Abstract
While neuronal cell types display an astounding degree of phenotypic diversity, most if not all neuron types share a core panel of terminal features. However, little is known about how pan-neuronal expression patterns are genetically programmed. Through an extensive analysis of the cis-regulatory control regions of a battery of pan-neuronal C.elegans genes, including genes involved in synaptic vesicle biology and neuropeptide signaling, we define a common organizational principle in the regulation of pan-neuronal genes in the form of a surprisingly complex array of seemingly redundant, parallel-acting cis-regulatory modules that direct expression to broad, overlapping domains throughout the nervous system. These parallel-acting cis-regulatory modules are responsive to a multitude of distinct trans-acting factors. Neuronal gene expression programs therefore fall into two fundamentally distinct classes. Neuron type-specific genes are generally controlled by discrete and non-redundantly acting regulatory inputs, while pan-neuronal gene expression is controlled by diverse, coincident and seemingly redundant regulatory inputs.
INTRODUCTION
The differential expression of neuron-type specific combinations of effector genes defines the vast array of neuron types in a nervous system. However, there are cellular and molecular features shared by all neuron types throughout the nervous system. For example, biochemical and genetic analyses have defined many pan-neuronally expressed proteins that localize to synaptic vesicle and play key roles in the synaptic vesicle cycle to ensure neuron-neuron communication (Sudhof, 2004). However, remarkably little is known about how the expression of such pan-neuronal genes is controlled in any organism. This is in striking contrast to the substantial knowledge that has been accumulated on how neuron type-specific genes are controlled. Genetic loss-of-function studies have revealed a plethora of transcription factors that control the expression of neuron type-specific features, such as genes involved in the synthesis of a specific neurotransmitter system. Some of this genetic analysis, particularly loss-of-function analysis conducted in Caenorhabditis elegans, has revealed a notable theme in the control of neuron type-specific identity features in the form of terminal selector transcription factors that initiate, coordinate and maintain terminal differentiation programs in mature neuron types (Hobert, 2011; Hobert et al., 2010). Terminal selectors control the expression of many and perhaps all neuron-type specific identity features of a neuron, but in none of the many cases examined (in both C. elegans and mice) do they control the expression of broad or pan-neuronally expressed genes (Altun-Gultekin et al., 2001; Doitsidou et al., 2013; Hobert, 2011; Hobert et al., 2010; Kratsios et al., 2011; Uchida et al., 2003). In other words, the adoption of neuron type-specific identity features can be genetically decoupled from the adoption of broad or pan-neuronally expressed genes.
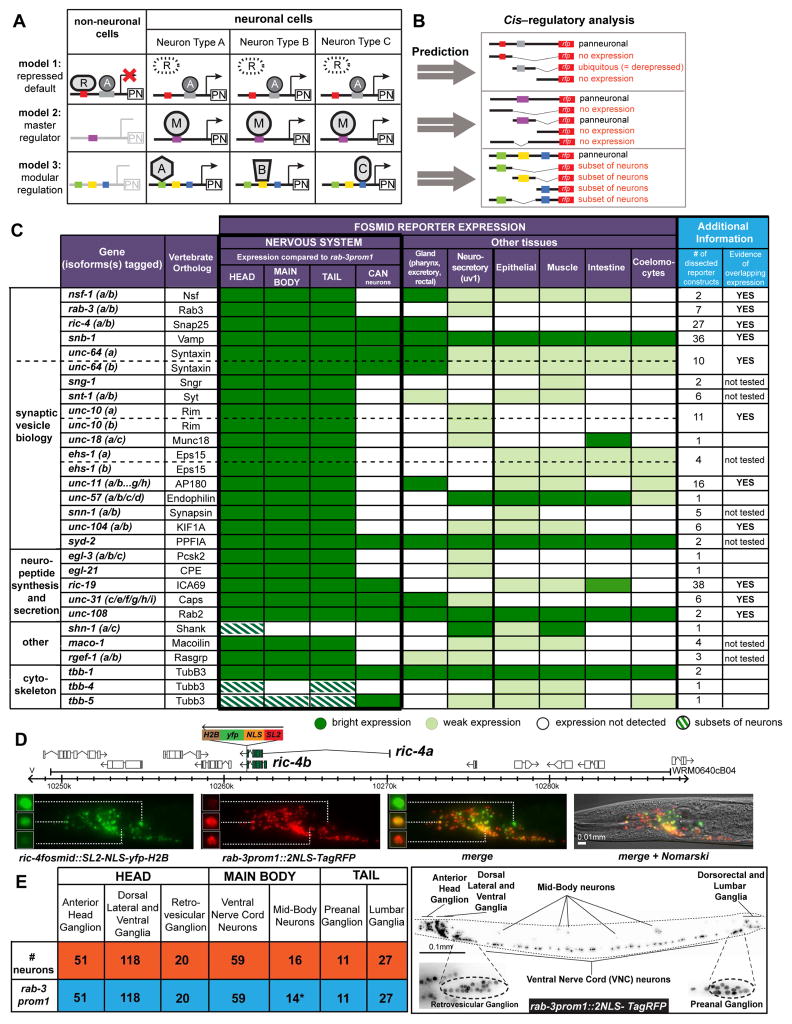
Three different mechanistic models for how pan-neuronal gene expression is regulated can easily be envisioned (Fig. 1A). There is some evidence in support of each of these three mechanisms, but in all cases, the experimental evidence is limited. In model #1, pan-neuronal genes may be controlled by ubiquitously acting transcriptional activators, but their expression is restricted to the nervous system by repressors that act outside the nervous system. This model was brought forward by the identification of the vertebrate REST/NRSF transcription factor, a repressor protein expressed in non-neuronal cells that can bind to a large set of neuronally expressed genes and supposedly downregulates their expression outside the nervous system (Schoenherr and Anderson, 1995). Even though some gene derepression effects have been observed in non-neuronal cells in REST/NRSF mutant mice, it is not clear how extensively pan-neuronal gene expression is indeed derepressed in these mutant mice (Aoki et al., 2012; Chen et al., 1998). In model #2, a pan-neuronally expressed master regulatory factor may activate expression of pan-neuronal genes throughout the nervous system. This model is supported by a number of bioinformatic studies that identified conserved sequence motifs in proximity to many pan-neuronally expressed genes (Kusakabe et al., 2004; Liu et al., 2009; Ruvinsky et al., 2007). However, the functional relevance of these presumptive cis-regulatory motifs for gene expression in vivo is unclear and binding factors are not known. Lastly, in model #3, pan-neuronal gene expression may be controlled in a modular manner in which distinct neuron types use distinct combinations of transcription factors. The one line of evidence in support of this model is the identification of a cis-regulatory element in the ric-4/SNAP25 locus that is activated by a neuron-type specific gene activator complex in C. elegans (Hwang and Lee, 2003). Distinct pan-neuronal genes may each employ distinct mechanisms and combinations of these three mechanisms can also be envisioned.
Fig. 1. Probing Pan-neuronal Gene Expression in C. elegans.
A: Schematic representation of 3 possible models for regulation of pan-neuronal gene expression. PN = pan-neuronal gene, R = non-neuronal repressor, A = activator, M = master regulator, A,B,C = different transcription factors in different neuron types.
B: Different possible outcomes of our cis-regulatory analysis based on the three predicted models in panel A.
C: Summary of the expression patterns of the fosmid reporters of the 26 genes under study. For genes that have isoforms with alternative 3′ ends, more than one fosmid reporter was made to tag these different isoforms. 23 genes (all except for shn-1, tbb-4 and tbb-5) are expressed in a pan-neuronal manner, as compared to rab-3prom1 pan-neuronal expression. The two columns on the right summarize additional reporter constructs made for each gene in this study and whether these additional reporter constructs provided evidence of overlapping expression, meaning more than one element show expression in the same domains. Expression of the unc-10fosmid reporter can also be observed in very few cells in the very anterior head part of C. elegans (supported by smFISH in Fig. 2H).
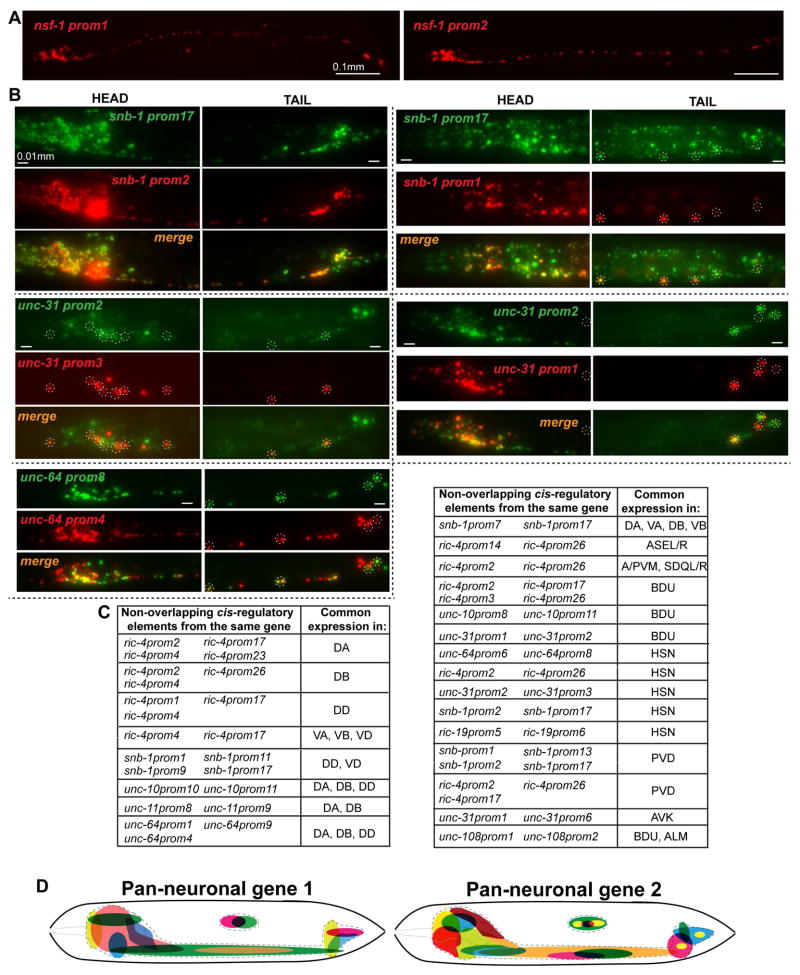
D: Schematic representation of the ric-4 fosmid reporter (top) and expression of ric-4 fosmid reporter in the head neurons (bottom). Fosmid reporter expression patterns (YFP) are always scored in comparison to the reference rab-3prom1 reporter (RFP). Expression intensity varies in distinct neurons also in comparison to the rab-3 expression. Three representative examples are shown: the neuron shown on top expresses high YFP but low RFP levels. The neuron in the middle has equal levels of expression of YFP and RFP. The neuron at the bottom has low YFP but high RFP expression levels. Schematic representation of all fosmid reporters and fluorescent worm images for each reporter are shown in supplemental Fig. S1A, B and expression intensity variability for snb-1 and unc-31 fosmids in supplemental Fig. S1C and Fig. S1D respectively.
