Abstract
Viral infections elicit anti-viral antibodies and have been associated with various chronic diseases. Detection of these antibodies can facilitate diagnosis, treatment of infection and understanding of the mechanisms of virus associated diseases. In this work, we assayed anti-viral antibodies using a novel high density-nucleic acid programmable protein array (HD-NAPPA) platform. Individual viral proteins were expressed in situ directly from plasmids encoding proteins in an array of microscopic reaction chambers. Quality of protein display and serum response was assured by comparing intra- and inter- array correlation within or between printing batches with average correlation coefficients of 0.91 and 0.96, respectively. HD-NAPPA showed higher signal to background (S/B) ratio compared with standard NAPPA on planar glass slides and ELISA. Antibody responses to 761 antigens from 25 different viruses were profiled among patients with juvenile idiopathic arthritis (JIA) and type 1 diabetes (T1D). Common as well as unique antibody reactivity patterns were detected between patients and healthy controls. We believe HD-viral-NAPPA will enable the study of host-pathogen interactions at unprecedented dimensions and elucidate the role of pathogen infections in disease development.
Keywords: anti-viral antibodies, HD-NAPPA, juvenile idiopathic arthritis, type 1 diabetes
1 Introduction
Viral infections not only elicit acute symptoms but have been implicated in a variety of chronic illnesses including autoimmune diseases and cancers [1, 2]. One direct consequence of viral infections is the elicitation of antibodies against viral proteins [3]. Detection of these antibodies can facilitate diagnosis and treatment of viral infections [3]. It can further help elucidate the roles of viral infections and the role of specific viral antigens in disease development [4].
ELISA is a traditional method to study anti-viral antibodies; however, it is typically limited to only one or a few protein antigens and often requires significant investment in optimizing antigen production [5]. Even within the same virus, different antigens display markedly different immunogenicity. Sero-reactivity to these antigens may correlate with different clinical parameters and have different clinical utility [6]. The opportunity to gather the information of antibody responses to an entire viral proteome will enable the understanding of the relationship between individual anti-viral antibody responses and clinical parameters and measurements. It is very common that more than one virus has been epidemiologically associated with certain diseases [7]. Comprehensive studies of complete viral proteomes for multiple viruses are impractical with the traditional one-antigen-at-a-time approach. An assay platform which can examine responses to whole proteomes of many viruses could generate a comprehensive overview of responses to viral infections, providing biological log files of past infections, and possibly unveil viral associations with autoimmune diseases or cancers.
Protein arrays provide an ideal tool to profile antibodies in blood against thousands of proteins on a microscopic format [8]. Traditional protein array technology is based on expressing, purifying and printing thousands of different proteins. This is scientifically challenging and labor intensive. NAPPA circumvented these inherent problems by printing plasmids with cDNA encoding each protein instead of the proteins themselves [9]. NAPPA has been successfully used in novel autoantibody biomarker discovery and protein functional studies [10–15].
Standard NAPPA involves just-in-time in situ protein expression from printed cDNAs using in vitro transcription and translation (IVTT)-coupled cell lysates [9, 16, 17]. Expressed tagged proteins are captured on a planar glass surface by co-printed anti-tag antibodies. When feature densities are increased, mRNAs and proteins from one feature start to diffuse to the neighboring features during protein expression resulting in mixed protein display [18]. Diffusion prohibits standard NAPPA from achieving densities higher than 2,500 features per array. HD-NAPPA overcame these challenges by expressing and capturing proteins in arrays of isolated sealed ‘nanowells’ to prevent diffusion and cross-talk between spots [18]. The utility of HD-NAPPA has been demonstrated in commercial antibody target detection and protein-protein interactions [18].
Connections between viral infections and JIA and T1D were supported at the epidemiological, serological and molecular levels [7, 19]. Parvovirus B19 (PB19) and coxsackievirus B4 (CVB) were isolated directly from the synovial tissue of a patient with severe arthritis and the pancreas of a child with diabetic ketoacidosis, respectively [20, 21]. PCR, in-situ hybridization (ISH) and immunohistochemistry (IHC) were employed to detect viral genomes or proteins among JIA and T1D patients [22, 23]. Other immunological methodologies, including plaque, ELISA and complement fixation assays, were applied to measure antibodies specific to viral antigens from various biological samples such as serum, plasma and synovial fluid [24–26]. All these immunoassays depended on the detection of anti-viral antibodies to the intact whole virus or a limited number of viral proteins. This precluded us from acquiring a complete picture of viral infections in JIA and T1D [27, 28].
To characterize the advantages and demonstrate the utility of HD-viral-NAPPA to document past viral infections, we profiled anti-viral antibodies to 761 viral antigens from 25 different viruses in the two most common juvenile autoimmune diseases: JIA and T1D. HD-viral-NAPPA enabled studying anti-viral antibodies in JIA and T1D patients at unprecedented breadth and depth. We first showed high reproducibility of protein display and serum profiling on HD-viral-NAPPA. We further proved HD-viral-NAPPA greatly improved sensitivity in detecting anti-viral antibodies compared to standard NAPPA and ELISA. Unique and common signatures of antiviral antigen antibodies were found. We have clearly demonstrated that HD-viral-NAPPA is a flexible, sensitive and high-throughput platform enabling quantitative measurements of anti-viral antibody levels in infectious and autoimmune diseases.
2 Materials and methods
2.1 Serum samples
T1D samples were collected at the University of Florida with written informed consent and approval of institutional review board (IRB) at the University of Florida. Peripheral blood samples were obtained from T1D patients diagnosed within three months according to the American Diabetes Association (ADA) criteria. Serum was prepared and stored as aliquots at −80°C. T1D controls were age/gender matched family members of patients and tested to be negative for the known T1D autoantibodies (GADA, IA-2A and ZnT8A). JIA samples were collected at Queen’s University of Belfast with the Office for Research Ethics Committees Northern Ireland (ORECNI) approval (ORECNI 408/03). JIA patients and JIA patients with uveitis were matched with disease subtypes and antinuclear antibodies (ANA) status in addition to age/gender. Healthy controls were only age/gender matched to JIA patients or JIA patients with uveitis. Uveitis is the inflammation of the uvea which is regarded as a severe symptom in JIA patients [29]. The sample information is characterized in Table 1.
