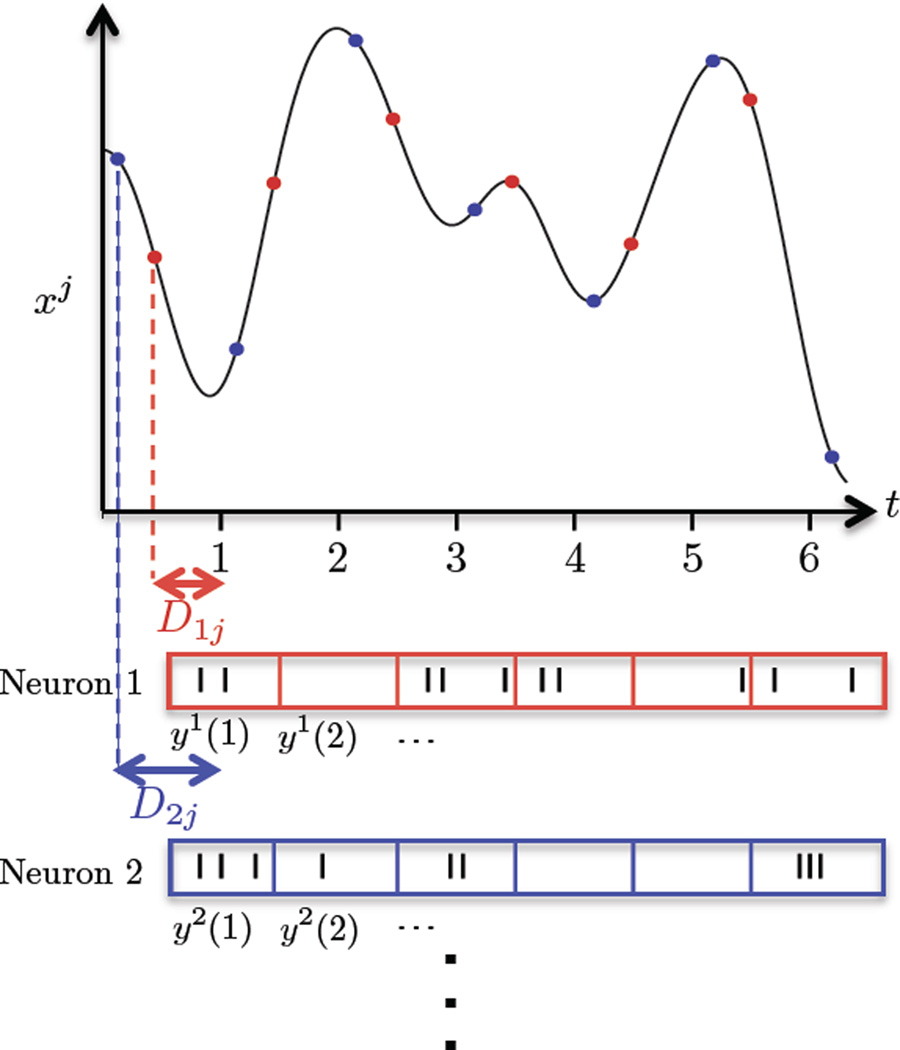
The TD-GPFA model can be summarized as follows. Each observed variable yi (i = 1 … q) is a linear combination of p latent variables xj (j = 1 … p). The key innovation is the introduction of a time delay Di,j between the ith observed variable and the jth latent variable such that yi reflects the value of xj on a grid of T time points. Because each observed variable yi can have a different time delay Di,j with respect to a latent variable xj, these grids are shifted in time with respect to each other. Therefore, for q observed variables, the value of each latent variable must be estimated along q such grids (Figure 2). We use an independent Gaussian process for each latent variable to weave these qT points along a smooth function which represents the temporal evolution of the latent variable.
Figure 2.
Illustration of the TD-GPFA observation model. y1 and y2 are two time series observed at times t = 1 … 6. The first observed variable y1 reflects the values of the jth latent variable xj at points on a grid (red dots) shifted by D1j. The second observed variable y2 reflects the values of xj at points on a grid (blue dots) shifted by D2j, and so on. For example, y1 and y2 could be binned spike counts of neurons, with the bins centered at times t = 1 … 6.
We specify a fixed time delay Di,j between the ith observed variable yi and the jth latent variable xj, such that yi(t) reflects the values of xj(t − Di,j). We then relate the latent and observed variables by the observation model defined in Equation (2), and specify the observation noise to be Gaussian.
| (3) |
where εi(t) ∈ ℝ.
For convenience of notation, let denote yi(t), and let denote xj(t − Di,j). In words, is the value of the jth latent variable that drives the ith observed variable at time t. We form the vector by stacking the values of the p latent variables that drive the ith observed variable at time t. Then, collecting ci,j from Equation (2) into a vector ci ∈ ℝp, we have
| (4) |
We embed the vectors ci in a matrix of zeros and collect the noise terms into a vector εt. This allows us to write the observation model as a matrix product:
| (5) |
| (6) |
We can then stack these vectors for each observed variable i = 1 … q to form a vector Equations (5) and (6) can then be compactly written as
| (7) |
Here C ∈ ℝq×pq is a structured, sparse matrix that maps the latent variables onto the observed variables. The vector d ∈ ℝq is an offset term that captures the non-zero mean of the observed variables, where the ith element is di. As in FA we assume the noise covariance matrix R ∈ ℝq×q is diagonal, where the ith diagonal element is Ri,i as defined in Equation (2). We collect all the delays Di,j into a matrix D ∈ ℝq×p. This completes our specification of the TD-GPFA observation model parameters C, d, R and D.
We now desire a way to describe latent variables that evolve continuously in time, so that we can specify the elements of for any real valued setting of the delays in D. In addition, we seek a latent variable model that can capture the underlying temporal dynamics of the time series. To this end, we model the temporal dynamics of the jth latent variable, (j = 1, 2, … p) by a separate Gaussian process (GP) (Rasmussen and Williams, 2006).
| (8) |
In Equation (8), is a vector formed by stacking the vectors for all times t = 1 … T. Kj ∈ ℝqT×qT is the covariance matrix for this GP. Choosing different forms for the covariance matrix provides different smoothing properties of the latent timecourses. In this work, we construct the elements of Kj using the commonly-used squared exponential (SE) covariance function (Rasmussen and Williams, 2006). Given two points along the jth latent variable, and ,
| (9) |
| (10) |
The characteristic timescale τj ∈ ℝ+ specifies a degree of smoothness for the jth latent variable, while the delays Di1,j and Di2,j along with t1 and t2 specify the temporal locations of the pair of points. δΔt is the Kroneker delta, which is 1 if Δt = 0 and 0 otherwise. is the GP signal variance, while is the noise variance. The SE is an example of a stationary covariance; other stationary and non-stationary GP covariances (Rasmussen and Williams, 2006) can be seamlessly incorporated into this framework. This completes our specification of the TD-GPFA latent variable model.
3.2 Fitting the TD-GPFA model
Given observations Y and a prescribed number of latent dimensions p, we seek to fit model parameters θ = {C, d, R, D, τ1, …, τp}. We initially sought to learn the model parameters using the Expectation Maximization (EM) algorithm (Dempster et al., 1977). The EM algorithm iteratively seeks model parameters θ that maximize the data likelihood P(Y|θ), by performing alternating Expectation (E) steps and Maximization (M) steps. The E-step updates the latents X keeping the model parameters fixed, by computing P(X|Y) using the most recent parameter values. The M-step updates the model parameters by maximizing f(θ) = E[log P(X, Y|θ)] with respect to the parameters θ, where the expectation in f(θ) is taken with respect to the distribution P(X|Y) found in the E-step. However, the delays in D moved very little in each iteration. This motivated us to consider a variant of the EM algorithm, known as the Expectation/Conditional Maximization Either (ECME) algorithm (Liu and Rubin, 1994). The ECME algorithm uses the same E-step as EM, but replaces the M-step in each EM iteration with S Conditional Maximization (CM) steps. Each CM step maximizes either the expected joint probability f(θ) or directly the data likelihood P(Y|θ), with the constraint that some function of θ, say gs(θ), s = 1, 2 … S remains fixed. Different choices for gs(θ), and different choices of whether to optimize f(θ) or P(Y|θ) in each CM step result in different ECME algorithms. If the gs(θ) span the θ space as described in (Liu and Rubin, 1994), then ECME guarantees monotone convergence of the data likelihood.