E: The 302 neurons of the adult hermaphrodite C. elegans (orange) are distributed in different ganglia in the head, main body and tail of the worm (see Table S2 for list of these neurons). The rab-3prom1 transcriptional reporter (schematically shown in Fig. S3) is expressed in all neurons (blue) except for the CAN (*) mid-body neuronal pair. Right panel: Expression pattern of the rab-3prom1 reporter transgene in the different ganglia. Lateral view where anterior is to the left and ventral is down. Scale bar for E is 0.1 mm.
In this study, we probe these different models of pan-neuronal gene expression by making use of the extremely well characterized nature of the C. elegans nervous system, its genetic amenability and the ability to examine on a large scale the cis-regulatory information content of a substantial number of distinct genetic loci. This large sample size allowed us to extract common regulatory principles of pan-neuronal gene expression, which are strikingly distinct from the regulatory principles of neuron-type-specifically expressed genes. Given the previous paucity of insights into the regulation of pan-neuronal gene expression, our study provides a substantial advance in our understanding of how neurons acquire their terminal properties.
RESULTS
Defining a pan-neuronal gene battery
We first set out to identify genes that may be expressed throughout the entire nervous system of C. elegans. Many previous studies have described genes with broad expression throughout the C. elegans nervous system (Table S1). However, these past studies have not systematically examined whether supposedly pan-neuronal genes are indeed expressed in all of the neurons of C. elegans. Due to sheer complexity, the question of whether there are proteins that are indeed shared by all neuron types in a nervous system and show either no, restricted or lower expression outside the nervous system has also not been systematically examined in vertebrate nervous systems. Notably, some proteins generally used as “generic neuronal markers” in the vertebrate nervous system are not expressed in some neuronal populations [e.g. TuJ1 (β-tubulin 3) is not expressed in all neuronal cells in the retina (Sharma and Netland, 2007), NeuN (Fox3) is not expressed in Purkinje and some neuronal retinal cells(Mullen et al., 1992)].
To probe the notion of “pan-neuronality”, we selected a set of 26 genes, including genes involved in synaptic vesicle biology (such as the genes encoding for synaptobrevin, syntaxin, synaptotagmin, synaptogyrin and others; 15 genes); genes involved in generic aspects of neuropeptide biology (such as dense core vesicle components and neuropeptide-processing enzymes; 5 genes); and a number of miscellaneous genes with reported broad neuronal expression in either C. elegans (e.g. the commonly used pan-neuronal marker rgef-1, a ras GTPase exchange factor) (Altun-Gultekin et al., 2001; Chen et al., 2011) or vertebrates (i.e. the C. elegans homologues of vertebrate β-tubulin 3 (TuJ1), which is a commonly used pan-neuronal marker in the mouse nervous system)(Fig. 1C; Table S1). For all these 26 genes we engineered reporter genes in the context of genomic fosmid clones (Tursun et al., 2009); such fosmid reporters usually encompass multiple genes up- and downstream of the locus of interest. In most cases reported so far, regulatory elements in C. elegans are located proximal to genes that they regulate and we are currently not aware of any instances where fosmid-based reporters have failed to capture regulatory elements (we will discuss below additional validation of expression patterns by single molecule (sm) FISH and antibody staining). To facilitate the assessment of expression in the nervous system, in all fosmid reporter constructs the fluorescent reporter gene was inserted at the 3′ end of the respective locus, separated from the locus with an SL2 trans-spliced leader sequence (Tursun et al., 2009). This allows the reporter protein to be produced independently of the usually subcellularly (e.g. synaptically) localized pan-neuronal protein. Through the addition of an NLS and a Histone (H2B) tag, the fluorescent reporter is then targeted to the nucleus, allowing for reliable quantification by counting the number of neuronal nuclei in different ganglia (Fig. 1D; Fig. S1A, B).
To be able to compare expression patterns systematically, we generated a reporter line that serves as a reference for expression of each yfp fosmid reporter line. To this end, we selected the rab-3 GTPase, a gene involved in controlling synaptic vesicle release, previously reported to be broadly expressed throughout the C. elegans nervous system (Nonet et al., 1997). We find that a fosmid-based rab-3 reporter gene construct, containing around 35kb of genomic sequences including neighboring genes, as well as a transcriptional reporter gene fusion containing 4.3kb sequences of upstream regions and the first intron, shown in Fig. S3) are both expressed in 99% (300/302) of all neurons of the adult nervous system (Fig. 1E). The only neurons in which we did not observe rab-3 expression are the canal-associated neurons (CAN), a neuron pair that was previously note for its scarcity of synaptic connections with other neurons (White et al., 1986).
We scored the expression of all 26 fosmid reporter lines relative to the transcriptional rab-3 reference reporter (rab-3prom1) and found that like rab-3prom1, 23 of the 26 examined reporters drive expression in all neurons of the nervous system (Fig. 1C; Fig. S1A, B) even though the intensity of expression in distinct neuron types may vary (Fig. 1D; Fig. S1C). Differences of relative expression levels of individual pan-neuronal genes compared to rab-3 are reproducible from animal to animal and reproducible across different lines. Single molecule fluorescence in situ hybridization (smFISH)(Ji and van Oudenaarden, 2012), described below in more detail, corroborates the notion of different expression levels of individual pan-neuronal genes in different neuron types, thereby ruling out transgene artifacts (see different number of ric-4 transcripts between different neuron types in Fig. 5P).
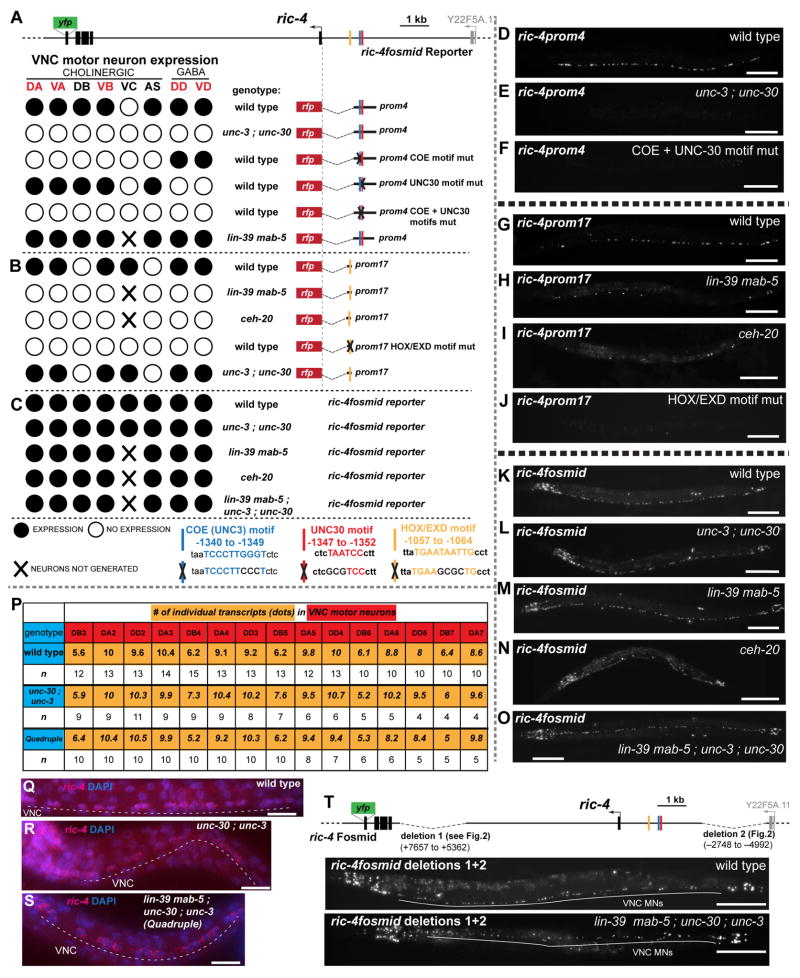
Fig. 5. Terminal selectors act in parallel to HOX Genes to regulate VNC MN expression.
A – C: ric-4 reporter gene expression in various genetic backgrounds. In a wild-type background, both ric-4prom4 and ric-4prom17 drive overlapping expression in VNC MNs. The terminal selectors unc-3 and unc-30 directly control ric-4prom4 expression in the cholinergic and GABAergic VNC MNs respectively (panel A; see also Fig. S6B, C), while ric-4prom4 expression does not depend on HOX genes. ric-4prom17 expression in the VNC MNs depends on HOX genes (lin-39, mab-5) and the HOX cofactor ceh-20, and is independent of unc-3 and unc-30 (B). In all panels, the reporter transgene (otIs490 for ric-4prom4, otIs414 for ric-4prom17 and otIs353 for ric-4fosmid) was crossed into the respective mutant background. ric-4 fosmid reporter (schematic shown again on top) VNC expression is unaffected in the unc-3 ; unc-30 mutants, HOX mutants and in the quadruple mutant background (C). VC neurons are not generated in HOX mutants. Additional data and quantification are provided in supplemental Fig. S6.