Table 1.
Sample information: (A) T1D sample characteristics including age, gender and known T1D autoantibodies (AAb) status; (B) JIA sample characteristics including age, gender, number of joints affected and antinuclear antibodies (ANA) status (GADA, IA-2A and ZnT8A).
| (A) | |||
|---|---|---|---|
| Characteristics | New-Onset Patients (n=20) | Healthy Controls (n=20) | |
| Age | Mean±SD | 13.2±4.96 | 13.3±4.94 |
| Median | 13.5 | 13.5 | |
| Gender | Male(%) | (14)70% | (14)70% |
| AAb Status | GADA positive | (14)70% | 0 |
| IA-2A positive | (14)70% | 0 | |
| ZnT8A positive | (11)55% | 0 | |
| (B) | ||||
|---|---|---|---|---|
| Characteristics | JIA (n=10) | JIA with Uveitis (n=10) | Healthy controls (n=10) | |
| Age | Mean± SD | 5.83±3.86 | 4.78±3.43 | 6.17±6.46 |
| Median | 3.86 | 3.51 | 2.71 | |
| Gender | Male(%) | (1)10% | (1)10% | (1)10% |
| Disease Subtype | Oligo | 7 | 7 | NA |
| Ext.Oligo | 1 | 1 | NA | |
| Poly | 2 | 2 | NA | |
| ANA status | Negative | (5)50% | (5)50% | NA |
2.2 HD-viral-NAPPA Fabrication
2.2.1 DNA preparation
Viral genes were cloned into the T7-based mammalian expression vector pANT7_cGST [30–33]. All genes were sequence verified and are publicly available at https://dnasu.org/DNASU/ [31]. The detailed viral gene list was shown in Table S1. Plasmid DNA was extracted and quality assured as described [16, 17]. DNA concentration was normalized to 100 ng/uL before printing.
2.2.2 Silicon nanowell (SiNW) substrate preparation
All SiNW substrates were fabricated at Arizona State University Center for Solid State Electronics Research (CSSER). The detailed procedure for nanowell production was described in [18]. Briefly, isotropic wet etching was used to produce circular nanowells with flat surface at the bottom. Nanowells were 250 μm in diameter and 70 μm in depth. The etched silicon wafers were diced to yield the SiNW substrates the same size as a standard microscope slide. A silicon dioxide layer was thermally grown on the surface and later coated with a 3-Aminopropyltriethoxysilane monolayer for NAPPA chemistry.
2.2.3 Piezoelectric printing in nanowells
HD-viral-NAPPA was printed by au302 piezoelectric dispensing system (Engineering Arts LLC, Tempe, AZ) with integrated alignment system. “On the fly” non-contact dispensing with 16-pin dispensing head was used to dispense DNA/printing mix at 175 mm/sec speed. Each nanowell was filled with 1,200 picoliters of printing mix followed by 300 picoliters of DNA. Each SiNW substrate was equally divided into four sub-arrays. Each viral gene was printed in duplicate within the sub-arrays. Printed arrays were stored desiccated in a nitrogen atmosphere at room temperature until use.
2.3 Protein expression
After printing, SiNW substrates were blocked using SuperBlock TB (Thermo Scientific, Waltham, MA) for 30 min. The substrates were rinsed, dried and placed in an airtight chamber [18] with a flexible film above the substrates. Air in the chamber was removed by vacuum from a fluid gap between the substrate and the flexible film. Human HeLa cell lysate-based IVTT system was injected into the fluid gap by syringe thus filling nanowells with lysate. Excess reagent was swept away from the substrate by flowing pressurized viscous liquid over the flexible film. Individual nanowells were thus sealed into isolated reaction chambers by the flexible film and pressurized viscous liquid. The chamber was placed in an incubator (EchoTherm chilling incubator, Torrey Pines Scientific, Carlsbad, CA) for protein expression at 30°C for 2 hrs and capture at 15°C for 1 hr. Displayed proteins were detected with a monoclonal anti-GST antibody (Cell signaling Inc., Danvers, MA) and Alexa Fluor® 647 goat anti-mouse IgG (H+L) (Life technologies, Carlsbad, CA). Substrates were washed, dried and scanned by Tecan PowerScanner™ (Tecan Group Ltd, Männedorf, Switzerland).
2.4 Serum profiling on HD-viral-NAPPA
Following protein expression, substrates were blocked in 5% milk-PBST (0.2% Tween) for 1 hr and later incubated with serum samples in proplate 4-well tray set (Grace Bio-Labs, Bend, OR) at 4°C overnight. Next day, the substrates were washed and detected by Alexa Fluro® 647-conjugated goat anti-human IgG (Jackson ImmunoResearch, West Grove, PA). Finally, substrates were washed, dried and scanned as described.
2.5 RAPID ELISA
RAPID ELISA was performed to verify the sero-reactivity to viral proteins [34]. Briefly, each well of 96-well ELISA plates (Corning life science, Union City, CA) was coated with 50 μL 10 μg/mL anti-GST antibody (GE Healthcare Life Sciences, Pittsburgh, PA) in coating buffer (0.5 M sodium bicarbonate buffer, pH 9.6) at 4°C overnight. Next day, coated plates were washed with PBST and blocked with 5% milk-PBST (0.2% Tween) for 1.5 hrs. Meanwhile, 40 ng/μL plasmids encoding viral antigens were expressed in the human HeLa cell lysate-based IVTT system at 30°C for 1.5 hrs. Viral proteins were diluted 1:200 in milk-PBST and captured in the wells. Then plates were washed with PBST, incubated with serum samples diluted at 1:1,000, washed with PBST again and incubated with peroxidase AffiniPure goat anti-human IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). Finally, the plates were detected by 1-Step Ultra TMB - ELISA Substrate (Thermo scientific, Waltham, MA) for 10 min and then 2 M sulfuric acid was used to stop the reaction. OD450 was measured by Envision Multilabel Reader (Perkin Elmer, Waltham, MA).