We now describe an ECME algorithm with two CM steps, where the first CM step maximizes the expected joint probability f(θ) over all parameters keeping the delays fixed, and the second CM step directly maximizes the data likelihood P(Y|θ) over the delays keeping all the other parameters fixed. This is outlined in Algorithm 1. Here, the vec operator converts a matrix into a column vector, while the diag operator extracts the diagonal elements of a matrix into a column vector. Although it is often possible to maximize P(Y|θ) directly with respect to a subset of parameters while keeping all other parameters fixed (as is done here), this does not imply that it is possible to readily maximize P(Y|θ) with respect to all parameters jointly.
Algorithm 1.
ECME algorithm to learn TD-GPFA model parameters
| Initialize θ0; | |
| n ← 1; | |
| repeat | |
| E-step: Compute P(X|Y) according to θn; | |
| CM-step 1: Define g1(θn) = {vec(Dn)} and maximize | |
| f(θn) = E[log P(X, Y|θn)] wrt θn keeping g1 (θn) fixed; | |
| CM-step 2: Define and | |
| maximize L(θn) = P(Y|θn) wrt θn keeping g2(θn) fixed; | |
| n ← n + 1 | |
| until θn has converged or n = max number of iterations; | |
Because X and Y are jointly Gaussian, the conditional in the E-step can be calculated in closed form. In CM-step 1, we can find analytical solutions for C, d and R, while the timescales for each GP, τ1 … τp, are estimated using an iterative gradient based approach, for which the gradient can be computed analytically. In CM-step 2, the delays in D are updated using an iterative gradient based approach, where the gradient can be computed analytically as well. The mathematical details for the parameter updates are provided in Appendix A. Partitioning the parameters θ in this way satisfies the requirements for valid gs(θ) specified in Equation 3.6 of (Meng and Rubin, 1993), and we are guaranteed monotone convergence to a local optimum of the data likelihood.
Under this formulation, the delays D are unconstrained. However, we may wish to constrain the time delays in D to lie within some desired range. This can be motivated by two reasons: i) in some applications, only delays that lie within a known range may be physically realizable, and we may desire to use this domain knowledge to constrain the delays learned by the model, and ii) if the delays are allowed to be arbitrarily large (e.g. much larger than the observation duration T) the model can use completely non-overlapping segments from each latent variable to drive the observations. Since we want to identify latents that are shared by the observed variables, we need to enforce some overlap. We can constrain the delays to lie within a desired range by re-parameterizing Di,j using a logistic function. If we want −Dmax ≤ Di,j ≤ Dmax, we can define
| (11) |
This re-parameterization converts the constrained optimization for Di,j into an unconstrained optimization over using a change of variable. For the results presented in this paper, we set .
The TD-GPFA model as defined above exhibits a degeneracy in the delays - shifting a latent variable xj in time can be compensated for by appropriately subtracting out the shift from the delays between xj and all observed variables, i.e. from all elements in the jth column of D. We temporally align all the latents to the first observed variable by fixing all elements in the first row of the delay matrix D at 0 during model fitting. The delays in D are therefore specified with respect to the first observed variable.
The scale of the latents as defined by the Kj in Equation (8) is arbitrary, since any scaling of can be compensated by appropriately scaling ci,j such that the scale of remains unchanged. Following (Yu et al., 2009), and by analogy to factor analysis, we fix the scale of the latents by fixing k(0) = 1, where k(Δt) is defined in Equation (9). This can be achieved by setting , where to ensure that is non-negative. We fixed to a small quantity (10−6), which allowed us to extract smooth latent timecourses.
Since the ECME algorithm is iterative and converges to a local optimum, initialization of the model parameters is important. We initialized our model parameters using M-SISA, an extension of Shift Invariant Subspace Analysis (SISA) (Morup et al., 2007) that we developed to handle multiple time-series where each time-series may correspond, for example, to a different experimental trial. We chose to extend SISA as it identifies delays between latent and observed variables, and has low computational demands for fitting model parameters. However, SISA does not take into account temporal dynamics in the latent space, which motivates the use of TD-GPFA. A detailed description of M-SISA is provided in Appendix B. Another reasonable initialization would be to use the parameters fit by GPFA and initialize all delays in D to 0. Although in this work we present results obtained by initializing with M-SISA, we found that in some cases initializing with GPFA can yield higher cross-validated log-likelihood. We describe the computational requirements of the ECME algorithm in Appendix C.
3.3 Estimating latent dimensionality
We have thus far presented an algorithm to extract a given number of latent variables from high dimensional observations. In practice however, we do not know the true dimensionality of the latent space. In order to estimate the unknown latent dimensionality, we compute the cross-validated log likelihood (CV-LL) of the data for a range of candidate latent dimensionalities. Although in principle, we want to select the latent dimensionality at which the CV-LL curve peaks, in practice the CV-LL curve often begins to flatten at a lower latent dimensionality. Even though the corresponding CV-LL is lower, it is often close to the peak and this latent dimensionality can be selected to provide a more parsimonious representation of the data. Hence we also report the latent dimensionality at which each CV-LL curve begins to flatten, which we define as the latent dimensionality at which the curve reaches 90% of its total height, where the height of each curve is the difference between its maximum and minimum values over the range of dimensionalities we tested. We call this point the “elbow”. In all subsequent sections, the CV-LL is computed using 4-fold cross-validation.
3.4 Reach task and neural recordings
All animal handling procedures were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. We trained an adult male monkey on a center-out reaching task. At the beginning of each trial, a start target (circle, radius = 14mm) appeared in the center of the workspace. The monkey needed to acquire the start target with the center of the cursor (circle, radius = 15mm) and hold the cursor within the start target for a duration that is randomly selected uniformly between 250 and 500 ms. Then, one of 8 possible peripheral targets appeared (circle, radius = 14 mm, 125 mm from centre of workspace, separated by 45°). The monkey needed to move the cursor to the peripheral target and hold there for a time randomly selected uniformly between 200 and 300 ms. The monkey received a liquid reward when he successfully held the cursor on the peripheral target. We monitored hand movements using an LED marker (PhaseSpace Inc.) on the hand contralateral to the recording array.
While the monkey was performing this reaching task, we recorded from the shoulder region of the primary motor cortex using a 96-channel electrode array (Blackrock Microsystems) as the monkey sat head-fixed in a primate chair. The neural activity was recorded 10 days after array implantation. At the beginning of each session, we estimated the root-mean-square voltage of the signal on each electrode while the monkeys sat calmly in a darkened room. We then set the spike threshold at 3.0 times the root-mean-square value for each channel and extracted 1.24 ms waveform snippets (31 samples at 25 kHz) that began 0.28 ms (7 samples) before the threshold crossing. We manually sorted the waveforms recorded on each electrode using the waveforms themselves or the low-dimensional components that resulted from applying PCA to the waveforms. In this work, we used waveforms that were from well isolated neurons.
We analyzed the activity of 45 simultaneously-recorded neurons. For each experimental trial, we analyzed neural activity for a duration of 520 ms that started 100 ms before movement onset. We considered the first 700 successful trials of the experiment for which the movement times (i.e., from movement onset to movement end) were longer than 420 ms. For these trials, the mean movement time was 510 ms (standard deviation = 82 ms).
4 Results
We compared the performance of TD-GPFA to that of GPFA on two sets of simulated data, as well as on neural spike train recordings. GPFA was chosen as our benchmark for comparison for two reasons: First, the TD-GPFA and GPFA models are nominally identical, and differ only in that the former accounts for time delays. This comparison therefore highlights the effects of accounting for time delays while analyzing neural data. Second, for the analysis of single-trial neural spike train data recorded from the motor cortex of a monkey during a reaching task, GPFA has been shown to outperform two-stage approaches which involve kernel smoothing spike trains and then applying PCA, PPCA, or FA (Yu et al., 2009). For both simulated and real data, we fit the TD-GPFA model by running our ECME algorithm for 200 iterations. GPFA was fit by running the EM algorithm described in (Yu et al., 2009) for 200 iterations.