D – O: Fluorescent worm images of the data shown in panel A–C. Animals are shown in late L4 larval or young adult stages.
P – S: Detection of endogenous ric-4 transcripts show no changes in expression levels of ric-4 among wildtype, unc-30 ; unc-3 and quadruple mutant backgrounds. P: The average number of transcripts (yellow) for each embryonic VNC MN (red), in the three different genetic backgrounds (blue) is shown. Small variations in the average number of transcripts for each neuron are not statistically significant, as assessed by a three-way ANOVA statistical analysis. Note the difference in expression levels of the DB neurons (~6 transcripts/neuron) in comparison to the DA and DD neurons (~10 transcripts/neuron) that verifies endogenous variability of expression in different neuron types. Fluorescent images are shown for wild type (Q), unc-30 ; unc-3 (R) and quadruple (S) mutant backgrounds.
T: The ric-4 fosmid reporter construct with two deleted regions that contain information for VNC expression [deletion 1 (ric-4prom1 + ric-4prom2) and deletion 2 (ric-4prom26 + ric-4prom27) see Fig. 3B] is shown on top. Fluorescent images of young adult worms show that this construct is still able to drive VNC MN expression in a wild type and lin-39 mab-5 ; unc-30 ; unc-3 quadruple mutant background. Quantification is shown in Fig. S6I. Scale bars are 0.1 mm, except in Q, R, S, where scale bars are 0.01 mm.
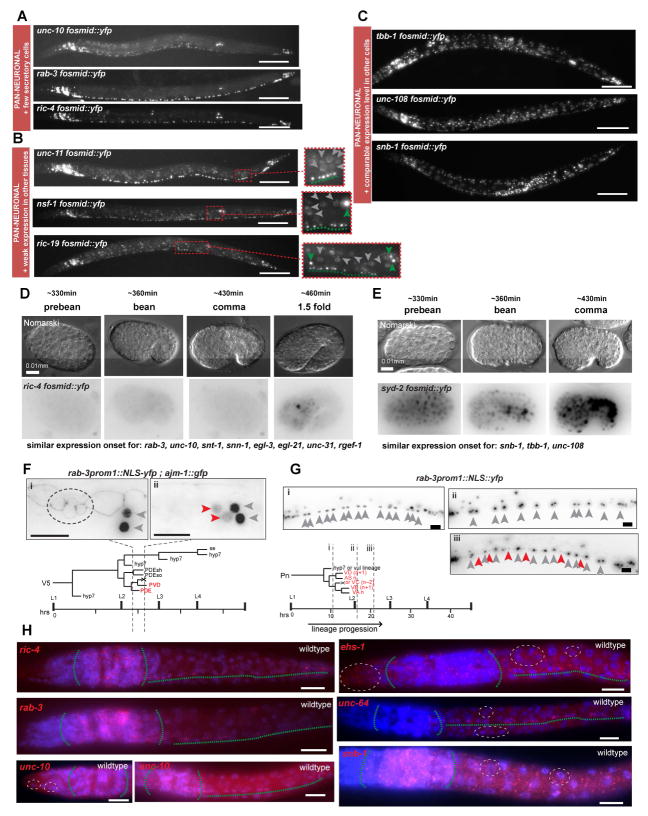
The expression of the pan-neuronal battery of 23 genes is not entirely restricted to the nervous system. Some members of this gene battery are expressed in neurons and a small number of neurosecretory cells, some are expressed in a restricted number of non-ectodermal cells and a few are ubiquitously expressed (Fig. 1C, Fig. 2A – C). Non-neuronal reporter expression is generally significantly lower than the expression in the nervous system (Fig. 2B), with the exception of four cases (snb-1, syd-2, unc-108, tbb-1) in which we detected uniform expression throughout all tissues (Fig. 2C). Genes expressed strongly both in neurons and many non-neuronal cells also show a distinct onset of embryonic expression compared to mostly neuron-restricted genes. The former category shows broad neuronal and non-neuronal expression during the proliferative phase in the developing embryo (Fig. 2D) while the latter category did not show any expression prior to cell cycle exit (Fig. 2E). The onset of expression of these largely neuronal-restricted pan-neuronal genes usually rather coincides with postmitotic phases of neuronal maturation in both the 1.5 to 2-fold stage of embryonic development (460–470 minutes of development; most neurons have terminally divided by 330 min of development, Fig. 2D) and in postembryonically born neurons (Fig. 2F, G).
Fig. 2. Different Categories of Pan-neuronal Genes.
Pan-neuronal genes can be grouped in three categories based on their expression in non-neuronal cells (panels A – C).
A: Expression in all neurons and only few non-neuronal secretory cells.
B: Expression in all neurons and weaker expression in other tissues. Expanded boxes show better the difference in levels between neurons and non-neuronal cells. Green arrowheads indicate neurons, dashed greens line underlines ventral nerve cord motorneurons (VNC MNs) and grey arrowheads indicate non-neuronal cells.
C: Expression in all neurons and equally bright expression in all other tissues. Fluorescent images of L4/young adult worms of selected fosmid reporter for each category are shown. For description of spatial expression patterns of all fosmid reporters see Fig. 1C. Temporal onset of expression of pan-neuronal genes differs between genes that belong in different categories (panels D – G).
D: Embryonic expression onset of the fosmid reporter of ric-4, a pan-neuronal gene that is more restricted to the nervous system. Expression at first is detected at the comma stage, when all neurons have already been born. Other pan-neuronal genes that are also mainly nervous system restricted (listed below) have similar temporal expression pattern.
E: Embryonic onset of expression of the fosmid reporter of syd-2, a pan-neuronal gene that is expressed broadly in non-neuronal cell types. Broad expression is detected in very early embryonic stages when neurons are not yet born. Other pan-neuronal genes that are also expressed broadly outside the nervous system (listed below) have similar temporal expression pattern.
F – G: Onset of expression of the neuronal restricted rab-3 in post-embryonically born neurons. In F, the V5 postembryonic lineage gives rise to two neurons, PDE and PVD, two glial cells and epidermal cells. rab-3prom1::2xNLS-yfp expression is detected only in mature postmitotic PDE and PVD neurons (ii), but not at an earlier stage in the “young” postmitotic PDE neuron and the PVD progenitor (i). Also in ii, the YFP expression levels in PDE and PVD (red arrowheads) are lower in comparison to neighboring neurons SDQL and PVM (grey arrowheads) that are born in the embryo. In later larval and adult stages PDE and PVD expression of rab-3prom1 is similar to the expression in SDQL and PVM. ajm-1::gfp is an apical junction marker that is used to follow the different stages of progression of the V5 lineage. In (i) the dashed circle indicates the ajm-1::gfp expression in 4 cells at the corresponding stage (i) indicated in the lineage diagram. One of these 4 cells is the “young” PDE neuron. In G, the Pn postembryonic lineage gives rise to different VNC MN types. Expression of rab-3prom1::2xNLS-yfp is not detected in the neural progenitors (i), or even at a stage when the neurons have just been born (ii). YFP expression in the postembryonic VNC MNs (read arrowheads) is detected only at a later stage (iii) and is initially weaker in comparison to YFP expression of the embryonically born VNC neurons (grey arrowheads). In later larval stages and adult worms all VNC neurons have more similar rab-3prom1::2xNLS-yfp expression levels. In F and G, grey arrows indicate embryonic neurons and red arrows indicate postembryonic neurons.
H: Single molecule in situ hybridization (smFISH) verifies expression patterns of selected pan-neuronal genes. C. elegans larvae were fixed and hybridized at the L1 stage. In red the labeled smFISH probes and in blue is DAPI staining. smFISH for ric-4, rab-3 and unc-10 (left column) shows neuronally restricted fluorescent signals. smFISH for unc-10 recapitulates the unc-10 fosmid reporter expression in just a few cells in the tip of the head (dashed white circle). smFISH for ehs-1, unc-64 and snb-1 shows more broad staining in cells outside the nervous system corroborating the fosmid reporter results. Green dashed lines outline nervous system (head ganglia and VNC). White dashed-line circles outline examples of expression in non-neuronal cells.
Scale bars are 0.1 mm in A – C and 0.01 mm in D – H.
The reporter expression results are validated by independent approaches. The expression pattern of 18 of the 26 examined pan-neuronal genes had previously been examined by antibody staining (Table S1) revealing broad expression throughout the nervous system, corroborating our reporter results. Antibody staining revealed either predominant or exclusive expression in the nervous system but since these proteins are subcellularly localized, antibody staining patterns are difficult to interpret in regard to potential neuron type specificity of expression. Therefore, as a further independent assessment of expression patterns we examined the expression of 6 genes using smFISH. The smFISH analysis for unc-10, ric-4, snb-1, unc-64, rab-3 and ehs-1 validates the expression in and outside of the nervous system we observed with our transcriptional fosmid reporters. unc-10, rab-3 and ric-4 transcription is largely restricted to the nervous system, while snb-1, unc-64 and ehs-1 transcription is observed throughout all tissue types (Fig. 2H). This ubiquitous transcription contrasts the apparently neuron-restricted antibody staining. This may simply be because in non-neuronal cells SNB-1, UNC-64, and EHS-1 proteins may localize much more diffusely thereby given a false impression of nervous system restriction; alternatively, these genes may be posttranscriptionally regulated. As the main focus of this study is to assess transcriptional regulatory mechanisms, we did not pursue this observation further.