2.6 Statistical analysis
Sample characteristics including age, gender and known autoantibody status were presented as proportion, median and mean with standard deviations. Signal intensities of protein display and serum profiling were extracted by Array-Pro Analyzer (MediaCybernetics, Rockville, MD). Raw signal intensities of protein display were log transformed before comparing their intra-array, intra-batch and inter-batch correlations. To calculate the S/B ratios on the array platform, the background signal was represented by the median of the raw signal intensities of all the antigens on an array probed with a serum sample. For ELISA, the background signal was represented by the median signal of all the antigens for a serum sample probed on the same day. Wilcoxon rank-sum test was used to test the difference of sero-reactivity among subgroups in the JIA sample set. Heatmaps were generated in MultiExperiment Viewer version 4.9 obtained from http://www.tm4.org/. Bar graphs and plots were generated in GraphPad Prism 6 (GraphPad software, La Jolla, CA).
3 Results
3.1 Quality of protein display on HD-viral-NAPPA
We collected 761 viral genes from 25 viral strains to build the HD-viral-NAPPA platform (Table 2). Among these, 16 viral strains had 100% of viral genes available in our collection. During printing, each array was divided into four sub-arrays and each viral gene was printed in duplicate on each sub-array. To confirm the quality of protein display on HD-viral-NAPPA, we compared protein display levels for all ~1,500 features on two sub-arrays from the same printing batch and one sub-array from a separate printing batch. Representative array images and scatter plots of signal intensities of protein display are shown in Figure S1. The average intra-array, intra-batch and inter-batch correlation coefficients of protein display calculated by the log transformed raw signal intensity were 0.92, 0.91 and 0.91, respectively (Figure S2).
Table 2.
Characteristics of viruses including names of virus species, abbreviations, virus family, genome type, the number of open reading frame (ORF) clones and percentage of complete ORFeome.
| Virus Species | Abbre. | Family | Genome | ORF clones | % of complete ORFeome |
|---|---|---|---|---|---|
| Human Cytomegalovirus | HCMV/HHV-5 | Herpesvirinae | dsDNA | 164 | 100% |
| Esptein-Barr virus | EBV/HHV-4 | Herpesvirinae | dsDNA | 85 | 100% |
| Coxsackievirus B | CVB | Picornaviridae | ssRNA+ | 12 | 100% |
| Rubella virus | RUBA | Togaviridae | ssRNA+ | 6 | 100% |
| Mumps virus | MuV | Paramyxoviridae | ssRNA− | 9 | 100% |
| Human endogenous retrovirus K | HERK | Retroviridea | ssRNA− | 4 | 100% |
| Rotaviruses | RV | Reoviridae | dsRNA | 12 | 100% |
| Parvovirus B19 | B19 | Parvoviridae | ssDNA | 6 | 100% |
| Hepatitis B virus | HBV | Herpesviridae | dsDNA | 10 | 100% |
| Human Papillomavirus16 | HPV16 | Papillomaviridae | dsDNA | 10 | 100% |
| Human Papillomavirus18 | HPV18 | Papillomaviridae | dsDNA | 9 | 100% |
| Chikungunya virus | CHIKV | Togaviridae | ssRNA+ | 9 | 100% |
| Semliki Forest virus | SFV | Togaviridae | ssRNA+ | 9 | 100% |
| Sindbis virus | SINV | Togaviridae | ssRNA+ | 9 | 100% |
| Influenza A virus (H1N1) | n/a | Orthomyxoviridae | ssRNA− | 10 | 100% |
| Influenza A virus (H3N2) | n/a | Orthomyxoviridae | ssRNA− | 10 | 100% |
| Varicella-zoster virus | VZV | Herpeviridae | dsDNA | 91 | 93.10% |
| Simian virus 40 | SV40 | Polyomaviridae | dsDNA | 6 | 85.71% |
| Vaccinia virus | VACV | Poxviridae | dsDNA | 167 | 74.90% |
| Yellow fever virus | YF | Flaviviridae | ssRNA+ | 11 | 71.42% |
| Measles virus, vaccine strain | MeV, vaccine | Paramyxoviridae | ssRNA− | 5 | 62.50% |
| Measles virus, WT strain | MeV, WT | Paramyxoviridae | ssRNA− | 5 | 62.50% |
| Herpes simplex virus 1 | HSV-1 | Herpesviridae | dsDNA | 83 | 61.90% |
| Adenovirus | n/a | Adenoviridae | dsDNA | 16 | 42.10% |
| Tioman virus | n/a | Paramyxoviridae | ssRNA− | 3 | 37.50% |
3.2 Quality of serum profiling on HD-viral-NAPPA
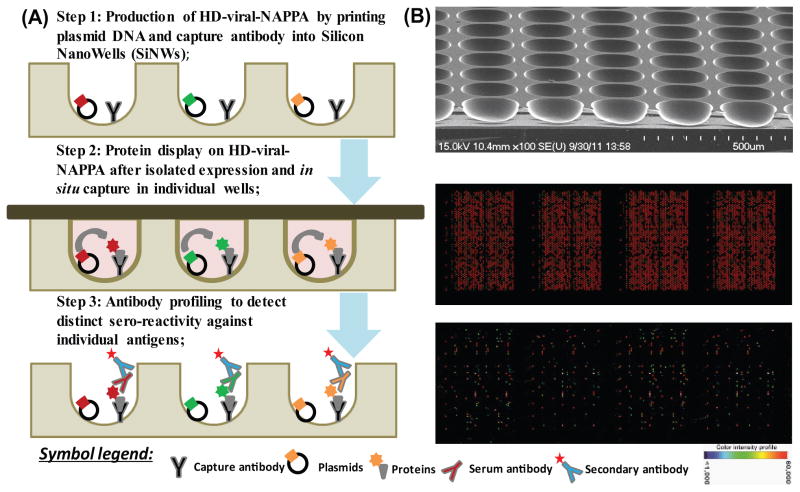
The work flow of serum profiling on HD-viral-NAPPA includes the production of HD-viral-NAPPA, protein display and antibody profiling to detect sero-reactivity against individual antigens (Figure 1). To demonstrate the quality of serum profiling, we applied the same pair of T1D patient and control samples on two arrays from the same printing batch and one array from a separate printing batch. Four sub-arrays of each array (1,522 features, 761 unique viral genes) were probed twice with the patient sample and twice with the control sample. The average correlation coefficients of intra-array, intra-batch and inter-batch for both samples were all above 0.95. The results were highly reproducible as shown by the representative array images and the correlation coefficients between sub-arrays in Figure S3.