4.1 Simulated Data
Simulation 1
Simulation 1 follows directly from our conceptual illustration in Figure 1. We constructed a 1-dimensional latent variable which exhibits a single hill of activity (Figure 3a). The height of this hill of activity was different for each of 120 simulated trials, but its location in time was the same. We then generated 10 observed variables sampled at 20 ms intervals for a total duration of 600 ms for each trial. These observed variables were generated according to Equation (7) such that each observed variable is a noisy, scaled and temporally shifted version of the latent variable. The 10 delays in D were obtained by randomly drawing integers from [−5, 5] and multiplying by 20 ms, the scale factors in C were all identically 5, and the offsets in d were all identically 2. All the diagonal elements of the observation noise R were set to 0.5. We then evaluated the performance of TD-GPFA and GPFA on this dataset.
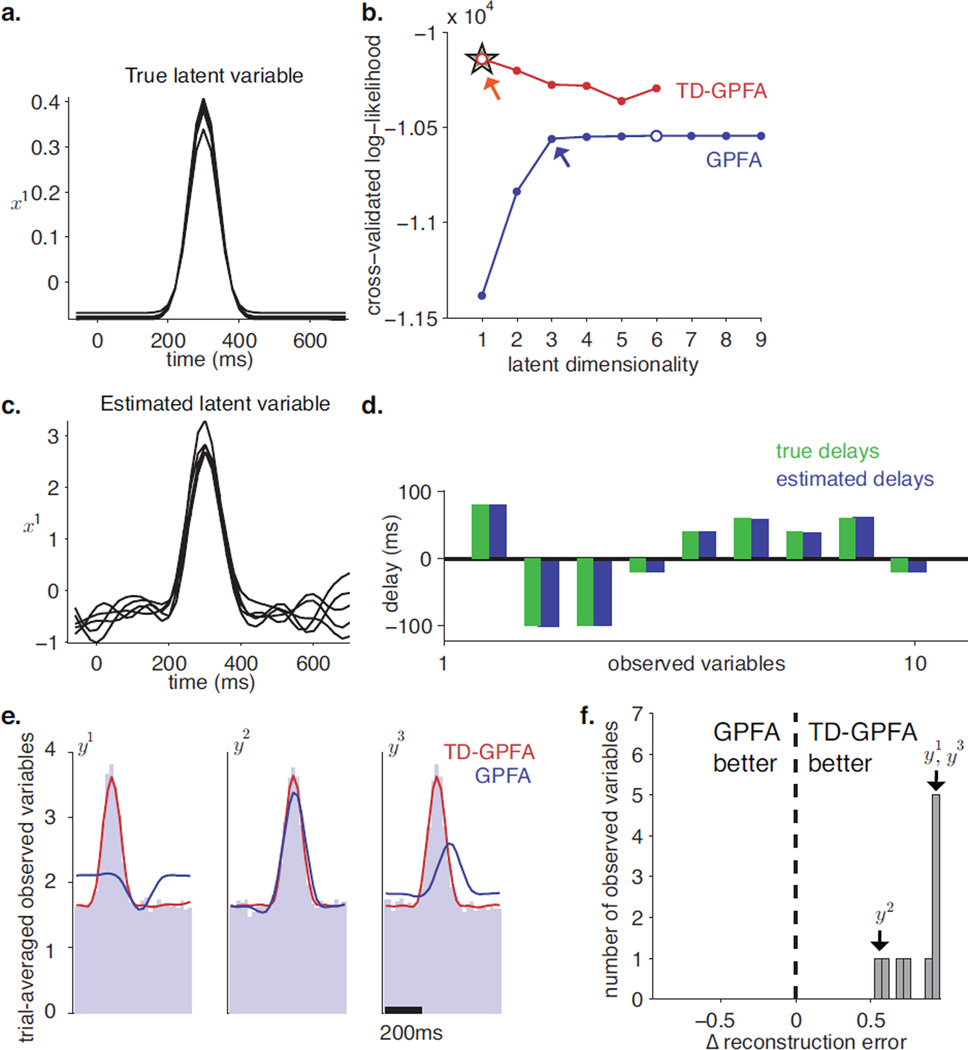
Figure 3.
Simulation 1. a. Simulated latent variable (5 trials shown). b. Cross-validated log-likelihood (CV-LL) plots versus latent dimensionality for TD-GPFA (red) and GPFA (blue). For each curve, the open circle indicates the peak while the arrow indicates the elbow. The green star indicates the CV-LL obtained by initializing ECME for TD-GPFA with ground truth parameters. c. Latent variable estimated by TD-GPFA for latent dimensionality 1 (same 5 trials as shown in panel a.). d. Ground truth (green) and estimated (blue) delays from the latent variable to all observed variables. e. Comparison of trial-averaged observed time courses for 3 representative observed variables (light blue) with those predicted by TD-GPFA (red) and GPFA (blue), with a single latent variable. f. Histogram of the normalized difference between GPFA and TD-GPFA reconstruction errors, computed separately for each observed variable. Observed variables to the right of 0 are better reconstructed by TD-GPFA, while those to the left of 0 are better reconstructed by GPFA.
We computed 4-fold cross-validated log-likelihood (CV-LL) for a range of candidate latent dimensionalities for both TD-GPFA and GPFA. For each method, we evaluated performance by comparing the peak values of the CV-LL curves (Figure 3b). We found that the CV-LL curve for TD-GPFA has a higher peak (red unfilled circle) than the corresponding peak for GPFA (blue unfilled circle). Next, we compared the compactness of the latent space required by each method to describe the data. In this simulation, the CV-LL curve for GPFA has an elbow at 3 dimensions (blue arrow) and reaches its maximum value at 6 dimensions (blue unfilled circle). However, when we account for delays, we are able to find a more compact model that describes the data better. This claim is supported by the following two findings: i) the CV-LL curve for TD-GPFA (red) attains its maximum value at one latent dimension (red unfilled circle), which agrees with the true latent dimensionality, and ii) the maximum CV-LL for TD-GPFA is higher than that of GPFA. Next, because ECME only guarantees convergence to a local optimum, we sought to evaluate how good the local optimum found by initializing ECME with M-SISA is. Since our simulated latent variables were not drawn from the TD-GPFA latent variable model (Equation (8)), we did not have ground truth values for the timescales, and we only knew C up to scale. Therefore, we could not directly evaluate CV-LL with the parameters used to simulate the data. Instead, we performed this comparison by running our ECME algorithm for 100 iterations initializing C, d, D and R at their simulated values, and initializing the timescales τ at 300 ms. We then computed the corresponding CV-LL (green star). We found that this CV-LL is close to the CV-LL attained by our algorithm (red unfilled circle is on top of green star).
The latent variable recovered by TD-GPFA (Figure 3c) captures the single hill of activity seen in the simulated latent variable (Figure 3a), even though it is determined only up to scale. There are 10 delays for the latent variable, one to each observed variable (Figure 3d). The estimated delays are in close agreement with the true delays, with a mean absolute error of 0.67 ms (standard deviation = 0.53 ms).
While the CV-LL curves provide a quantitative comparison of the two algorithms, we also sought to illustrate the benefit of incorporating delays into the dimensionality reduction framework. To this end, we compared the abilities of TD-GPFA and GPFA to capture the timecourse of the observations using the same latent dimensionality. For each method, we used the model parameters estimated from each training fold, and the recovered latent variable, E(X|Y) from the corresponding validation folds. We then used the corresponding observation models (Equation (7) for TD-GPFA, Equation (1) for GPFA) to reconstruct the high-dimensional observed variables. We show these reconstructions averaged across all trials for 3 representative observed variables, y1, y2 and y3, plotted on top of their actual trial-averaged values (Figure 3e). Note that GPFA (blue) misses the hill of activity for y1 and gets the location of the hill wrong for y3, while TD-GPFA (red) is in close agreement with the actual trial-average. Next, we asked if the reconstruction provided by TD-GPFA agrees better with the observed timecourses across all the observed variables. For each of the 4 cross-validation folds, we calculated the reconstruction error for each observed variable by computing the sum of squared errors between the observed and reconstructed values at each time point, and then averaging across trials. We then averaged this reconstruction error across the 4 folds. To interpret the difference in reconstruction errors between TD-GPFA and GPFA, we computed a normalized difference in reconstruction error for each observed variable by subtracting the reconstruction error of TD-GPFA from the reconstruction error of GPFA, and then dividing by the reconstruction error of GPFA (Figure 3e). A normalized difference in reconstruction error equal to 0 implies that TD-GPFA provides no improvement over GPFA, while a value of 1 implies that TD-GPFA provides perfect reconstruction. A negative value indicates that GPFA is better than TD-GPFA at reconstructing the observed variable. For this simulation, the normalized difference in reconstruction error is positive for all ten observed variables (0.82 ± 0.16, mean ± standard deviation), which means that TD-GPFA is better able to capture the timecourse for every observed variable.