Taken together, as illustrated by the color scheme in Fig. 1C, we have defined a battery of genes that are truly pan-neuronal, i.e. expressed in all cells of the nervous system. Most (but not all) pan-neuronal genes are also expressed in a variety of distinct patterns outside the nervous system, but usually always at much lower levels and often in just a very restricted set of highly secretory cells. A consistent overlap of expression of all of these genes is restricted to the nervous system.
Dissection of cis-regulatory elements defines organizational principles of pan-neuronal gene expression
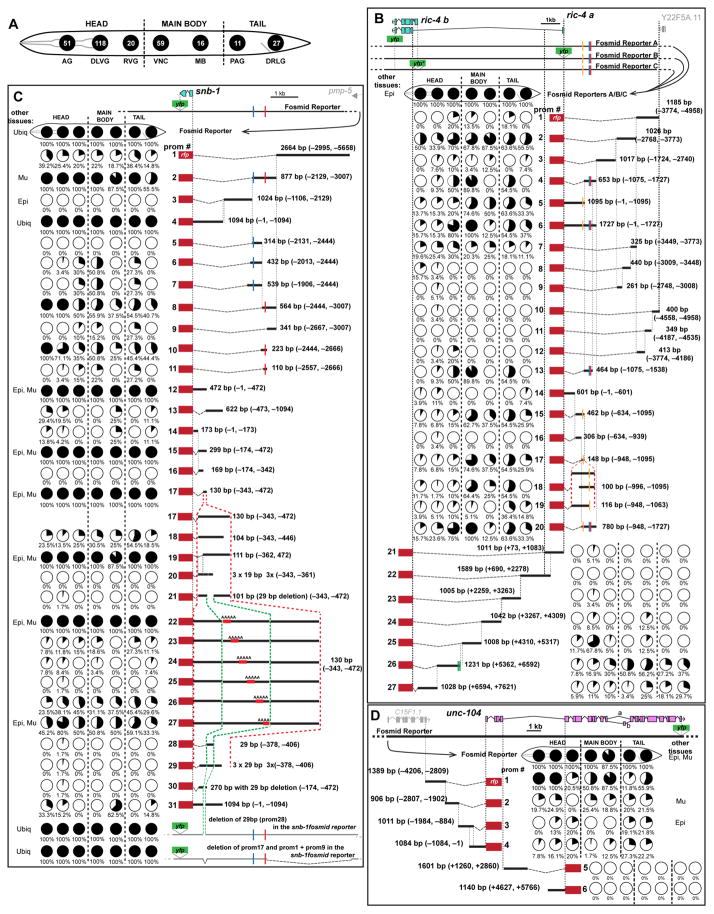
To decipher the logic of pan-neuronal gene expression we generated more than 500 transgenic lines containing 196 different reporter gene fusions, spanning from about 100 to 1500 base pairs, that interrogate the cis-regulatory information content of the 23 pan-neuronally expressed genes. For 19 of the 23 genes we generated multiple (up to 38) reporters that scan the cis-regulatory content of upstream and intronic regions of the respective genetic loci and for the remaining four genes (egl-3, egl-21, unc-18, unc-57) we generated 1kb fusions upstream of the respective gene (see Fig. 3 and Fig. S2 – 4 for all constructs generated). Using the rab-3prom1 reference transgene in the background, we carefully examined the expression of all these reporters throughout the entire nervous system, asking how the expression of these isolated elements compares to the expression of the respective fosmid reporters. We reasoned that the breadth and depth of this cis-regulatory analysis may provide evidence to distinguish the different models shown in Fig. 1A. As illustrated schematically in Fig. 1B, if expression of the respective gene locus were shaped by cis-regulatory elements that reduce or repress expression in cells outside the nervous system (model #1), at least some of the reporter fusions may lack such repressor elements, resulting in derepression outside the nervous system. Alternatively, if pan-neuronal expression were defined by a master-regulator and its cognate cis-regulatory element – such as the bioinformatically defined “N1 box” (model #2) (Ruvinsky et al., 2007) – only a small set of reporters that contain this pan-neuronal cis-regulatory element would show broad neuronal expression, while many other reporters would not show any expression. In contrast, if expression were controlled in a modular manner by distinct factors in distinct neuron types (model #3), we would observe that many of the reporters would reveal expression in subsets of neuron types.
Fig. 3. Modular Architecture of Cis-Regulatory Regions of Pan-neuronal Genes.
A: Schematic representation of the nervous system of C. elegans. Neurons belonging in the different ganglia or regions, also shown in Fig. 1E, are clustered together and represented by a black circle (numbers of neurons belonging in each ganglion are indicated inside the circle). In the ensuing panels the fraction of neurons of each ganglion expressing a reporter is indicated with a partially filled circle (pie-chart). AG = anterior head ganglion, DLVG = dorsal, lateral and ventral head ganglia, RVG = retrovesicular ganglion, VNC = ventral nerve cord motor neurons, MB = mid-body neurons, PAG = preanal ganglion, DRLG = dorsorectal and lumbar ganglia.
B – D: Dissection analysis of cis-regulatory regions of the ric-4, snb-1 and unc-104 loci. Schematics of the fosmid reporters are shown below gene schematics (YFP = pBALU23, YFP* = pBALUNI). The expression of each reporter construct is presented in the form of pie-charts that show % of neurons expressing in each of these different ganglia. For example, ric-4prom1 drives expression in 4 out of the 20 neurons of the Retrovesicular Ganglion (RVG, that this represented by the third circle, as shown in panel A), which translates into 20% of the neurons of the RVG. For each of these reporter constructs, 3 independent transgenic lines are scored (≥10 worms scored for each line); very little variation is observed across the three different lines. The % shown is an average of the average number of neurons for each line. The length in base pairs (bp) and the coordinates of each promoter fragment in relation to the translational start site are shown next to each construct. Expression in other tissues: ubiq = ubiquitous, Epi = epidermis, Mu = muscle, Int = intestine, Cc = coelomocytes. Functional binding motifs are shown as vertical colored lines: blue = COE (binding motif for unc-3) motif, red = UNC-30 motif, yellow = HOX/EXD motif, green = ASE motif (binding motif for che-1). Cis-regulatory analysis for all the other genes is shown in supplemental Fig. S2, Fig. S3 and Fig S4.
The evidence from examining 196 reporter constructs of the 23 pan-neuronal genes supports the modular control mechanism (model #3 in Fig. 1A). The data is shown in an exemplary manner for three genes in Fig. 3B – D and the evidence for all other genes is shown in Fig. S2 – 4. In virtually all cases examined, we could break pan-neuronal expression down to expression into smaller domains of the nervous system. In many cases (e.g. ric-4, unc-64, unc-10, unc-104 and unc-31 loci), modular control elements that drive expression in subdomains of the nervous system are spread over larger (ranging from 5kb to more than 10kb) intervals. In other cases (e.g. snb-1, unc-11, and ric-19 loci), small elements of between 130 - 300 bps in length still drive very broad or pan-neuronal expression; in these three cases, we undertook a deletion analysis to assess expression throughout the nervous system (in one case, ric-19, this included the generation of 29 deletion constructs with a scanning window size of 5bp). This deletion analysis resulted in the loss of expression of reporter constructs in various distinct domains of the nervous system, thereby further corroborating the concept of modularity of regulatory elements (Fig. 3C for snb-1; Fig. S2 for unc-11; Fig. S3 for ric-19).
The modular organization of regulatory elements that drive expression in restricted subsets of neuron types, disfavors the existence of pan-neuronal master regulatory molecules that operate throughout the nervous system to control pan-neuronal gene expression (model #2). Consistent with absence of pan-neuronal regulatory inputs, we could also not assign any pan-neuronal regulatory activity to the bioinformatically defined “N1 box”, a sequence motif found enriched in pan-neuronal loci and proposed to be involved in specifying pan-neuronal gene expression (Fig. S4B) (Ruvinsky et al., 2007).
Our extensive deletion analysis of cis-regulatory control regions also provided no substantial evidence for the existence of repressor elements, i.e. we never observed derepressed expression of individual cis-regulatory elements of any given gene, outside of tissues that this gene is initially expressed in. If repressor elements located in close proximity to activator elements or if multiple repressor elements were to act redundantly, such repressor motifs may have been hard to identify; however, considering the substantial number cis-regulatory elements analyzed, as well as fine-grained scanning deletion analysis that we performed on some pan-neuronal regulatory elements (e.g. ric-19), we do not favor the repressor model as being a major determinant of restriction of pan-neuronal gene expression.
We also probed the non-neuronal repressor model by examining the mutant phenotype of two genes, spr-3 and spr-4, which were previously suggested to code for the C. elegans homologs of REST/NRSF repressor protein (Lakowski et al., 2003; Lu et al., 2014). Null mutants of either gene alone, a spr-3; spr-4 double mutant or spr-1 null mutants, which eliminate the C.elegans ortholog of the cofactor of REST/NRSF, called CoREST (Jarriault and Greenwald, 2002), show no derepression of the pan-neuronally expressed ric-4 and rab-3 genes in any of the non-neuronal cells in which these genes are not normally expressed in (Fig. S4C, D).
We also examined the domains of expression of modular elements from each of the pan-neuronal genes, asking whether these domains define neuron types that show any specific relationship to one another. For example, it could be envisioned that these modules carry positional information, share a common lineage origin or are expressed in functionally related neurons. We find that such relationships are not readily apparent. Cis-regulatory modules from different pan-neuronal genes drive expression in neurons that are scattered throughout the nervous system (i.e. not clustered in specific ganglia), do not share a common lineage history and are not confined to sensory or motor neurons (i.e. no modular element drives specific expression in all sensory neurons). The only clustering of related neurons that we observed with any given module is a 62bp module from the ehs-1 cis-regulatory control region (ehs-1prom4), which drives expression in all pharyngeal neurons but no other neurons (Fig. S3).