Figure 1.
Antibody profiling on HD-viral-NAPPA. A. Step 1: Production of HD-viral-NAPPA by printing plasmid DNA and capture antibody into Silicon NanoWells (SiNWs); Step 2: Protein display on HD-viral-NAPPA after isolated expression and in situ capture in individual wells; Step 3: Antibody profiling to detect distinct sero-reactivity against individual antigens; B. Images of a SiNW substrate by scanning electron microscopy (top), proteins displayed on HD-viral-NAPPA as detected by anti-GST antibody (middle), and sero-profiling on HD-viral-NAPPA (bottom).
3.3 Higher sensitivity in detecting antibodies on HD-NAPPA
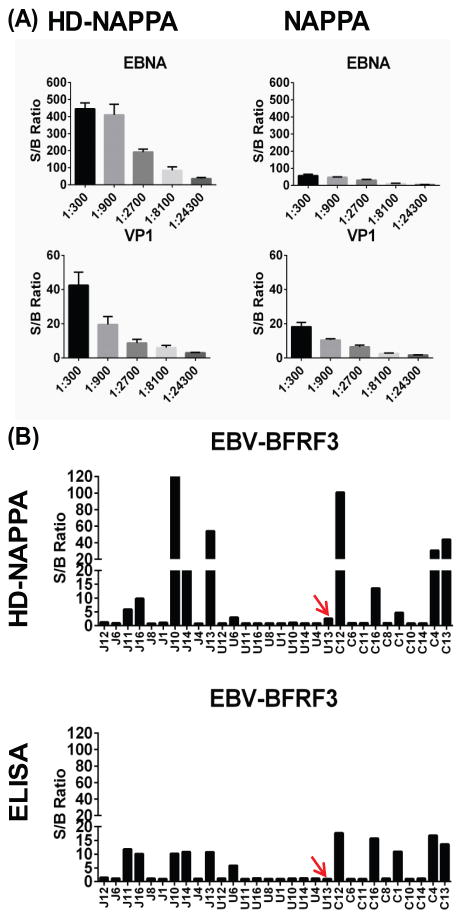
We compared assay sensitivity of sero-reactivity on HD-NAPPA and standard NAPPA. The same set of 190 unique genes was printed on both glass, using the standard NAPPA method [9, 17], and the SiNW surface, using the HD-NAPPA method [18]. We applied a serum sample at three-fold serial dilutions from 1:300 to 1:24,300 on both platforms. The S/B ratio was used to assess sero-positivity, which was the signal of any feature divided by the median signal on the same array, because the majority of features on the array were considered non-responses (see Materials and methods). We found an increase in the S/B ratio as high as 9 times for anti-EBNA1 response from Epstein-Barr virus (EBV) at 1:900 dilution and more than 2 times for anti-VP1 (CVB) response at 1:300 dilution when comparing HD-NAPPA with standard NAPPA (Figure 2A).
Figure 2.
Higher sensitivity in detecting anti-viral antibodies on HD-NAPPA. (A) S/B ratios of sero-reactivity to EBNA1 (EBV) and VP1 (CVB) at serial dilutions on HD-NAPPA and NAPPA; (B) S/B ratios of sero-reactivity to BFRF3 (EBV) among JIA sample set on HD-NAPPA and ELISA.
We then profiled sero-reactivity to the BFRF3 antigen of EBV among the JIA sample set on HD-viral-NAPPA and by ELISA to compare their ability to measure positive sero-reactivity. Overall, sero-positivity for all 30 samples agreed well on both platforms (Fig. 3B) except for sample U13, which showed S/B >2 on HD-viral-NAPPA but was not distinguishable from background by ELISA (red arrows on Figure 2B). Additional tests are needed to further confirm the sero-positivity. Sample J10 had more than 12 times higher S/B ratios on HD-viral-NAPPA than that of ELISA. The wide S/B ratio range made HD-viral-NAPPA a more sensitive immunoassay platform in distinguishing sero-positive from sero-negative samples than standard NAPPA or ELISA (Figure 2).
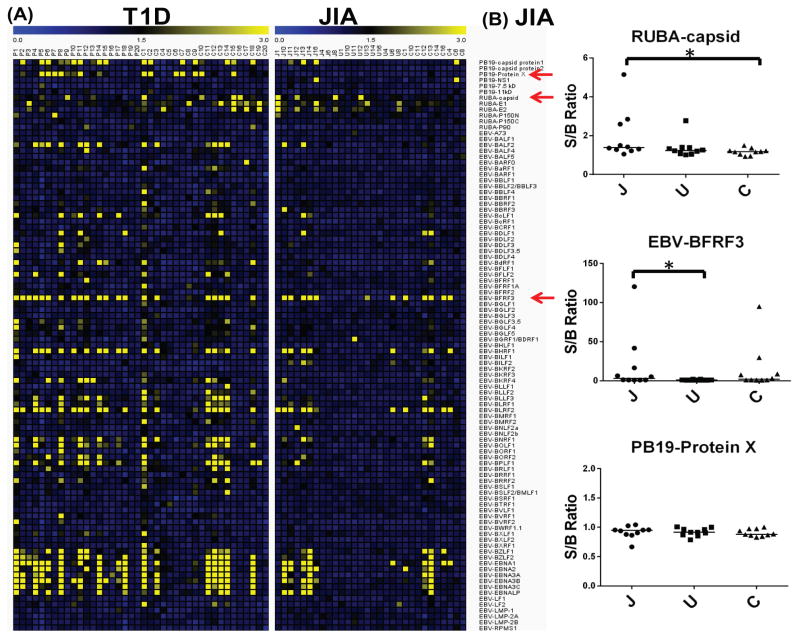
Figure 3.