Simulation 2
Our second simulation takes us a step closer to the neuroscience application, where we posit that neural population activity can be explained by latent variables that drive the neurons after different delays. To simulate a dataset with known delays, we first applied GPFA to extract latents from real neural data, then artificially introduced delays and constructed observed variables from these latents according to the TD-GPFA observation model (Equation (7)). We considered 140 experimental trials of neural activity of 45 neurons, where the monkey made movements towards one reach target. We applied GPFA to this data and extracted 2 latent dimensions (Figure 4a). Next, we generated TD-GPFA observation model parameters to simulate 45 observed variables. The delays D were integers randomly chosen from [−5, 5] and multiplied by 20 ms. The scale factors in C were integers randomly chosen from [1, 5]. All elements of d were set to 2, and the observation noise matrix R was the identity matrix. Using these parameters and the TD-GPFA observation model (Equation (7)), we then constructed 140 trials from the latent variables extracted from GPFA. Each trial was 520 ms long, discretized into 26 time steps at intervals of 20 ms.
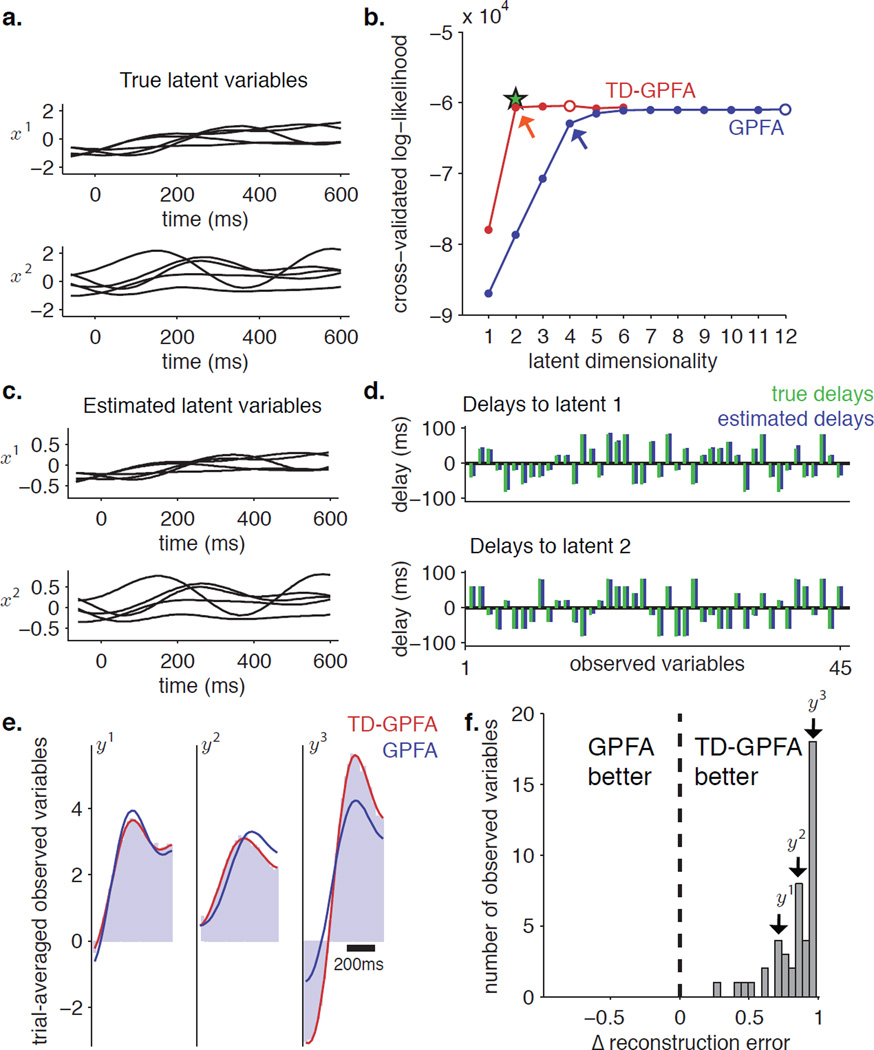
Figure 4.
Simulation 2. a. Simulated latent variables (5 trials shown). b. Cross-validated log-likelihood (CV-LL) plots versus latent dimensionality for TD-GPFA (red) and GPFA (blue). For each curve, the open circle indicates the peak while the arrow indicates the elbow. The green star indicates CV-LL obtained by initializing ECME for TD-GPFA with ground truth parameters. c. Latent variables estimated by TD-GPFA for latent dimensionality 2 (same 5 trials as shown in panel a.). d. Ground truth (green) and estimated (blue) delays from each latent variable to all observed variables. e. Comparison of trial-averaged observed time courses for 3 representative observed variables (light blue) with those predicted by TD-GPFA (red) and GPFA (blue), with a latent dimensionality of 2. f. Histogram of the normalized difference between GPFA and TD-GPFA reconstruction errors, computed separately for each observed variable. Observed variables to the right of 0 are better reconstructed by TD-GPFA, while those to the left of 0 are better reconstructed by GPFA.
We found that by accounting for delays, TD-GPFA describes the data using a smaller number of latent variables than GPFA (Figure 4b). The CV-LL curve for GPFA (blue) exhibits an elbow at 5 latent dimensions (blue arrow) and reaches its maximum at 12 latent dimensions (blue unfilled circle). In contrast, the CV-LL curve for TD-GPFA (red) has an elbow at 2 latent dimensions (red arrow) where 2 is the true latent dimensionality, although the peak is attained at 4 latent dimensions (red unfilled circle). As in Simulation 1, the maximum CV-LL attained by TD-GPFA is higher than that of GPFA. Next, we ran ECME for 100 iterations initializing C, d, D and R at their ground truth values and the timescales τ at the values estimated by GPFA for the simulated latents. We found that the CV-LL attained by TD-GPFA for 2 latent variables is close to the CV-LL obtained by this initialization (green star). The latent variables recovered by TD-GPFA for latent dimensionality 2 show timecourses that closely resemble those of the simulated latents (Figure 4c), even though the scale of the latent variables is different. Each latent variable has 45 delays, one to each observed variable (Figure 4d). The estimated delays are again very close to the true delays, with a mean absolute error of 1.84 ms (standard deviation = 1.79 ms).
As in Simulation 1, we show trial-averaged reconstructions achieved by each method using 2 latent variables for 3 representative observed variables y1, y2 and y3 (Figure 4e). Once again we see that TD-GPFA is able to capture the empirical trial average well. GPFA however misses out on key features of the activity timecourse. We then asked if this is true across all observed variables by computing the normalized difference in reconstruction error between TD-GPFA and GPFA (Figure 4f). We found that this difference is positive across all observed variables (0.84 ± 0.17, mean ± standard deviation), indicating that TD-GPFA reconstructed all 45 observed timecourses better than GPFA.
4.2 Neural Spike Train Data
We applied TD-GPFA and GPFA to neural spike train recordings obtained as described in Section 3.4. Our observations were spike counts taken in non overlapping 20 ms bins and then square-root transformed to stabilize the variance of the spike counts (Yu et al., 2009).