Modular elements contain redundant cis-regulatory information
Apart from the striking and pervasive theme of modularity, we consistently observed another major theme applicable to almost all cases in which we examined 2 or more constructs per gene: Discrete, non-overlapping regulatory regions from individual pan-neuronal genes drive expression in largely overlapping parts of the nervous system (Fig. 4). In some cases this is simply evidenced by the fact that separate, discrete elements of the same locus produce expression in >85% of the nervous system (for example, the cis-regulatory elements “prom2” and ”prom4” of snb-1, or the elements “prom1” and “prom2” of nsf-1, Fig. 3C and Fig. 4A; Fig. S2 respectively). We confirmed the redundancy of cis-regulatory information in several manners. First, for a number of cases, we generated reporters in which one discrete fragment from a locus is tagged with GFP and another non-overlapping fragment from the same locus is tagged with RFP. These reporters were then crossed together and overlaps in the expression pattern were examined systematically. As shown in Fig. 4B, discrete elements from the snb-1, unc-31 and unc-64 loci showed large domains of GFP/RFP overlaps. Second, we honed in on specific neuron types - mainly ventral nerve cord (VNC) motor neurons (MNs) and mid-body neurons but also some head neurons - and examined whether discrete, separate fragments from individual pan-neuronal loci would drive expression in these identified neuron types. We found this to happen in all cases examined (Fig. 4C). For example, four non-overlapping elements of the ric-4 locus drive expression in the DA motor neurons and four different elements of the snb-1 locus drive expression in the PVD sensory neurons. Taken together, we defined a common organizational principle of the regulatory architecture of all pan-neuronal genes analyzed, in the form of redundant modules that drive expression in overlapping domains of the nervous system. This theme is schematically illustrated in Fig. 4D.
Fig. 4. Modular Elements Contain Redundant Cis-Regulatory Information.
Overlapping expression can be evidenced in different ways. In panel A, nsf-1prom1 and nsf-1prom2 drive expression in >85% of the C. elegans nervous system and they obviously have overlapping expression in most of C. elegans neurons. In panel B, overlapping expression is directly visualized. In this case the non-overlapping fragments are tagged with fluorescent proteins of different colors and when subsequently crossed together they reveal neurons with overlapping expression (seen as orange/yellow neurons in the merge, also specific cases are outlined with dashed line circles). Finally in panel C, we identified specific neuron types (right column) in which there is overlapping expression from non-overlapping fragments of the same locus (left column). The temporal expression pattern of two elements from the ric-4 locus, ric-4prom4 and ric-4prom17, with overlapping expression in many VNC MNs also appears to be indistinguishable between the two (data is shown in supplemental Fig. S5A). D: Schematic summary of the redundant modular expression of pan-neuronal genes. Distinct cis-regulatory elements drive overlapping expression in different domains (colored) of the C. elegans nervous system (outlined with dashed line). Scale bars are 0.01 mm in A and 0.01 mm in B.
We considered the possibility that cis-regulatory elements that appear to show the same expression in a mature nervous system may display distinct onsets of expression. For example, one element may capture early, initiating phases of pan-neuronal gene expression, which may fade during adult life, whereas an apparent and seemingly “redundant” element may only capture a later transcriptional maintenance phase. To address this possibility we carefully examined the onset of expression of two non-overlapping elements from the ric-4 locus, which drive expression in VNC MNs (ric-4prom4 and ric-4prom17 in Fig. 3B) and found the onset and maintenance of expression to be indistinguishable (Fig. S5A). Generally, we also find that the expression levels of parallel-acting elements appear superficially similar.
Parallel-acting, redundant elements are controlled by distinct transcription factors
The observation of separable cis-regulatory regions driving expression in the same neuron types could be explained in two different ways. There may be multiple copies of the same regulatory motifs, recognized by the same cohort of transcription factor(s) and each separable element may contain copies of these motifs. Alternatively, discrete elements may be controlled by distinct control mechanisms. We tested this possibility by a combination of sequence motif analysis and the examination of candidate trans-acting factors. Specifically, we noted that small elements from the ric-4 and snb-1 loci that drove expression in VNC MNs contained conserved predicted binding sites (“COE motifs”) for the terminal selector of cholinergic VNC motor neuron identity, unc-3 (ric-4prom4 in Fig. 5A,D and snb-1prom7 in Fig. S7A,E). Terminal selectors like unc-3 are known to be required for the expression of many, most or all known neuron-type specific identity features of specific neuron types (Hobert, 2011; Kratsios et al., 2015; Kratsios et al., 2011). However, as assessed in many different cellular contexts, terminal selectors are not required for the expression of pan-neuronal identity features (Altun-Gultekin et al., 2001; Doitsidou et al., 2013; Hobert, 2011; Kratsios et al., 2011; Uchida et al., 2003). As such, the presence of unc-3 binding sites (COE motifs) in discrete elements from the ric-4 and snb-1 loci was unexpected. However, we do find that mutation of the COE motif in the context of these smaller regulatory elements from the ric-4 and snb-1 loci does abolish expression in cholinergic VNC MNs. Moreover, the expression of these isolated regulatory elements is lost if reporter transgenes are crossed into an unc-3 null mutant background (Fig. 5A,E,F; Fig. S6A, B; Fig. S7A, E, H). This is in striking contrast to expression of the fosmid-based ric-4 and snb-1 reporters: when crossed into an unc-3 null mutant background, expression is not affected (Fig. 5C,L; Fig. S6I; Fig. S7D, G, J).
Notably, other regions of the ric-4 and snb-1 loci, which also produce expression in VNC MNs, do not contain COE motifs and, when crossed into an unc-3 null mutant background, still drive reporter expression in VNC MNs (ric-4prom17 in Fig. 5B, and snb-1prom1 and snb-1prom17 in Fig. 3C; Fig. S7C). This data suggests that pan-neuronal genes in cholinergic VNC MNs are controlled by multiple, parallel-acting regulatory inputs, with one, but only one component of these inputs being a selector of terminal, neuron type-specific identity.
We tested the broadness of the concept of (a) distinct, parallel-acting regulatory inputs and (b) terminal selector involvement by examining several other neuron types: first, we considered another VNC MN class, the GABAergic, D-type motor neurons, which are controlled by the terminal selector unc-30 (Eastman et al., 1999; Jin et al., 1994). Here again, we find that discrete elements from the ric-4 and snb-1 loci (ric-4prom4 and snb-1prom11) show a genetic dependence on unc-30 and on the predicted UNC-30 binding site, i.e. reporter expression is lost in unc-30 mutants or upon mutation of the UNC-30 binding motif. Yet other elements of the same loci that also drive expression in GABAergic MNs do not show any unc-30 dependence (Fig. 5A,E,F; Fig. S6A,C; Fig. S7B, F, I). As is the case for unc-3, expression of the ric-4 and snb-1 fosmid based reporters is not affected in unc-30 null mutants (Fig. 3C,L; Fig. S6I; Fig. S7D, G, J).
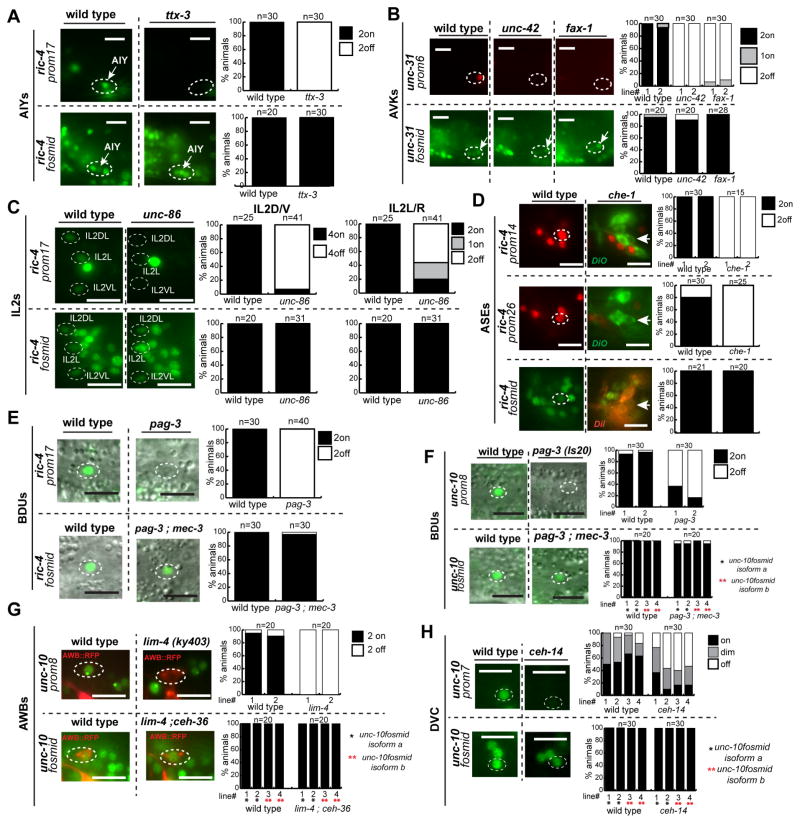
As shown by the examples in Fig. 6 (and Fig. S7L,M,N) and also summarized in Fig. 7A, the theme of redundancy and terminal selector inputs applies to neurons throughout the entire nervous system. For example, we find that in null mutants of pag-3, ceh-14 and lim-4, terminal selectors of BDU interneuron, DVC interneuron and AWB sensory neuron identity respectively (Nokes et al., 2009; Sagasti et al., 1999; Serrano-Saiz et al., 2013), the expression of the unc-10 fosmid reporter is unaffected. Yet individual, isolated and parallel-acting elements from the unc-10 locus do require pag-3, ceh-14 and lim-4 for the expression in BDU, DVC and AWB, respectively. Similarly, ric-4 fosmid gene expression is unaffected in the AIY interneurons of ttx-3 mutants or the ASE neurons of che-1 mutants, but individual, isolated elements from the ric-4 locus are ttx-3 or che-1-dependent in AIY or ASE, respectively.
Fig. 6. Multiple parallel inputs are a common theme for pan-neuronal gene regulation.