Anti-viral antibodies to the three common viruses (PB19, RUBA and EBV) in JIA and T1D sample sets. (A) Heatmaps of S/B ratios of sero-reactivity to viral proteins from the three viruses on HD-viral-NAPPA in T1D and JIA sample sets; (B) Jitter plots of sero-reactivity to representative viral antigens (RUBA-capsid, EBV-BFRF3 and PB19-protein X) on HD-viral-NAPPA in JIA sample set.
3.4 Anti-viral antibodies of three common viruses in JIA
We analyzed sero-reactivity to 761 viral antigens from 25 viruses by HD-viral-NAPPA using JIA and T1D samples (Table 2). The T1D samples included 20 new onset patients and 20 healthy controls. The percentage of males with T1D was 70%, which is slightly higher than the expected distribution, probably due to sampling error, but was appropriately matched in our controls. The JIA samples included 10 patients, 10 patients with the symptom of uveitis and 10 healthy controls. In the case of JIA, the percentage of males in the cases was 10%, reflecting the well-documented gender disparity of JIA, which primarily affects females [35]. The controls for JIA were appropriately gender matched. Among 25 viral strains, PB19, rubella virus (RUBA) and EBV are the most reported viruses associated with JIA. The heatmaps of sero-reactivity to the three viruses for both diseases were generated (Figure 3A). The power of a proteomic approach in discovering autoimmune disease-associated viral infections is the opportunity to test all possible viral antigens to see which one gives the best response. This could not be achieved by a more conventional one-antigen-at-a-time approach. In the JIA sample set, sero-reactivity to the capsid protein (RUBA) was significantly higher in JIA patients than healthy controls by Wilcoxon rank-sum test (p<0.05; Figure 3B); whereas, no difference of sero-reactivity to other proteins (E1, E2 and P150) from RUBA was observed. JIA patients with uveitis had less sero-reactivity to most EBV proteins compared to the other subgroups. In the T1D sample set, the prevalence of antibodies to EBV proteins was higher in patients than controls (Figure 3A).
3.5 Profiles of anti-viral antibodies in JIA and T1D samples
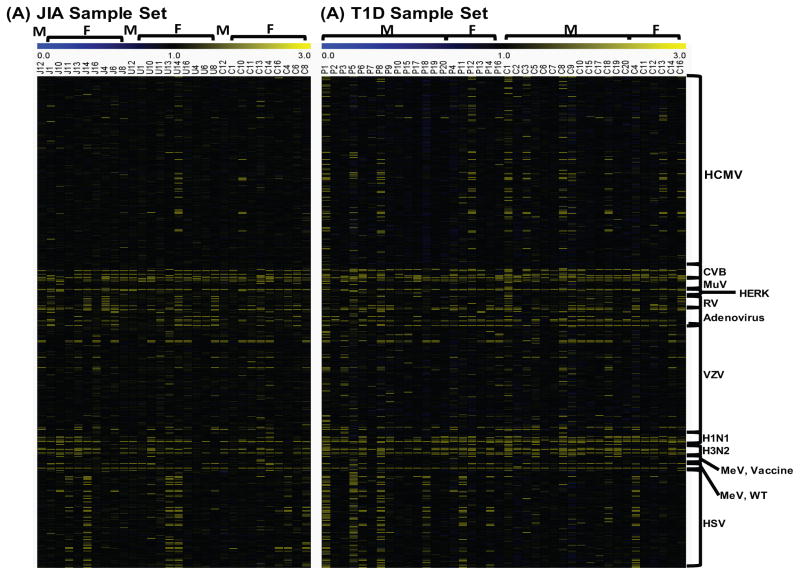
Profiles of anti-viral antibodies were generated on HD-viral-NAPPA and illustrated in the heatmaps (Figure 4). Overall, most samples (both cases and controls) were positive for CVB, RUBA, mumps virus (MuV), rotavirus (RV), adenovirus, influenza A virus and measles virus (MeV), while negative for Human endogenous retrovirus K (HERK) and varicella zoster virus (VZV). The JIA samples have less sero-reactivity to human cytomegalovirus (HCMV) proteins compared with T1D samples which may result from the age and geographic difference between these two sample collections. Interestingly, antibody response to herpes simplex virus (HSV) was found to be more prevalent in both JIA and T1D patients than healthy controls.
Figure 4.
Profiles of anti-viral antibodies to the other viruses in JIA and T1D patients. (A) JIA sample set, J represents JIA patients, U represents JIA patients with uveitis, C represents healthy controls; (B) T1D sample set, P represents T1D patients, C represents healthy controls. M stands for males, F stands for females.
4 Discussion
Host innate and adaptive immunity works cooperatively to fight against viral infections [11]. Antibodies recognizing antigen proteins are produced during this process. Detection and quantification of these antibodies will aid sero-diagnosis of infections, design of preventive vaccines, discovery of innovative therapeutics and monitoring of anti-viral treatments [3]. Traditional methods focusing on one-protein/one-virus at a time suffer greatly from the limitation of throughput. Protein arrays, as one of the key innovations in the era of functional proteomics, provide an ideal tool to profile antibody response to thousands of proteins on a microscopic slide in a multiplexed manner [8]. NAPPA, as a robust in situ cell free protein array platform, prints full length cDNAs on the arrays instead of purified proteins [9].
HD-NAPPA achieved higher density and less diffusion by expressing and capturing proteins in isolated sealed ‘nanowells’ [18]. It shares some common advantages with standard NAPPA. First, antigens are expressed by a HeLa cell lysate-based IVTT system, yielding high expression levels and functional conformations of displayed proteins. Second, the ability to reconfigure new arrays allows exploration of sero-reactivity to any antigens from new pathogens, new strains or new mutants in diseases as they emerge. This is especially important for microbial studies because conventional protein arrays for human and yeast, although expensive, are at least commercially available; but, similar arrays are not available for microorganisms. Third, both assays are highly reproducible and the turnaround time for one assay for thousands of proteins is as short as one day.