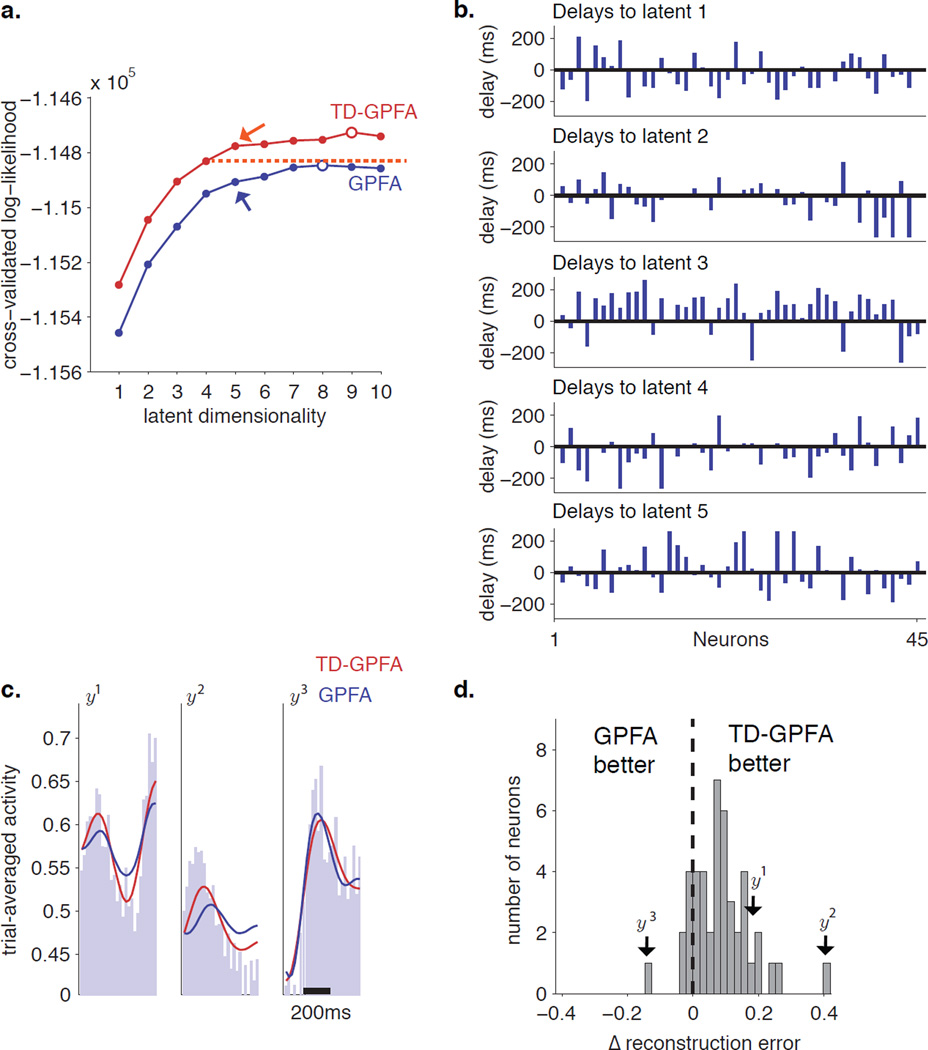
We compared the CV-LL versus latent dimensionality curve of TD-GPFA with that of GPFA (Figure 5a). The CV-LL curve of TD-GPFA is consistently higher than that of GPFA. Furthermore, with as few as 4 latent dimensions (red dotted line), TD-GPFA finds a better description of the data than GPFA with any latent dimensionality up to 10. This is the main result of this work, that TD-GPFA finds a model that is more compact yet can describe the neural recordings better than GPFA.
Figure 5.
TD-GPFA applied to neural activity recorded in the motor cortex. a. Cross-validated log-likelihood (CV-LL) plots versus latent dimensionality for TD-GPFA (red) and GPFA (blue). For each curve, the open circle indicates the peak while the arrow indicates the elbow. The red dotted line is the CV-LL of the TD-GPFA model with 4 latent dimensions. b. Estimated delays from each latent variable to all neurons. c. Comparison of empirical firing rate histograms (light blue) to firing rate histograms predicted by TD-GPFA (red) and GPFA (blue) for 3 representative neurons. The vertical axis has units of square-rooted spike counts taken in a 20 ms bin. Five latent variables were used for both TD-GPFA and GPFA. d. Histogram of the normalized difference between GPFA and TD-GPFA reconstruction errors, computed separately for each neuron. Neurons to the right of 0 are better reconstructed by TD-GPFA, while those to the left of 0 are better reconstructed by GPFA.
We present a closer look at the model parameters returned by TD-GPFA with 5 latent dimensions, which is the latent dimensionality at the elbow of the CV-LL curve. We found that most of the recovered delays are non-zero (Figure 5b). This indicates that TD-GPFA makes use of time delays to obtain a more compact, yet better description of the neural activity. As in our simulations, we sought to illustrate the benefit of incorporating delays in capturing the neural activity. We show trial-averaged reconstructions using 5 latent variables for 3 representative neurons (y1, y2 and y3) plotted on top of the empirical trial average (Figure 5c). TD-GPFA (red) is better able to capture the firing rate profiles of neurons y1 and y2 as compared to GPFA (blue), which tends to underestimate the hills of activity. We also show a neuron (y3) for which GPFA is better than TD-GPFA at capturing the trial-averaged activity. Finally, to compare TD-GPFA and GPFA for all neurons, we computed the normalized difference in reconstruction error (Figure 5d). We found that this difference is positive for 38 out of 45 neurons (0.09 ± 0.09, mean ± standard deviation). Thus, TD-GPFA provided overall better reconstructions than GPFA (Paired t-test, P < 0.001). Additionally, we verified that TD-GPFA with 5 latent dimensions is able to provide better reconstructions than GPFA with 6–10 latent dimensions, consistent with our findings from the CV-LL curves (Figure 5a).
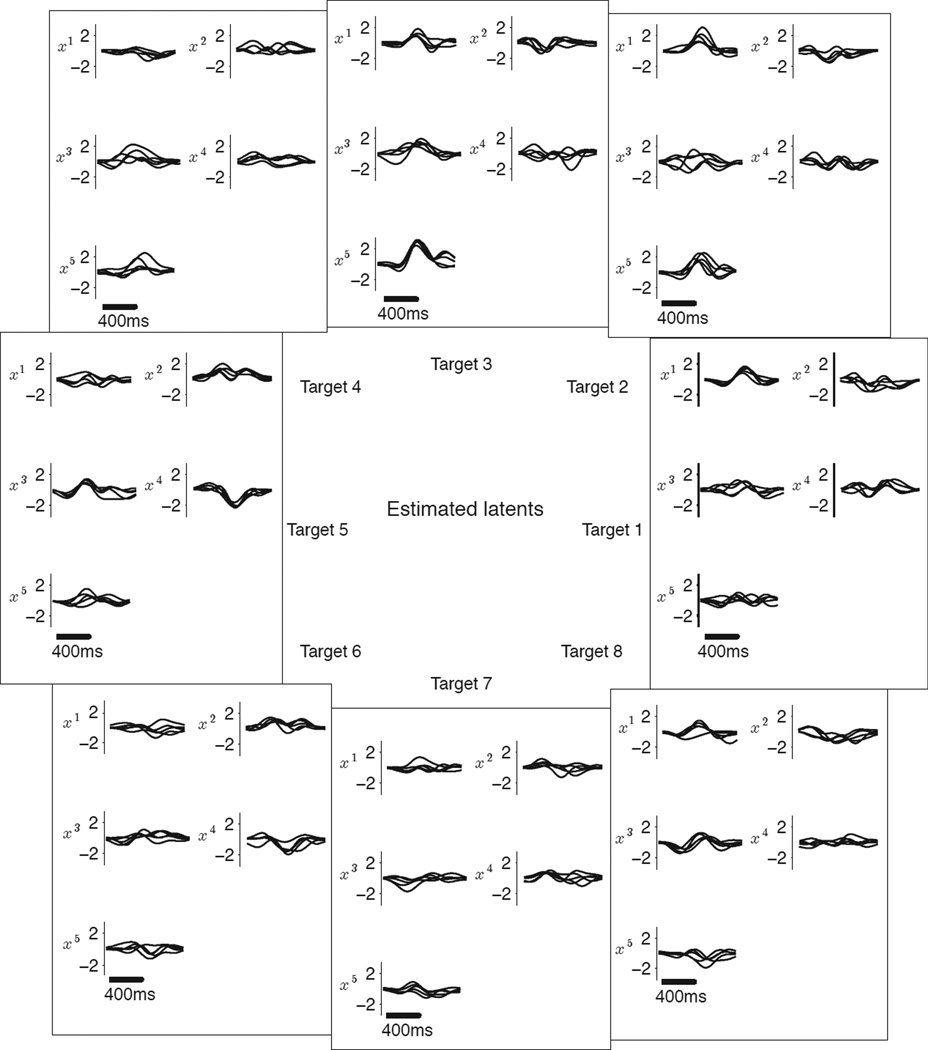
The timecourses of the estimated latent variables (Figure 6) can be interpreted as timecourses of latent drivers that influence neurons at different time delays. Although we leave a detailed analysis of the estimated latent variables for future scientific studies, we can verify here that the extracted timecourses of the latent variables vary systematically with reach target, as would be expected from neurons with smooth tuning curves. For example, the first latent variable (x1) shows a pronounced peak for rightward reaches, and this peak becomes less pronounced for directions that are further from the right. Similarly, the fifth latent variable (x5) shows a pronounced peak for upward reaches. Note that while the shape of the latent timecourses are meaningful, the absolute position in time for each latent variable is arbitrary. Intuitively, if we have a number of observed variables that reflect a latent driver after different delays, we can only determine the delays of the observed variables with respect to each other, but not the absolute delay between the latent driver and the observed variables. Thus, a latent variable (i.e., all the timecourses for the latent variable) can be shifted arbitrarily forward or backward in time by correspondingly adding or subtracting the shift to all the delay values of that latent variable in D.
