Abstract
Fatty acid biosynthesis is essential to life and represents one of the most conserved pathways in Nature, preserving the same handful of chemical reactions over all species. Recent interest in the molecular details of the de novo fatty acid synthase (FAS) has been heightened by demand for renewable fuels and the emergence of multidrug resistant bacterial strains. Central to FAS is the acyl carrier protein (ACP), a protein chaperone that shuttles the growing acyl chain between catalytic enzymes within the FAS. Human efforts to alter fatty acid biosynthesis for oil production, chemical feedstock or antimicrobial purposes has been met with limited success in part due to a lack of detailed molecular information behind the ACP-partner protein interactions inherent to the pathway. This review will focus on recently developed tools for the modification of ACP and analysis of protein-protein interactions, such as mechanism-based crosslinking, and the studies exploiting them. Discussion specific to each enzymatic domain focuses first on mechanism and known inhibitors, followed by available structures and known interactions with ACP. While significant unknowns remain, new understandings into the intricacies of FAS point to future advances in manipulating this complex molecular factory.
Keywords: Acyl Carrier Protein, biosynthesis, crosslinkers, fatty acids, inhibitors
1. Overview of Fatty Acid Biosynthesis
The de novo fatty acid synthase (FAS) is ubiquitous in nature and critical for life. New technologies have emerged to coincide with renewed interest in FAS because of recent societal demands. One such demand is the sustainable production of hydrocarbon fuels and feedstocks, due to dwindling supplies of easily accessible fossil fuels. As the central metabolic foundry for hydrocarbon production in Nature, the FAS is seen by many as the first target for engineering diverse hydrocarbon production via synthetic biology. Another pressing concern is the need for new antibiotics. Enzymes within the FAS pathway offer untapped targets for future inhibitor development, as the arms race against antibiotic resistance requires new drugs against such linchpin targets. Luckily, tools for interrogating and engineering the FAS have emerged in recent years to help address bottlenecks in our understanding. While significant work in the 20th century identified and investigated the proteins involved in FAS, only recently has it become possible, through structural biology combined with mechanistic probes and advanced kinetics, to interrogate discrete enzymes during productive interactions. This review will focus on such new tools and progress to date that will inform meaningful future developments.
As a very well-studied system, much of the work discussed in this review will focus on the Escherichia coli FAS, with care taken to denote other organisms as necessary. A thorough understanding of FASs will allow for new engineering and exploitation, and many of the tools discussed here should also prove useful in the study of FAS-related secondary metabolites from polyketide synthases (PKS). PKSs are beyond the scope of this review, and we refer to recent reviews from Khosla and Keatinge-Clay.[1–7]
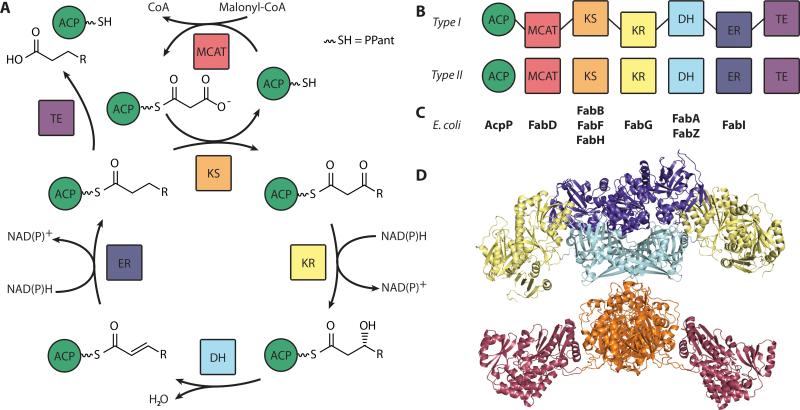
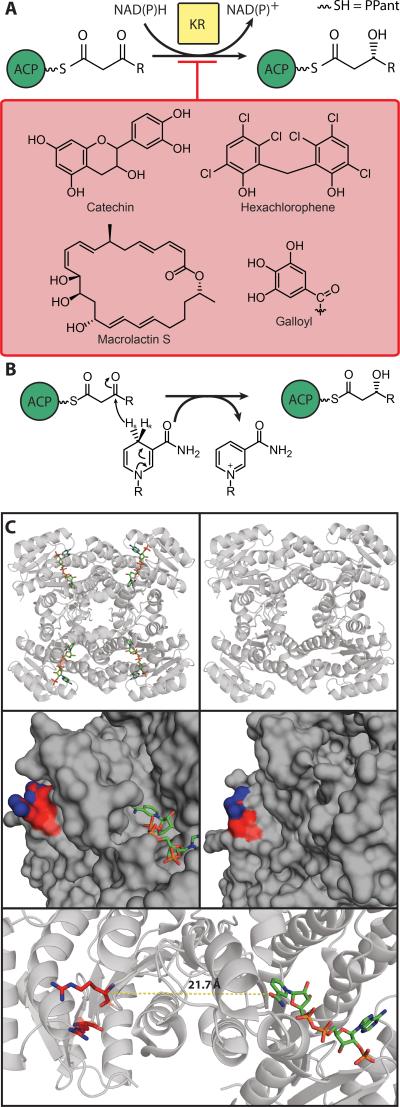
Fatty acid biosynthesis (FAB) is an iterative series of enzyme reactions (Figure 1a) in which an acyl chain is extended by two carbon units with each cycle. Initiation occurs when an acyl carrier protein (ACP) is charged with a two-carbon starting unit by a transacylase. A ketosynthase (KS) elongates the chain by two carbons via decarboxylative addition using malonyl-ACP as a substrate. The resulting ketone is reduced to an (R)-alcohol by a ketoreductase (KR), which is then eliminated to a trans-alkene by a dehydratase (DH). A final reduction step by an enoyl reductase (ER) yields an elongated acyl chain, which can then be subjected to the same cycle again until the desired chain length is reached. Termination is achieved via chain release by either a thioesterase (TE) or chain transfer by an acyl transferase (AT) for incorporation into fats, lipids and other metabolites.