In addition, HD-NAPPA introduces some unique advantages. First, based on its high density feature, as many as 24,000 proteins can be tested simultaneously compared to only 2,500 proteins by standard NAPPA on each array. Second, the high density nature reduces the sample amount needed for assaying each protein, thus preserving precious resources. Third, here we demonstrated that HD-NAPPA had higher sensitivity and better S/B in detecting antibody responses compared with standard NAPPA and ELISA. Fourth, HD-NAPPA protocols use much less DNA per printing batch when compared to standard NAPPA, which means that many more samples can be tested based on a single round of DNA preparation and reducing cost of array production. Fifth, HD-NAPPA has negligible diffusion between neighboring spots reducing false positives during serum screening.
Aside from the above advantages, there are some limitations of NAPPA technology. Although it is easier and more robust to prepare and print DNA than to express, purify and print proteins, it is nevertheless beyond the means for most general research labs to work with thousands of genes/proteins at the genome/proteome level. Collaboration or fee-for-service (such as http://nappaproteinarray.org/) might make more sense in this setting. Fast and accurate noncontact piezoelectric dispensing systems capable of targeting nanowells are required for printing HD-NAPPA but are inaccessible to most researchers. An affordable user-friendly non-contact printing instrument is currently in development to allow widespread adoption of HD-NAPPA. Finally, it should be noted that this platform is well-suited for screening for possible interactions or responses at the proteome level. But, all candidates that emerge from such screens must be confirmed by orthogonal methods and, where relevant, with in vivo assays.
In this study, HD-NAPPA showed superior S/B ratio to standard NAPPA from a direct comparison of sero-reactivity to the same set of genes on these two platforms. On average, we obtained more than a seven fold increase in the S/B ratio of anti-EBNA1 sero-reactivity at various sample dilutions (Figure 2A). The S/B ratio of HD-NAPPA also compares favorably to reported serum antibody studies using purified protein arrays [36] or Luminex bead arrays [37]. Thus, this tool detects serum antibodies at high sensitivity and can potentially improve the accuracy of clinical studies.
To demonstrate the great utility of HD-NAPPA, we constructed HD-viral-NAPPA containing 761 protein antigens from 25 viral strains. There are over 10,000 nanowells per SiNW array in our current configuration [18]. ORFs for all 761 viral antigens were printed in the nanowells, in duplicate, four times to produce four sub-arrays on one array. This allowed for the profiling of the sero-reactivity against all viral antigens of four serum samples on one array in parallel. The quality of protein display was evaluated by array-to-array reproducibility. We observed 0.64%, 4.16 % and 3.81% of features with CVs higher than 20% using the log transformed raw signal intensities when comparing intra-slide, inter-slide and inter-batch correlations, respectively. Preliminary studies suggest that this may be due to imperfect dispensing of the same amount of DNA into the nanowells, resulting in inconsistent protein display on arrays from two different arrays. We are currently modifying our piezoelectric printing protocols to achieve higher dispensing reproducibility. Nonetheless, this affected a very small fraction of the features on the array and was mitigated somewhat by having duplicate spots for each gene on the array.
We profiled anti-viral antibodies in the two most common juvenile autoimmune diseases, JIA and T1D, as a demonstration of HD-NAPPA in serological studies. It has long been suspected that certain types of viral infections are involved in the development of these two autoimmune diseases [7, 38]. T1D controls were collected in the United States (US) as T1D patient samples, and JIA controls were collected in the United Kingdom (UK) as JIA patient samples. In our antiviral antibody profile, the prevalence of sero-reactivity to HSV was higher in both JIA and T1D patients than healthy controls while the sero-reactivity to EBV was more prevalent only in T1D patients. The percentage of males is lower than females in JIA, it is the opposite in T1D. No obvious differences of anti-viral antibody profiles were seen between the genders. Although our sample size is too small to draw any statistical conclusions, this may suggest a potential role of these viral infections in JIA or T1D development. Interestingly, we found the antibody responses to EBV proteins were higher in JIA patients and healthy controls than JIA patients with uveitis. It has been reported that JIA patients went into remission after EBV infection [39]. Therefore, the potential protection of EBV infection against uveitis in JIA patients warrants future investigation.
Our protein array platform detects multiplexed antibody responses to viral antigens from the whole viral proteome. Many viruses undergo latency after active/lytic infection and may subsequently become reactivated after a latent stage [3]. Different viral antigens are preferentially expressed depending on these different states. Thus, both the antibody response and its magnitude to specific viral antigens may provide clues to the different stages of viral infection [40]. In cases where the lifecycle of a virus is well understood, a viral proteome array can be revealing. For example, EBV commonly enters latency in adults and in that state continues to produce the EBNA protein, which may explain the strong IgG response observed to that protein in adults [40]. The prevalence of strong anti-EBNA responses is much less common in children (unpublished observation). For less understood viruses, the inclusion of the whole viral proteome allows the investigation of every viral antigen that is linked to either active/lytic or latent stages of viral infection. Furthermore, IgM is often used to detect acute infection while IgG detects past infection. Our array platform allows multiplexed detection of IgG and IgM using secondary antibodies labeled with two different fluorescent dyes. In summary, the inclusion of all viral antigens from the complete viral proteome and the ability of multiplexed detection of IgM and IgG may enable the discrimination between active/lytic or latent viral infection.
In summary, we have successfully demonstrated the utility of HD-viral-NAPPA in profiling antiviral antibodies in diseases. The small sample size used in this study may preclude reaching strong statistical conclusions. However, we generated high quality serological profiles in 70 subjects; observed reactivity differences among them and confirmed results with orthogonal ELISA data. We believe HD-NAPPA may be applied to large scale anti-microbial antibody studies and help us better understand the role of microbial infections in various autoimmune diseases.
Supplementary Material
Figure S1. Quality of protein display on HD-viral-NAPPA. (A) Visual representative images of HD-viral-NAPPA protein display on slides within the same printing batch and across different printing batches; (B) Scatter plot of log transformed signal intensities for protein display on slides within the same printing batch and across different printing batches.
Figure S2. Correlation coefficients of protein display from sub-arrays printed within the same batch and across separated batches.
Figure S3. Quality of serum profiling on HD-viral-NAPPA. (A) Visual representative images for serum profiling on two arrays from the same printing batch and one array from a different printing batch; (B) Correlation coefficients for intra-array and inter-array correlation within the same printing batch, inter-array correlation across different printing batch. Sample#1 is a T1D patient sample, sample#2 is a healthy control sample.