Figure 6.
Latent variable timecourses extracted by TD-GPFA from neural activity recorded in the motor cortex. Results shown for latent dimensionality 5 and plotted separately for each latent dimension (x1, x2, x3, x4 and x5) and each reach target (5 trials shown per target).
5 Discussion
In this work we have presented a novel method (TD-GPFA) that can extract low-dimensional latent structure from noisy, high-dimensional time series data in the presence of a fixed time delay between each latent and observation variable. TD-GPFA unifies temporal smoothing and dimensionality reduction in a common probabilistic framework, using Gaussian processes to capture temporal dynamics in the latent space. We then presented an ECME algorithm that learns TD-GPFA model parameters from data. Next, we verified using simulated data that TD-GPFA is able to recover the time delays and the correct dimensionality of the latent space, while GPFA, which is nominally identical but does not consider delays needs a latent space of higher dimensionality to explain the same observations. Finally, we applied TD-GPFA to neural activity recorded in the motor cortex during a center-out reaching task. We found that i) the 45-dimensional neural activity could be succinctly captured by TD-GPFA using 5 latent dimensions; ii) TD-GPFA finds a more compact description of the latent space that better describes the neural activity, as compared to GPFA.
Modeling temporal dynamics of the latent variables using Gaussian processes (GPs) offers us the following advantages: i) Using GPs allows us to tractably learn time delays over a continuous domain. In our formulation, the delays appear only in the GP covariance kernel. During parameter learning using the ECME algorithm, these delays can be optimized using any gradient descent method; ii) GPs offer an analytically tractable yet expressive way to model time-series in cases where the true latent dynamics are unknown, as is often the case in exploratory data analysis. This is because we only need to specify the covariance kernel for each GP. The extracted latent variables can guide further parametric modeling of latent dynamics, such as using a dynamical system (Smith and Brown, 2003; Yu et al., 2006; Kulkarni and Paninski, 2007; Macke et al., 2011; Petreska et al., 2011; Pfau et al., 2013); iii) The GPs smooth the latent dynamics according to the specified covariance kernel, and can be thought of as performing regularization on the latent space, which is helpful in analyzing time series data (Rahimi et al., 2005); and iv) Our approach allows us to combine smoothing of the noisy time series with dimensionality reduction in a unified probabilistic framework. This has been shown to be advantageous in analyzing neural spike train data, as opposed to previous methods that smooth the data as a preprocessing step using a kernel of fixed width (Yu et al., 2009). The key insight is that performing smoothing as a preprocessing step assumes a single timescale of evolution of neural activity, while our approach allows each latent variable to evolve with its own timescale. This benefit may translate well to other domains as well, where underlying latent variables evolve with different timescales.
In neuroscience, dimensionality reduction methods have proven useful in studying various neural systems, including the olfactory system (Mazor and Laurent, 2005; Broome et al., 2006; Saha et al., 2013), the motor system (Briggman et al., 2005; Yu et al., 2009; Churchland et al., 2012; Sadtler et al., 2014), decision-making (Harvey et al., 2012; Mante et al., 2013) and working memory (Machens et al., 2010; Rigotti et al., 2013). However, these and other previous studies do not consider delays between latent variables and neurons. Because information in the brain can flow across multiple pathways to reach the recorded neurons, physical conduction delays as well as differences in the information processing times along each pathway can give rise to delays. For example, a latent process originating in the visual cortex may influence some motor cortical neurons (M1) through parietal cortex, but may also influence some M1 neurons through a longer path involving parietal cortex and dorsal premotor cortex. When we applied TD-GPFA to M1 neurons recorded during a reaching task, TD-GPFA recovered different non-zero delays, which is suggestive of latent drivers influencing neurons at different delays. Future experiments are needed to establish the connection between latent variables and upstream brain areas. Another property of the TD-GPFA framework that makes it suitable to application in neuroscience is its ability to perform single-trial analysis. If neural activity varies substantially on nominally identical trials, common approaches like averaging responses across trials may mask the true underlying processes that generate these responses. TD-GPFA allows us to leverage the statistical power of recording from an entire neural population to extract a succinct summary of the activity on a trial-by-trial basis.
While our metrics show that the description of the neural activity extracted by TD-GPFA is more accurate than one extracted without considering delays, it remains to be seen whether these delays correspond to actual neural processes, and if so, to understand their physical underpinnings. Additionally, TD-GPFA may provide an attractive tool for analyzing data recorded simultaneously from multiple brain areas, where a driving process may manifest in neurons belonging to each area after different delays (Semedo et al., 2014).
Several extensions to the TD-GPFA methodology can be envisaged. It may be possible to i) allow each latent variable to drive an observation at multiple delays, which might be a more realistic assumption for some systems; ii) allow non-linear relationships between latent and observed variables; iii) use non stationary GP covariances. Software for TD-GPFA is available at http://bit.ly/1HAjyBl.
Acknowledgments
This work was funded by NIH NICHD CRCNS R01-HD071686 (B.M.Y. and A.P.B.), the Craig H. Neilsen Foundation (B.M.Y. and A.P.B.), NIH NINDS R01-NS065065 (A.P.B.), Burroughs Wellcome Fund (A.P.B.), NSF DGE-0549352 (P.T.S.) and NIH P30-NS076405 (Systems Neuroscience Institute).
Appendix A: ECME update equations
Given observations y1:T and a prescribed number of latent dimensions p < q, we seek to fit model parameters θ = {C, d, R, D, τ1, …, τp}. We first define ỹ ∈ ℝqT and x̃ ∈ ℝpqT by stacking observed variables yt and latent variables (as defined in Equations (5–7)) across all time points.
| (A.1) |
This allows us to write the latent and observation variable models across all time as follows:
| (A.2) |
| (A.3) |
where C̃ ∈ ℝqT×pqT and R̃ ∈ ℝqT×qT are block diagonal matrices comprising of T blocks of C and R respectively. d̃ ∈ ℝqT is constructed by stacking T copies of d. The elements of K̃ ∈ ℝpqT×pqT can be computed using the covariance kernel defined in Equation (9). From Equations (A.2) and (A.3), we can write the joint distribution of x̃ and ỹ as
| (A.4) |
A.1 E-step
Using the basic result of conditioning for jointly Gaussian random variables,
| (A.5) |
Thus, the extracted latent variables are
| (A.6) |
A.2 CM-step 1
In CM-step 1 we maximize f(θ) = E[log P(x̃, ỹ|θ)] with respect to {C, D, R, γ1 … γp} keeping g1(θ) = {vec(D)} fixed. The expectation in f(θ) is taken with respect to the distribution P(x̃|ỹ) found in Equation (A.5). Even though this is a joint optimization with respect to all the parameters, their optimal values are dependent on only a few or none of the other parameters.