Terminal Selectors affect pan-neuronal gene expression only in the context of isolated cis-regulatory elements but not in the context of the fosmid reporters (A – H). Data of panels (A – H) are summarized in Fig. 7A. Quantification is shown on the right. Y- axis always shows % of animals with expression of the respective reporter. Data are shown in the same way for panels B – H. Double mutant backgrounds (pag-3; mec-3 and lim-4; ceh-36) were used in several cases to avoid homeotic identity transformations (Gordon and Hobert, 2015; Sagasti et al., 1999).
Scale bars are 0.01 mm.
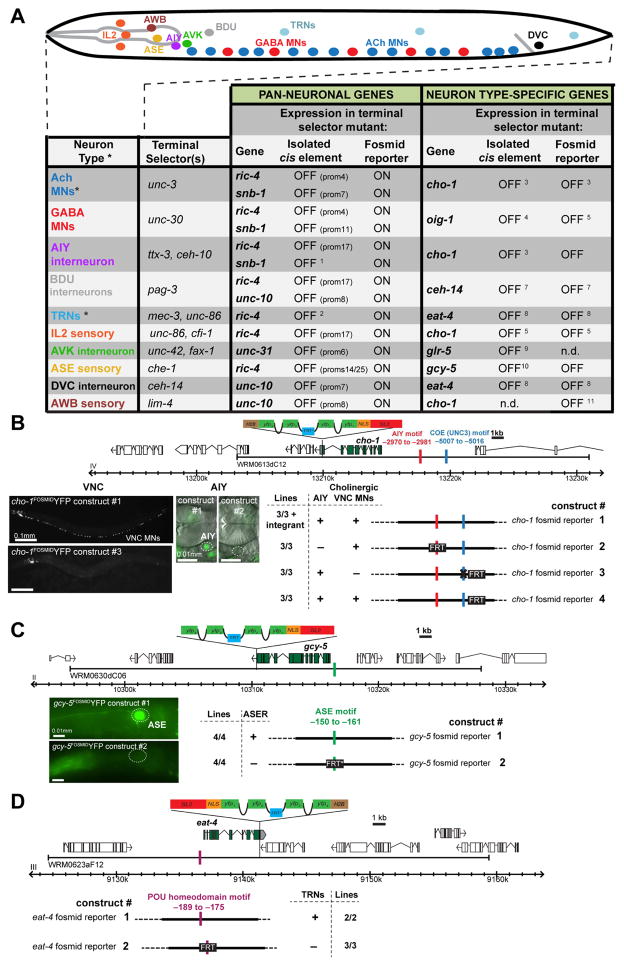
Fig. 7. Distinct Regulation of Pan-neuronal and Neuron-Type Specific Identity Features.
A: Summary of distinct regulatory effects of terminal selectors on neuron-type specific and pan-neuronal genes. Mutagenesis of Terminal Selector motifs in neuron type-specific gene fosmid reporters abolishes expression in the respective neuron types, shown in panels B, C and D. Primary data for A is shown in Fig 6A – H; Fig. S7L, M, N except for cases with footnotes* MNs = motor neurons, TRN = light touch receptor neurons. n.d. = not determined. 1: (Wenick and Hobert, 2004), 2: (Hwang and Lee, 2003), 3: (Kratsios et al., 2011), 4: (Eastman et al., 1999), 5: (Howell et al., 2015), 6: (Zhang et al., 2014), 7: (Gordon and Hobert, 2015), 8: (Serrano-Saiz et al., 2013), 9: (Wightman et al., 2005), 10: (Chang et al., 2003) 11: Pereira et al., in preparation.
B: cho-1/ChT (choline transporter) fosmid reporter expression in the cholinergic VNC MN and the head interneuron AIY is controlled by the terminal selector unc-3 (Kratsios et al., 2011) and ttx-3 (Altun-Gultekin et al., 2001). Mutagenesis of the AIY motif (replacement by FRT) and of the COE motif (GG to CC substitution) in the nuclear cho-1fosmid::SL2::NLS::yfp::H2B reporter abolishes expression in AIY and VNC MNs respectively. Mutagenesis in the fosmid reporters was done by recombineering an FRT sequence in the place of a binding site (Tursun et al., 2009). A control cho-1fosmid reporter containing only the FRT scar, without the mutations in the COE and AIY motif, drives expression in AIY and VNC MNs same as the not mutated cho-1fosmid reporter.
C: gcy-5 expression in the ASER neuron depends on the ASE terminal selector che-1 (Uchida et al., 2003). Mutagenesis of the ASE motif (replacement by FRT*) of the gcy-5fosmid reporter, abolishes expression in ASER.
D: eat-4 expression in the Touch Receptor Neurons (TRN) depends on the terminal selector unc-86 (Serrano-Saiz et al., 2013). Mutagenesis of the POU homeodomain motif (replacement by FRT) of the eat-4fosmid reporter abolishes expression in the TRNs.
HOX transcription factors provide parallel regulatory inputs
To investigate the nature of the multiple, parallel-acting control mechanisms, we honed in on the ric-4 locus. We noted a conserved HOX/EXD binding site (Mann and Affolter, 1998) in the 148 bp cis-regulatory element ric-4prom17 (Fig. S6F); this element does not require the unc-3 and unc-30 terminal selectors for its expression in VNC MNs (Fig. 5B). Like in vertebrates, C. elegans HOX genes are expressed in the context of the nervous system predominantly in motor neurons along the ventral/spinal nerve cord (Kenyon et al., 1997). We first examined the VNC MNs of the midbody region, which are known to express the lin-39/HOX gene, the C. elegans homolog of Scr and Dfd (Kenyon et al., 1997). We find that VNC MN expression of the ric-4prom17 element is severely reduced in lin-39 mutants (Fig. S6D,E). Animals that lack the Antennapedia-type HOX gene mab-5, which is expressed in a partially overlapping midbody domain with lin-39 (Kenyon et al., 1997)(Kratsios and Hobert, unpubl. data) do not show a reduction in ric-4prom17 expression (Fig. S6D, E). However, lin-39 mab-5 double null mutants show a stronger downregulation of expression than lin-39 single mutants (Fig. 5B, H; Fig. S6D,E). The phenotype of lin-39 mab-5 double null mutants is not completely penetrant and we considered whether the Labial ortholog ceh-13, known to be coexpressed with lin-39 and mab-5 in VNC MNs (Streit et al., 2002), may also contribute to ric-4prom17 expression. We indeed find this to be the case (Fig. S6H). At the posterior end of the VNC, the AbdB ortholog egl-5 affects expression of ric-4prom17 in neurons of the preanal ganglion (Fig. S6G). As expected from these results, genetic removal of the HOX cofactor ceh-20, an Extradenticle/Pbx ortholog, results in a similar, strong reduction of ric-4prom17 expression (Fig. 5B, I; Fig. S6D, E).
All of these interactions may be direct since upon deletion of the predicted HOX/EXD binding site in ric-4prom17 the VNC MN expression of the reporter gene is completely lost (Fig. 5B,J). Strikingly, expression of the ric-4fosmid reporter was completely unaffected in HOX gene mutant backgrounds (Fig. 5C,M,N; Fig. S6I, J), thereby mirroring the situation with terminal selectors, which affect the expression of individual modules but not the expression of fosmid-based reporters. The unaffected fosmid reporter expression in HOX mutants also demonstrates that the lack of expression of individual cis-regulatory elements in HOX mutants is not merely a consequence of developmental loss of the VNC MNs.
The redundancy of the HOX and terminal selector (unc-3, unc-30) inputs can be recapitulated, by “stitching back together” the terminal selector-dependent ric-4prom4 module with the HOX-dependent ric-4prom17 module. Mutating the terminal selector or HOX binding site (which are essential for expression of either module alone) in this construct does not result in loss of expression of this reporter (Fig. S5B).
Intriguingly, the redundancy of pan-neuronal ric-4 regulation is not restricted to terminal selectors and HOX genes. In mutant animals in which we removed both terminal selectors (unc-30 and unc-3) together with the HOX genes lin-39 and mab-5, pan-neuronal expression of the ric-4 fosmid reporter is still unaffected (Fig. 5O; Fig. S6I) and the expression level appears to be unaltered, as assessed by smFISH analysis (Fig. 5P,Q, R, S). Hence, there are more than two parallel inputs into ric-4 regulation. We deleted four other elements in the ric-4 locus which, in isolation, produced VNC MN expression (ric-4prom1, ric-4prom2, ric-4prom26 and ric-4prom27 in Fig. 3B) and that may constitute response elements to parallel-acting factors. Deleting these elements from the fosmid reporter construct did not result in a loss of VNC MN expression, confirming that these elements are in isolation sufficient, but not required for VNC MN expression. We crossed this mutated fosmid reporter into unc-3, unc-30, lin-39 mab-5 quadruple mutant to also eliminate the combined terminal selector and HOX input and find that this reporter still provides expression of ric-4 in 60% of VNC MNs (Fig. 5T, Fig. S6K). To address the possibility that other two HOX factors that are expressed in VNC MNs, ceh-13 and egl-5, might be compensating for loss of lin-39 and mab-5, we also deleted the HOX binding site from the ric-4 fosmid reporter (in addition to the previous deletions). Again, expression in the VNC was not affected in a wild-type background and still more than 60% of VNC MNs were expressing in a unc-3 ; unc-30 ; lin-39 mab-5 quadruple mutant background (data not shown).
Mirroring the example of ric-4 regulation, deletion of three elements from the snb-1 fosmid reporter that each drive VNC expression in isolation (snb-1prom17, snb-1prom1 and snb-1prom9), does not affect expression of snb-1 fosmid in the VNC MNs, even when crossed into the unc-3 ; unc-30 ; lin-39 mab-5 quadruple mutant background (Fig. S7K). These observations are a testament to the extreme redundancy of regulatory control mechanisms that direct pan-neuronal gene expression.