Acknowledgments
This work is partially funded by grant number 10489084 awarded by the National Institutes of Health to Engineering Arts LLC and partially funded by the Juvenile Diabetes Research Foundation (JDRF) innovative grant 5-2012-537. The authors wish to thank Dr. Roivainen Merja (National Institute of Health and Welfare, Finland), Dr. Tom Hobman (University of Alberta, Canada), Dr. Biao He (University of Georgia, USA), Dr. Marc Vidal (Harvard Medical School, USA), Dr. David E Hill (Dana-Farber Cancer Institute, USA), Dr Jürgen Haas (University of Edinburgh, UK), Dr. James M. Pipas (University of Pittsburgh, USA) and Dr. Vincent Lotteau (Université de Lyon, France) for kindly providing viral templates/clones. We acknowledge Desmond Schatz and Mark Atkinson (University of Florida, Gainesville, FL) for valuable discussion of clinical and biology characteristics of T1D samples.
Abbreviations (non-standard)
- HD - NAPPA
high density-nucleic acid programmable protein array
- S/B
signal to background
- JIA
juvenile idiopathic arthritis
- T1D
type 1 diabetes
- IVTT
in vitro transcription and translation
- PB19
Parvovirus B19
- CVB
Coxsackievirus B4
- ISH
in-situ hybridization
- IHC
Immunohistochemistry
- IRB
Institutional review board
- ADA
American Diabetes Association
- ORECNI
Office for Research Ethics Committees Northern Ireland
- ANA
Antinuclear Antibodies
- SiNW
Silicon NanoWell
- CSSER
Center or Solid State Electronics Research
- RAPID ELISA
Rapid antigenic protein in situ display ELISA
- EBV
Epstein-Barr virus
- RUBA
Rubella virus
- MuV
Mumps virus
- RV
Rotavirus
- HERK
Human endogenous retrovirus K
- VZV
Varicella zoster virus
- HCMV
Human cytomegalovirus
- HSV
Herpes simplex virus
Footnotes
Conflict of interest statement
All authors declare no financial/commercial conflicts of interest.
References
- 1.Fujinami RS, von Herrath MG, Christen U, Whitton JL. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clin Microbiol Rev. 2006;19:80–94. doi: 10.1128/CMR.19.1.80-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Odumade OA, Hogquist KA, Balfour HH., Jr Progress and problems in understanding and managing primary Epstein-Barr virus infections. Clin Microbiol Rev. 2011;24:193–209. doi: 10.1128/CMR.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cepok S, Zhou D, Srivastava R, Nessler S, et al. Identification of Epstein-Barr virus proteins as putative targets of the immune response in multiple sclerosis. J Clin Invest. 2005;115:1352–1360. doi: 10.1172/JCI23661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Athmaram TN, Saraswat S, Misra P, Shrivastava S, et al. Optimization of Dengue-3 recombinant NS1 protein expression in E. coli and in vitro refolding for diagnostic applications. Virus Genes. 2013;46:219–230. doi: 10.1007/s11262-012-0851-5. [DOI] [PubMed] [Google Scholar]
- 6.Schaade L, Kleines M, Hausler M. Application of virus-specific immunoglobulin M (IgM), IgG, and IgA antibody detection with a polyantigenic enzyme-linked immunosorbent assay for diagnosis of Epstein-Barr virus infections in childhood. J Clin Microbiol. 2001;39:3902–3905. doi: 10.1128/JCM.39.11.3902-3905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun HS, Yoon JW. A new look at viruses in type 1 diabetes. Ilar J. 2004;45:349–374. doi: 10.1093/ilar.45.3.343. [DOI] [PubMed] [Google Scholar]
- 8.Sutandy FX, Qian J, Chen CS, Zhu H. Overview of protein microarrays. Curr Protoc Protein Sci. 2013;Chapter 27(Unit 27):21. doi: 10.1002/0471140864.ps2701s72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, et al. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008;5:535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceroni A, Sibani S, Baiker A, Pothineni VR, et al. Systematic analysis of the IgG antibody immune response against varicella zoster virus (VZV) using a self-assembled protein microarray. Mol Biosyst. 2010;6:1604–1610. doi: 10.1039/c003798b. [DOI] [PubMed] [Google Scholar]
- 11.Anderson KS, Sibani S, Wallstrom G, Qiu J, et al. Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J Proteome Res. 2011;10:85–96. doi: 10.1021/pr100686b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson DS, Qiu J, Mendoza EA, Barker K, et al. Circulating and synovial antibody profiling of juvenile arthritis patients by nucleic acid programmable protein arrays. Arthritis Res Ther. 2012;14:R77. doi: 10.1186/ar3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright C, Sibani S, Trudgian D, Fischer R, et al. Detection of Multiple Autoantibodies in Patients with Ankylosing Spondylitis Using Nucleic Acid Programmable Protein Arrays. Molecular & Cellular Proteomics. 2012;11 doi: 10.1074/mcp.M9.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miersch S, Bian X, Wallstrom G, Sibani S, et al. Serological autoantibody profiling of type 1 diabetes by protein arrays. J Proteomics. 2013;94:486–496. doi: 10.1016/j.jprot.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Yu X, Woolery AR, Luong P, Hao YH, et al. Click chemistry-based detection of global pathogen-host AMPylation on self-assembled human protein microarrays. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M114.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu J, LaBaer J. Nucleic acid programmable protein array a just-in-time multiplexed protein expression and purification platform. Methods Enzymol. 2011;500:151–163. doi: 10.1016/B978-0-12-385118-5.00009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miersch S, LaBaer J. Nucleic Acid programmable protein arrays: versatile tools for array-based functional protein studies. Curr Protoc Protein Sci. 2011;Chapter 27(Unit 27):22. doi: 10.1002/0471140864.ps2702s64. [DOI] [PubMed] [Google Scholar]