We first define the following notation. Given a vector v,
| (A.7) |
| (A.8) |
where the expectations can be found from Equation (A.5).
C, d update : Maximizing f(θ) with respect to C and d yields joint updates for each row of C and the corresponding element of d, such that for i ∈ 1, 2 … q,
| (A.9) |
R update : Maximizing f(θ) with respect to R yields
| (A.10) |
The updated C, d found from Equation (A.9) should be used in Equation (A.10).
τ1 … τp update: Although there is no analytic form of the update equations for the timescales, we can learn them using any gradient optimization technique.
| (A.11) |
where
| (A.12) |
| (A.13) |
This is a constrained optimization problem because the GP timescales need to be positive. We can turn this into an unconstrained optimization problem by optimizing with respect to log τj using a change of variable.
A.3 CM-step 2
In CM-step 2 we directly optimize the log-likelihood L(θ) = P(ỹ|θ) with respect to each Di,j. P(ỹ|θ) can be computed easily since
| (A.14) |
The delays, like the GP timescales can also be learnt using any gradient optimization technique. If Σ = C̃K̃C̃T + R̃,
| (A.15) |
where
| (A.16) |
| (A.17) |
| (A.18) |
| (A.19) |
where Δt, i1, i2 are defined in Equation (10).
The derivations in this section are presented for a single time-series (corresponding to a single experimental trial) of duration T. In many cases we wish to learn the model given multiple time-series, each of which may have a different T. It is straightforward to extend Equations (A.5–A.15) to account for N time-series by considering instead of ∂f(θ)/∂θ. This assumes that each time-series is independent given the model parameters, meaning that we do not explicitly constrain the latent variables to follow similar time courses on different trials. However, the latent variables are assumed to all lie within the same low-dimensional state space with the same timescales and delays.
B Parameter Initialization for ECME
Shift Invariant Subspace Analysis (SISA) (Morup et al., 2007) is a technique that was developed to address the anechoic blind source separation problem. It uses the fact that a delay Di,j in the time domain can be approximated by multiplication by the complex coefficients e−ιωDi,j in the frequency domain, where and , where k = 1 … T and T is the number of time steps in the time series. While SISA was originally designed to handle a single time series, in some applications observations are collected as multiple time series as opposed to a single long time series, as in the neural application considered in this work. We developed an extension M-SISA that can handle multiple time series.
If Yi is the Fourier transform of yi, then we can rewrite the anechoic mixing model as a matrix product in the frequency domain. We first center our observed variables at mean 0 by subtracting out the mean computed across all time points in all time-series. For convenience of notation, in the rest of this section y will denote the mean 0 observed variables.
Then, for each time series, we have
| (B.20) |
where i ∈ 1 … q, j ∈ 1 … p. Taking the Fourier transform of each side yields
| (B.21) |
We can collect the terms of ci,j into a matrix Č ∈ ℝq×p, where the elements of Č are the same as the non-zero elements of the sparse matrix C ∈ ℝq×pq defined in Equation (7). If , where ◦ denotes Hadamard’s (element-wise) product and the exponential is computed element-wise for the matrix D ∈ ℝq×p, we can write this for each time series indexed by n in matrix notation as
| (B.22) |
where Yk{n},Ek{n} ∈ ℂq×1 and Xk{n} ∈ ℂp×1. We can now alternatingly solve for Č, X and D to minimize the least square error in both time and frequency domain (Morup et al., 2007). Briefly, at each iteration of the M-SISA algorithm, Č and X are computed using a pseudo-inverse, while D is computed using Newton-Raphson iterations. The detailed update equations are presented below.
X update: For each time series indexed by n,
| (B.23) |
For X to be real valued, we only update the first ⌊T/2⌋ + 1 elements according to Equation (B.23) and set the remaining elements such that where * denotes the complex conjugate.
Č update: Let be the delayed version of the latent to the ith observed variable. Then, for each time series indexed by n,
| (B.24) |
| (B.25) |
We form ỹi ∈ ℝ1×NT, X̃i ∈ ℝp×NT and ε̃i ∈ ℝ1×NT by horizontally stacking and as follows:
| (B.26) |
| (B.27) |
| (B.28) |
Then,
| (B.29) |
This allows us to update each row of Č as follows
| (B.30) |
D update: Following (Morup et al., 2007), the least square error for the single time series (Equation (B.22)) is given by
| (B.31) |
Analytical forms for the gradient g{n} and the Hessian H{n} of ls{n} with respect to the delays D can be computed (Morup et al., 2007). Therefore for multiple time series indexed by n,
| (B.32) |
| (B.33) |
| (B.34) |
We performed Newton-Raphson using the gradient g and the Hessian H to update the delays D.
M-SISA itself is prone to local optima, and is initialized using a multistart procedure. Briefly, we ran 15 iterations of M-SISA initialized at 30 randomly drawn settings of the model parameters. Of these 30 draws, we selected the draw with the lowest least square error and ran 100 more iterations.
Once Č and D are computed, we can compute the residuals ε such that
| (B.35) |
We then form R such that the diagonal elements of R are equal to the diagonal terms of the covariance matrix of ε across all time points and time-series. C is formed by rearranging the elements of Č as required in Equation (7). The initial value of the offset d is the mean of all observations, computed across all time steps in all the observed time-series. We set the initial values of all the GP timescales τ j, j = 1 … p to 100 ms. We fix the GP noise variance to a small positive value (= 10−6) as in (Yu et al., 2009).
C Computational Requirements
This section summarizes the computational time required to fit TD-GPFA model parameters and extract the latent variables. The computation time depends on the latent dimensionality p, number of observed variables q, number of trials, length of each trial T and the number of ECME iterations.
In the ECME algorithm, the most expensive equation to compute is the E-step (Equation (A.5)), which involves the inversion of a qT × qT matrix and multiplication with matrices of size pqT × pqT. Large parts of this computation are constant for a fixed p, q and T and therefore they can be performed once for all trials of the same length T, as they do not depend on the values of the observed variables in each trial. Each gradient computation in CM-step 2 (Equation (A.15)) is fast, but because this computation is performed thousands of times it accounts for a large fraction of the total running time.
Our results were obtained on a Linux 64-bit workstation with a 4 core Intel Xeon CPU (X5560), running at 2.80 GHz and equipped with 50 GB RAM. To fit the parameters for 45 observed variables and 140 trials of length 520 ms (i.e. T= 26 with 20 ms time bins), our ECME algorithm (for 200 iterations) took 3 hours and 14 minutes for 2 latent dimensions, and 9 hours and 54 min for 6 latent dimensions. These results were obtained using an exact implementation without harnessing any possible advantages of using approximate techniques. We can also take advantage of parallelization to reduce runtime, especially since during each ECME iteration, the delays and timescales can be learned independently for each latent variable. Lower runtimes may also be obtained by running fewer overall ECME iterations (in practice, the parameters often converge within the first 100 ECME iterations), as well as relaxing convergence criteria for gradient optimization of the delays (Equation (A.15)).