Comparing the regulatory architecture of pan-neuronal genes with shadow enhancers
Seemingly redundant regulatory elements, driving similar expression in the same cells or tissues of an animal have been documented in the literature for numerous developmental patterning genes (Frankel, 2012). In a number of these cases, the redundant regulatory elements have been coined “shadow enhancers” (Hong et al., 2008; Lagha et al., 2012). By the nature of their discovery (and reflected in their naming), shadow enhancers refer to regulatory elements bound by the same set of transcription factors (Hong et al., 2008; Perry et al., 2010). This is different from the cases described here in which distinct elements are bound by distinct factors. Shadow enhancers have been shown to confer robustness of gene expression under fluctuating environmental conditions and have been also found to ensure the correct timing of expression (Hong et al., 2008; Lagha et al., 2012). These features also do not appear to apply to the redundant control mechanisms of pan-neuronal gene expression. As mentioned above, a close examination of two redundant, independently controlled cis-elements from the ric-4 locus that drive expression in VNC MNs shows indistinguishable onsets of expression (Fig. S5A). As assessed by YFP fluorescence produced from a ric-4 fosmid reporter and as assessed by counting endogenous ric-4 mRNA levels with smFISH, we furthermore find ric-4 expression to be unaffected in animals in which we removed two of the parallel, redundant regulatory inputs (unc-3 and unc-30 terminal selector mutants combined with HOX gene mutants), even if we subject animals to various stressor (heat, starvation, gamma irradiation, oxidative stress, dauer formation, ethanol shock; data not shown). Therefore, the regulatory architecture that we describe here for pan-neuronal genes may differ on several levels from at least some of the previously described features of shadow enhancers. First, the multiplicity of parallel inputs that we observed in pan-neuronal expression control is unusual (as assessed by the deletion analysis described in the previous section); second, the factors controlling distinct cis-regulatory elements are different; third, there are no measurable differences in the timing and level of expression of redundant regulatory elements under the same type of stressful environmental conditions that were shown to be buffered by shadow enhancers.
Fundamental differences in the control of pan-neuronal and neuron-type specific gene expression
Our data suggests a fundamental difference between the mechanisms that control neuron-type specific genes and pan-neuronal genes. Whereas the expression of pan-neuronal genes depends on multiple parallel regulatory inputs, conferred by terminal selectors plus additional regulatory factors, neuron-type specific genes depend solely on terminal selector transcription factors (schematized in Fig. 8). This is evidenced by the fact that fosmid reporter expression of a number of neuron type-specific terminal identity genes is abolished in terminal selector mutants of the respective neuron type, as summarized in Fig. 7A. For example, a fosmid-based reporter for the choline transporter cho-1, which is exclusively expressed in cholinergic neurons, is controlled by: (i) the terminal selector unc-3 in the VNC MNs, (ii) the terminal selector ttx-3 in the cholinergic interneuron AIY and (iii) the terminal selector lim-4 in the olfactory neuron AWB (Fig. 7A). To further solidify the exclusive and non-redundant contribution of terminal selectors, we mutated individual terminal selector binding sites in fosmid reporters (TTX-3/CEH-10 and COE motif in cho-1 fosmid, UNC-86/MEC-3 motif in eat-4 and ASE motif in the gcy-5 fosmid). Introduction of single motif mutations resulted in loss of expression of the fosmid reporter in the specific neuron type (Fig. 7B–D). In additional support to that notion, a previous study has shown that a single nucleotide mutation (retrieved by a forward genetic screen) in the cis-regulatory region of the ASEL neuron-type specific miRNA lsy-6, affects an ASE motif and results in loss of lsy-6 expression in ASEL (Sarin et al., 2010); similarly, a loss of function allele of the vesicular acetylcholine transporter (unc-17) is defined by a point mutation in the binding site for the UNC-3 terminal selector (J. Rand, pers. comm.).
Fig. 8. Regulatory architecture of pan-neuronal genes and neuron-type specifi genes.
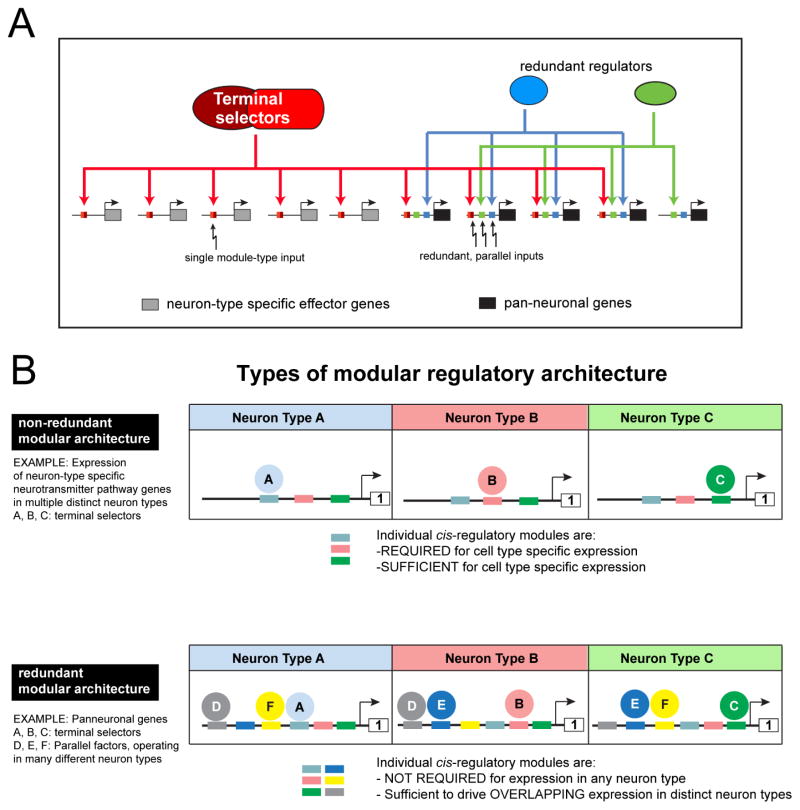
A: Neuron-type specific effector genes are controlled by combinations of terminal selectors (which differ in different neuron types) while pan-neuronal genes are controlled by many parallel-acting transcription factors, including terminal selectors, through modular regulatory elements. As deduced by our cis-regulatory analysis, the redundant regulators may be expressed in many different cell types.
B: Different types of modular regulatory architectures.
DISCUSSION
While considerable efforts have been made in various systems to understand how the cellular specificity of expression of neuron-type-specific genes is controlled, the control of pan-neuronal gene expression has received very little attention and, hence, no coherent theme about their regulation has emerged so far. We have sought to overcome this dearth of insight through an in-depth analysis of the regulation of a wide range of pan-neuronal gene in the nervous system of C.elegans. Our studies reveal a multitude of novel, direct regulators of pan-neuronal gene expression, including HOX genes, which have not previously been implicated in directly controlling terminal neuronal identity features. However, the most notable aspect of our study is the discovery of a common organizational principle shared by a large cohort of terminal differentiation genes that define features shared by all neuron types. The landmark of this organizational principle is the multiplicity of independent, parallel-acting and seemingly redundant regulatory inputs. The redundancy of regulation of pan-neuronal gene expression is not anecdotal, but a pervasive theme in the regulation of all pan-neuronal genes that we examined. This redundancy is possibly distinct from other previously described cases of regulatory redundancy, as exemplified by shadow enhancers (Hong et al., 2008; Lagha et al., 2012). Shadow enhancers are essentially duplicated regulatory control elements that respond to similar trans-acting factors (Hong et al., 2008; Lagha et al., 2012). In contrast, the redundant elements that we describe here integrate distinct trans-acting inputs and, in contrast to shadow enhancers, do not seem to be required to ensure robustness of gene regulation. Moreover, the redundancy of pan-neuronal gene expression appears to be more extensive than that of shadow enhancers of developmental control genes. For example, in the cases of ric-4 and snb-1, we can infer the existence of at least four distinct, parallel regulatory inputs for expression in VNC MNs (Fig. 5T; Fig. S7K). However, both the study of shadow enhancer and the regulatory elements that we describe here need to proceed to greater depth before definitive comparative conclusions can be drawn.
The key conceptual advance of our study lies in the revelation of fundamentally distinct features of the transcriptional control mechanisms in the nervous system, with two distinct organizational design principles emerging. Neuron-type specific genes, such as sensory receptors, ion channels and neurotransmitter synthesizing enzymes, are subject to control by a comparatively simple cis-regulatory architecture composed of discrete regulatory elements responsive to neuron-identity-defining terminal selector proteins (schematized in Fig. 8A). These elements act in a strictly non-redundant manner. In striking contrast, the coherent theme of pan-neuronal gene expression control is defined by a convergence of multiple, parallel-acting and seemingly redundant transcriptional regulatory inputs (Fig. 8). One way to illustrate the difference in the organization of regulatory control elements of pan-neuronal and neuron-type-specific genes is from the perspective of their modular organization (Fig. 8B). Both pan-neuronal and neuron-type-specific genes contain a modular array of regulatory elements, but the individual modules of neuron-type specific genes harbor discrete elements that are required and sufficient to drive expression of, for example, the vesicular glutamate transporter VGLUT in distinct classes of glutamatergic neurons (Serrano-Saiz et al., 2013) or acetylcholine-synthesizing enzymes and transporters in distinct classes of cholinergic neurons (Kratsios et al., 2011; Wenick and Hobert, 2004; Zhang et al., 2014) (our unpubl. data). In contrast, and as illustrated schematically in Fig. 4D and in Fig. 8, even very small cis-regulatory modules from pan-neuronal genes tend to be very broadly expressed, showing extensive, but not necessarily complete overlap in expression with other cis-regulatory modules from the same locus. Importantly, the dichotomy between pan-neuronal and neuron-type specific gene regulation is not anecdotal, but holds for scores of pan-neuronal and neuron-type specific genes.