- 18.Takulapalli BR, Qiu J, Magee DM, Kahn P, et al. High density diffusion-free nanowell arrays. J Proteome Res. 2012;11:4382–4391. doi: 10.1021/pr300467q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franssila R, Hedman K. Viral causes of arthritis. Best Pract Res Cl Rh. 2006;20:1139–1157. doi: 10.1016/j.berh.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Yoon JW, Austin M, Onodera T, Notkins AL. Isolation of a virus from the pancreas of a child with diabetic ketoacidosis. N Engl J Med. 1979;300:1173–1179. doi: 10.1056/NEJM197905243002102. [DOI] [PubMed] [Google Scholar]
- 21.Dijkmans BA, van Elsacker-Niele AM, Salimans MM, van Albada-Kuipers GA, et al. Human parvovirus B19 DNA in synovial fluid. Arthritis Rheum. 1988;31:279–281. doi: 10.1002/art.1780310218. [DOI] [PubMed] [Google Scholar]
- 22.Pak CY, Eun HM, McArthur RG, Yoon JW. Association of cytomegalovirus infection with autoimmune type 1 diabetes. Lancet. 1988;2:1–4. doi: 10.1016/s0140-6736(88)92941-8. [DOI] [PubMed] [Google Scholar]
- 23.Foy CA, Quirke P, Williams DJ, Lewis FA, et al. A Search for Candidate Viruses in Type-1 Diabetic Pancreas Using the Polymerase Chain-Reaction. Diabetic Med. 1994;11:564–569. doi: 10.1111/j.1464-5491.1994.tb02036.x. [DOI] [PubMed] [Google Scholar]
- 24.Gamble DR, Kinsley ML, FitzGerald MG, Bolton R, Taylor KW. Viral antibodies in diabetes mellitus. Br Med J. 1969;3:627–630. doi: 10.1136/bmj.3.5671.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banatvala JE, Bryant J, Schernthaner G, Borkenstein M, et al. Coxsackie B, mumps, rubella, and cytomegalovirus specific IgM responses in patients with juvenile-onset insulin-dependent diabetes mellitus in Britain, Austria, and Australia. Lancet. 1985;1:1409–1412. doi: 10.1016/s0140-6736(85)91843-4. [DOI] [PubMed] [Google Scholar]
- 26.Oikarinen S, Tauriainen S, Hober D, Lucas B, et al. Virus Antibody Survey in Different European Populations Indicates Risk Association Between Coxsackievirus B1 and Type 1 Diabetes. Diabetes. 2014;63:655–662. doi: 10.2337/db13-0620. [DOI] [PubMed] [Google Scholar]
- 27.Craig ME, Nair S, Stein H, Rawlinson WD. Viruses and type 1 diabetes: a new look at an old story. Pediatr Diabetes. 2013;14:149–158. doi: 10.1111/pedi.12033. [DOI] [PubMed] [Google Scholar]
- 28.Clarris BJ. Viral arthritis and the possible role of viruses in rheumatoid arthritis. Aust N Z J Med. 1978;8(Suppl 1):40–43. doi: 10.1111/j.1445-5994.1978.tb04783.x. [DOI] [PubMed] [Google Scholar]
- 29.Thurn J. Human parvovirus B19: historical and clinical review. Rev Infect Dis. 1988;10:1005–1011. doi: 10.1093/clinids/10.5.1005. [DOI] [PubMed] [Google Scholar]
- 30.Yu X, Bian X, Throop A, Song L, et al. Exploration of panviral proteome: high-throughput cloning and functional implications in virus-host interactions. Theranostics. 2014;4:808–822. doi: 10.7150/thno.8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seiler CY, Park JG, Sharma A, Hunter P, et al. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 2014;42:D1253–1260. doi: 10.1093/nar/gkt1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cormier CY, Park JG, Fiacco M, Steel J, et al. PSI:Biology-materials repository: a biologist’s resource for protein expression plasmids. J Struct Funct Genomics. 2011;12:55–62. doi: 10.1007/s10969-011-9100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Festa F, Steel J, Bian X, Labaer J. High-throughput cloning and expression library creation for functional proteomics. Proteomics. 2013;13:1381–1399. doi: 10.1002/pmic.201200456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson KS, Ramachandran N, Wong J, Raphael JV, et al. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J Proteome Res. 2008;7:1490–1499. doi: 10.1021/pr700804c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thierry S, Fautrel B, Lemelle I, Guillemin F. Prevalence and incidence of juvenile idiopathic arthritis: a systematic review. Joint Bone Spine. 2014;81:112–117. doi: 10.1016/j.jbspin.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Robinson WH, DiGennaro C, Hueber W, Haab BB, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 37.Wong J, Sibani S, Lokko NN, LaBaer J, Anderson KS. Rapid detection of antibodies in sera using multiplexed self-assembling bead arrays. J Immunol Methods. 2009;350:171–182. doi: 10.1016/j.jim.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravelli A, Martini A. Juvenile idiopathic arthritis. Lancet. 2007;369:767–778. doi: 10.1016/S0140-6736(07)60363-8. [DOI] [PubMed] [Google Scholar]
- 39.Kawada J, Ito Y, Torii Y, Kimura H, Iwata N. Remission of juvenile idiopathic arthritis with primary Epstein-Barr virus infection. Rheumatology (Oxford) 2013;52:956–958. doi: 10.1093/rheumatology/kes299. [DOI] [PubMed] [Google Scholar]
- 40.Klutts JS, Ford BA, Perez NR, Gronowski AM. Evidence-based approach for interpretation of Epstein-Barr virus serological patterns. J Clin Microbiol. 2009;47:3204–3210. doi: 10.1128/JCM.00164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Quality of protein display on HD-viral-NAPPA. (A) Visual representative images of HD-viral-NAPPA protein display on slides within the same printing batch and across different printing batches; (B) Scatter plot of log transformed signal intensities for protein display on slides within the same printing batch and across different printing batches.
Figure S2. Correlation coefficients of protein display from sub-arrays printed within the same batch and across separated batches.
Figure S3. Quality of serum profiling on HD-viral-NAPPA. (A) Visual representative images for serum profiling on two arrays from the same printing batch and one array from a different printing batch; (B) Correlation coefficients for intra-array and inter-array correlation within the same printing batch, inter-array correlation across different printing batch. Sample#1 is a T1D patient sample, sample#2 is a healthy control sample.