References
- Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nature Methods. 2013;10:413420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- Aoki M, Havenner A. Lecture notes in statistics. Springer; 1997. Applications of computer aided time series modeling. [Google Scholar]
- Barliya A, Omlor L, Giese MA, Flash T. An analytical formulation of the law of intersegmental coordination during human locomotion. Experimental Brain Research. 2009;193(3):371–385. doi: 10.1007/s00221-008-1633-0. [DOI] [PubMed] [Google Scholar]
- Be’ery E, Yeredor A. Blind separation of superimposed shifted images using parameterized joint diagonalization. IEEE Transactions on Image Processing. 2008;17:340–353. doi: 10.1109/TIP.2007.915548. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Abarbanel HDI, Kristan WB., Jr Optical imaging of neuronal populations during decision-making. Science. 2005;307:896–901. doi: 10.1126/science.1103736. [DOI] [PubMed] [Google Scholar]
- Broome BM, Jayaraman V, Laurent G. Encoding and decoding of overlapping odor sequences. Neuron. 2006;51:467–482. doi: 10.1016/j.neuron.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Churchland MM, Cunningham JP, Kaufman MT, Foster JD, Nuyujukian P, Ryu SI, Shenoy KV. Neural population dynamics during reaching. Nature. 2012;487:51–56. doi: 10.1038/nature11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JP, Yu BM. Dimensionality reduction for large-scale neural recordings. Nature Neuroscience. 2014 doi: 10.1038/nn.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the em algorithm. Journal of The Royal Statistical Society, series b. 1977;39(1):1–38. [Google Scholar]
- Everitt BS. An Introduction to Latent Variable Models. Netherlands: Springer; 1984. [Google Scholar]
- Harshman R, Hong S, Lundy M. Shifted factor analysis - part i: Models and properties. Journal of Chemometrics. 2003a;17:363–378. [Google Scholar]
- Harshman R, Hong S, Lundy M. Shifted factor analysis - part ii: Algorithms. Journal of Chemometrics. 2003b;17:379–388. [Google Scholar]
- Harvey CD, Coen P, Tank DW. Choice-specific sequences in parietal cortex during a virtual-navigation decision task. Nature. 2012;484:62–68. doi: 10.1038/nature10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni J, Paninski L. Common-input models for multiple neural spike-train data. Network: Computation in Neural Systems. 2007;18:375–407. doi: 10.1080/09548980701625173. [DOI] [PubMed] [Google Scholar]
- Liu C, Rubin D. The ecme algorithm: A simple extension of em and ecm with faster monotone convergence. Biometrika. 1994;81(4):633–648. [Google Scholar]
- Machens CK, Romo R, Brody CD. Functional, but not anatomical, separation of what and when in prefrontal cortex. J Neurosci. 2010;30:350–360. doi: 10.1523/JNEUROSCI.3276-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke JH, Buesing L, Cunningham JP, Yu BM, Shenoy KV, Sahani M. Advances in Neural Information Processing Systems. Vol. 24. Curran Associates, Inc.; 2011. Empirical models of spiking in neural populations; pp. 1350–1358. [Google Scholar]
- Mante V, Sussillo D, Shenoy KV, Newsome WT. Context-dependent computation by recurrent dynamics in prefrontal cortex. Nature. 2013;503:78–84. doi: 10.1038/nature12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor O, Laurent G. Transient dynamics versus fixed points in odor representations by locust antennal lobe projection neurons. Neuron. 2005;48:661–673. doi: 10.1016/j.neuron.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Meng X-L, Rubin DB. Maximum likelihood estimation via the ecm algorithm: A general framework. Biometrika. 1993;80(2):267–278. [Google Scholar]
- Morup M, Hansen LK, Arnfred SM, Lim HM, Madsen KH. Shift-invariant multilinear decomposition of neuroimaging data. NeuroImage. 2008;42:1439–1450. doi: 10.1016/j.neuroimage.2008.05.062. [DOI] [PubMed] [Google Scholar]
- Morup M, Madsen KH, Hansen LK. Shifted independent component analysis. ICA2007. 2007;42:89–96. [Google Scholar]
- Omlor L, Giese M. Anechoic blind source separation using wigner marginals. Journal of Machine Learning Research. 2011;12:1111–1148. [Google Scholar]
- Pearson K. On lines and planes of closest fit to systems of points in space. Philiosophical Magazine. 1901;2(6):559–572. [Google Scholar]
- Petreska B, Yu BM, Cunningham JP, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Advances in Neural Information Processing Systems. Vol. 24. Curran Associates, Inc.; 2011. Dynamical segmentation of single trials from population neural data; pp. 756–764. [Google Scholar]
- Pfau D, Pnevmatikakis EA, Paninski L. Advances in Neural Information Processing Systems 26. MIT Press; 2013. Robust learning of low-dimensional dynamics from large neuronal ensembles. [Google Scholar]
- Posadas AM, Vidal F, de Miguel F, Alguacil G, Peña J, Ibañez JM, Morales J. Spatial-temporal analysis of a seismic series using the principal components method: The antequera series, spain, 1989. Journal of Geophysical Research: Solid Earth. 1993;98(B2):1923–1932. [Google Scholar]
- Rahimi A, Recht B, Darrell T. IN COMPUTER VISION AND PATTERN RECOGNITION (CVPR. CVPR; 2005. Learning appearance manifolds from video; pp. 868–875. [Google Scholar]
- Rasmussen CE, Williams C. Gaussian processes for machine learning. MIT Press; 2006. [Google Scholar]
- Rigotti M, Barak O, Warden MR, Wang XJ, Daw ND, Miller EK, Fusi S. The importance of mixed selectivity in complex cognitive tasks. Nature. 2013;497:585–590. doi: 10.1038/nature12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roweis S, Ghahramani Z. A unifying review of linear gaussian models. Neural Computation. 1999;11(2):305–345. doi: 10.1162/089976699300016674. [DOI] [PubMed] [Google Scholar]
- Sadtler PT, Quick KM, Golub MD, Chase SM, Ryu SI, Tyler-Kabara EC, Yu BM, Batista AP. Neural constraints on learning. Nature. 2014;512:423–426. doi: 10.1038/nature13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D, Leong K, Li C, Peterson S, Siegel G, Raman B. A spatiotemporal coding mechanism for background-invariant odor recognition. J Neurosci. 2013;16:1830–1839. doi: 10.1038/nn.3570. [DOI] [PubMed] [Google Scholar]
- Santhanam G, Yu BM, Gilja V, Ryu SI, Afshar A, Sahani M, Shenoy KV. Factor-analysis methods for higher-performance neural prostheses. Journal of Neurophysiology. 2009;102(2):1315–1330. doi: 10.1152/jn.00097.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semedo J, Zandvakili A, Kohn A, Machens C, Yu B. Extracting latent structure from multiple interacting neural populations. Advances in Neural Information Processing Systems. 2014;27:2942–2950. [Google Scholar]
- Siddiqi S, Boots B, Gordon G. A constraint generation approach to learning stable linear dynamical systems. Advances in Neural Information Processing Systems. 2007 [Google Scholar]
- Smith A, Brown E. Estimating a state-space model from point process observations. Neural Computation. 2003;15:965–991. doi: 10.1162/089976603765202622. [DOI] [PubMed] [Google Scholar]
- Spearman C. General intelligence objectively determined and measured. American Journal of Psychology. 1904;15:201–293. [Google Scholar]
- Tipping ME, Bishop CM. Mixtures of probabilistic principal component analyzers. Neural Comput. 1999:443–482. doi: 10.1162/089976699300016728. [DOI] [PubMed] [Google Scholar]
- Torkkola K. Time-delay estimation in mixtures. IEEE Workshop on Neural Networks for Signal Processing. 1996;4:423–432. [Google Scholar]
- Turk M, Pentland A. Eigenfaces for recognition. J. Cognitive Neuroscience. 1991:71–86. doi: 10.1162/jocn.1991.3.1.71. [DOI] [PubMed] [Google Scholar]
- Vidne M, Ahmadian Y, Shlens J, Pillow JW, Kulkarni J, Chichilnisky EJ, Simoncelli E, Paninski L. A common-input model of a complete network of ganglion cells in the primate retina. Computational and Systems Neuroscience. 2010 doi: 10.1007/s10827-011-0376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeredor A. Blind source separation with pure delay mixtures. The 3rd International Workshop on Independent Component Analysis and Blind Source Separation (ICA) 2001 [Google Scholar]
- Yeredor A. Time-delay estimation in mixtures. Acoustics, Speech, and Signal Processing. 2003;5:237–240. [Google Scholar]
- Yu BM, Afshar A, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Advances in Neural Information Processing Systems 18. MIT Press; 2006. Extracting dynamical structure embedded in neural activity. [Google Scholar]
- Yu BM, Cunningham JP, Santhanam G, Ryu SI, Shenoy KV, Sahani M. Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity. Advances in Neural Information Processing Systems. 2009;21:1881–1888. doi: 10.1152/jn.90941.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]