Our study also reveals HOX genes and terminal selectors transcription factors as direct regulators of pan-neuronal genes. Previous genetic analysis of terminal selector-type transcription factors revealed that their loss results in loss of neuron-type-specific identity features, but no apparent effects on pan-neuronal features (Altun-Gultekin et al., 2001; Doitsidou et al., 2013; Hobert, 2011; Hobert et al., 2010; Kratsios et al., 2011; Uchida et al., 2003). However, our present analysis demonstrates that terminal selectors do participate, in a parallel, redundant manner, in the regulation of pan-neuronal gene expression. This means that even though cis-regulatory regions of pan-neuronal and neuron-type-specific effector genes are organized in a fundamentally different manner, the regulation of both types of effector genes involves the same set of regulatory factors, demonstrating the coupling of the acquisition of pan-neuronal and neuron-type-specific features (Fig. 8). This dichotomous theme of terminal selector function is apparent in many cell types throughout the nervous system.
The fundamental difference of the regulatory organization of pan-neuronal and neuron-type-specific genes may be a testament to the evolutionary history of gene expression profiles in the nervous system. The relative simplicity of neuron-type specific gene regulation may be a reflection of the relative rapid evolvability of neuronal type-specificity of gene expression programs. In contrast, the expression of pan-neuronal genes, which originated very early in nervous system evolution, necessitates stability and may have accumulated over time responsiveness to various transcriptional regulatory factors present in a mature neuron type.
EXPERIMENTAL PROCEDURES
Generation of Reporter Transgenes and Scoring of Expression
All fosmid reporter constructs were generated using λ-Red-mediated recombineering in bacteria as previously described (Tursun et al., 2009). For all fosmid reporters, an SL2 spliced, nuclearly localized YFP::H2B sequence was engineered right after the stop codon of the respective locus (most cases) are at the 5′ end of the locus. More detailed information on fosmid generation is provided in the Supplementary Experimental Procedures. All reporter gene fusions for cis-regulatory analysis (except rab-3prom1 transcriptional reporter) were generated using a PCR fusion approach (Hobert, 2002) using nuclearly localized 2xNLS-TagRFP coding sequence. All reporters injected into a pha-1(e2123) mutant background strain (Granato et al., 1994), resulting in transgenic arrays with very little mosaicism. A list of transgenes generated in this study, as well as a list of other strains used, is provided in Supplementary Methods.
Expression of all reporters were scored relative to a chromosomally integrated, panneuronally expressed “reference” reporter (rab-3prom1::NLS-TagRFP: otIs356 or rab-3prom1::NLS-yfp: otIs287IV or otIs291V). At least three different lines for each fosmid reporter were tested (≥5 worms from each line); generally, very little variation was observed across the three different lines. Fluorescent pictures were acquired for all the worms and expression of the pan-neuronal gene fosmid reporters throughout the nervous system was then scored by direct comparison/co-localization of the fosmid YPF to the “reference” RFP expression for all neurons in all different ganglia. For each reporter construct we scored the number of neurons for each ganglion/group of ganglia as explained in Fig. 3A.
Single molecule fluorescence in situ hybridization
Single-molecule (sm) FISH was done as previously described (Ji and van Oudenaarden, 2012). Samples were incubated over night at 37°C during the hybridization step. All sets of probes were designed by using the Stellaris RNA FISH probe designer and were obtained, already conjugated and purified, from Biosearch Technologies. The ric-4, unc-64, ehs-1, rab-3, snb-1, ric-19 and snt-1 probes were conjugated to Quasar 670 and the unc-10 probes were conjugated to CAL Fluor Red 610.
Supplementary Material
Acknowledgments
We thank Qi Chen for expert assistance with microinjection, members of the Hobert lab for technical advice and helpful discussions, Tulsi Patel for generating gcy-5-tagged fosmid, Baris Tursun for helping generate pBALU23, Paschalis Kratsios, Laura Pereira, Kelly Howell and Pat Gordon for communicating unpublished results, Iva Greenwald and members of the Hobert lab for comments on the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was funded by the National Institutes of Health [R01NS039996-05 and R01NS050266-03] and the Howard Hughes Medical Institute.
Footnotes
AUTHOR’S CONTRIBUTIONS
N.S., I.C. and O.H. designed the experiments. N.S. and I.C. performed the experiments. N.S., I.C. and O.H. analyzed the data and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- Aoki H, Hara A, Era T, Kunisada T, Yamada Y. Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development. 2012;139:667–677. doi: 10.1242/dev.072272. [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Fu Y, Ren M, Xiao B, Rubin CS. A RasGRP, C. elegans RGEF-1b, couples external stimuli to behavior by activating LET-60 (Ras) in sensory neurons. Neuron. 2011;70:51–65. doi: 10.1016/j.neuron.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Doitsidou M, Flames N, Topalidou I, Abe N, Felton T, Remesal L, Popovitchenko T, Mann R, Chalfie M, Hobert O. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes Dev. 2013;27:1391–1405. doi: 10.1101/gad.217224.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman C, Horvitz HR, Jin Y. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J Neurosci. 1999;19:6225–6234. doi: 10.1523/JNEUROSCI.19-15-06225.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N. Multiple layers of complexity in cis-regulatory regions of developmental genes. Dev Dyn. 2012;241:1857–1866. doi: 10.1002/dvdy.23871. [DOI] [PubMed] [Google Scholar]
- Gordon P, Hobert O. A competition mechanism for a homeotic neuron identity transformation in C. elegans. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.04.023. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends in neurosciences. 2010;33:435–445. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell K, White JG, Hobert O. Spatiotemporal control of a novel synaptic organizer molecule. Nature. 2015 doi: 10.1038/nature14545. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SB, Lee J. Neuron cell type-specific SNAP-25 expression driven by multiple regulatory elements in the nematode Caenorhabditis elegans. J Mol Biol. 2003;333:237–247. doi: 10.1016/j.jmb.2003.08.055. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Greenwald I. Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans. Genes Dev. 2002;16:2713–2728. doi: 10.1101/gad.1022402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N, van Oudenaarden A. Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos. WormBook. 2012:1–16. doi: 10.1895/wormbook.1.153.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Hoskins R, Horvitz HR. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature. 1994;372:780–783. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ, Austin J, Costa M, Cowing DW, Harris JM, Honigberg L, Hunter CP, Maloof JN, Muller-Immergluck MM, Salser SJ, et al. The dance of the Hox genes: patterning the anteroposterior body axis of Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 1997;62:293–305. [PubMed] [Google Scholar]
- Kratsios P, Pinan-Lucarre B, Kerk SY, Weinreb A, Bessereau JL, Hobert O. Transcriptional Coordination of Synaptogenesis and Neurotransmitter Signaling. Curr Biol. 2015 doi: 10.1016/j.cub.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci. 2011;15:205–214. doi: 10.1038/nn.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe T, Yoshida R, Ikeda Y, Tsuda M. Computational discovery of DNA motifs associated with cell type-specific gene expression in Ciona. Dev Biol. 2004;276:563–580. doi: 10.1016/j.ydbio.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Eimer S, Gobel C, Bottcher A, Wagler B, Baumeister R. Two suppressors of sel-12 encode C2H2 zinc-finger proteins that regulate presenilin transcription in Caenorhabditis elegans. Development. 2003;130:2117–2128. doi: 10.1242/dev.00429. [DOI] [PubMed] [Google Scholar]
- Liu R, Hannenhalli S, Bucan M. Motifs and cis-regulatory modules mediating the expression of genes co-expressed in presynaptic neurons. Genome Biol. 2009;10:R72. doi: 10.1186/gb-2009-10-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Nokes EB, Van Der Linden AM, Winslow C, Mukhopadhyay S, Ma K, Sengupta P. Cis-regulatory mechanisms of gene expression in an olfactory neuron type in Caenorhabditis elegans. Dev Dyn. 2009;238:3080–3092. doi: 10.1002/dvdy.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:8061–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levin M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Ohler U, Burge CB, Ruvkun G. Detection of broadly expressed neuronal genes in C. elegans. Dev Biol. 2007;302:617–626. doi: 10.1016/j.ydbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 1999;13:1794–1806. doi: 10.1101/gad.13.14.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S, Bertrand V, Bigelow H, Boyanov A, Doitsidou M, Poole RJ, Narula S, Hobert O. Analysis of multiple ethyl methanesulfonate-mutagenized caenorhabditis elegans strains by whole-genome sequencing. Genetics. 2010;185:417–430. doi: 10.1534/genetics.110.116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, de la Cruz ED, Hobert O. Modular Control of Glutamatergic Neuronal Identity in C. elegans by Distinct Homeodomain Proteins. Cell. 2013;155:659–673. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RK, Netland PA. Early born lineage of retinal neurons express class III beta-tubulin isotype. Brain Res. 2007;1176:11–17. doi: 10.1016/j.brainres.2007.07.090. [DOI] [PubMed] [Google Scholar]
- Streit A, Kohler R, Marty T, Belfiore M, Takacs-Vellai K, Vigano MA, Schnabel R, Affolter M, Muller F. Conserved regulation of the Caenorhabditis elegans labial/Hox1 gene ceh-13. Developmental biology. 2002;242:96–108. doi: 10.1006/dbio.2001.0544. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- Wenick AS, Hobert O. Genomic cis-Regulatory Architecture and trans-Acting Regulators of a Single Interneuron-Specific Gene Battery in C. elegans. Dev Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London B Biological Sciences. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ebert B, Carmean N, Weber K, Clever S. The C. elegans nuclear receptor gene fax-1 and homeobox gene unc-42 coordinate interneuron identity by regulating the expression of glutamate receptor subunits and other neuron-specific genes. Dev Biol. 2005;287:74–85. doi: 10.1016/j.ydbio.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Zhang F, Bhattacharya A, Nelson JC, Abe N, Gordon P, Lloret-Fernandez C, Maicas M, Flames N, Mann RS, Colon-Ramos DA, et al. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development. 2014;141:422–435. doi: 10.1242/dev.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.