Figure 1.
A) Schematic diagram of the FAB cycle. B) Comparison of domain organization in type I and type II FAS systems. C) Nomenclature of E. coli Fab enzymes, listed underneath their domain activities. D) A color-coded diagram of the type I mammalian FAS, demonstrating an overall “gingerbread-man” topology (PDB: 2VZ8). The TE and the ACP are not observed due to dynamics.
FASs are classified into two categories, type I and type II (Figure 1b). Type I FAS systems use large, multidomain proteins arranged into a single complex such that a tethered ACP can access all required active sites for iteration and synthesis. Type I FAS complexes are common in animals and fungi. For example, the fungal type I FAS is encoded by two genes that assemble as a α6β6 heterododecamer reaching 2.6 MDa (PDB: 2UVB, 2UVC).[8] The mammalian type I FAS expresses as a single protein and assembles as a homodimer of 540 kDa (PDB: 2VZ8).[9]
In contrast, type II FASs employ many single domains expressed as discrete proteins in the cytosol. Type II FASs are common in bacteria and eukaryotic organelles, notably chloroplast and mitochondria. Large amounts of FAS proteins are observed in type II systems, with the central and well-characterized E. coli ACP[10] comprising up to 0.25% of soluble protein during logarithmic growth.[11] The ACP shuttles the growing chain through each step of the elongation cycle and must find the correct reaction partners in the crowded cytosol. This necessitates multiple, low affinity protein-protein interactions, involving over 30 partners once regulatory and other primary and secondary metabolic interactions are considered.[12,13]
FAB in archaea is largely uncharacterized, and its existence is controversial due to archaeal membrane phospholipids lacking fatty acids.[14] In 2012, homologs of components of bacterial FAS II were found in the archaeal genome, with the exception of ACP, suggesting the possibility of ACP-independent FAB.[15] In 2014, archaeal fatty acid biosynthesis was proposed to be performed by bacterial-type enzymes of fatty acid β-oxidation and an enzyme from the mevalonate biosynthesis pathway.[16]
2. The Acyl Carrier Protein – An Extended Toolkit
Due to ACP's central position in FAB, consideration of ACP-partner protein interactions is a critical component of contemporary studies of FASs. New tools to examine and modify ACPs have been developed to facilitate this over the past decade.
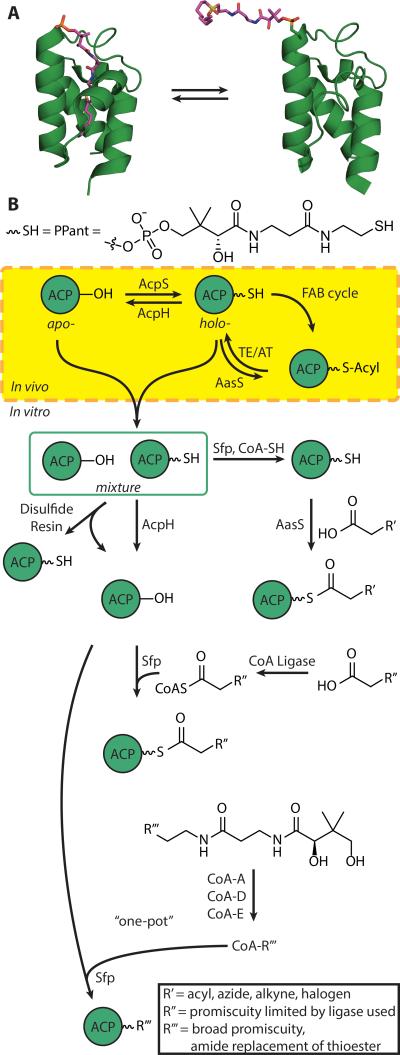
Classically, ACPs are small (~9 kDa) α-helical proteins. When part of a type I megasynthase, the ACP domain is attached by flexible N- and C-terminal peptide linkers allowing it to sample reaction partners for FAB. Computational work has suggested that the ACP's motion is stochastic in type I FASs.[17] Discrete type II ACPs must travel through the cytosol in bacteria to find reaction partners. The ACP is expressed as an inactive apo-ACP, which is known to be cytotoxic in E. coli.[18] Activation by post-translational attachment of a CoA-derived 4’-phosphopantetheine (PPant) to a conserved serine (Ser36 in E. coli) yields holo-ACP (Figure 2b). FAB occurs using thioester-linked intermediates attached to the terminal thiol of the PPant arm, forming acyl-ACP. Termination of FAS by TE or AT activity regenerates holo-ACP.
Figure 2.
A) Demonstration of chain flipping: at left is ACP sequestering a C7 chain (PDB: 2FAD) and at right is an ACP from the crosslinked complex with DH (PDB: 4KEH). B) Schematic of tools utilized to modify ACP naturally in vivo, and in vitro. Often, due to endogenous PPTase activity, heterologous expression of ACPs yields a mixture of apo- and -holo-ACP species . R’, R” and R’” are used to denote variability limited by the specificity of the pathway. R’ can be acyl chains, azides, alkynes, or halogens limited by the specificity of AasS. R” is limited by the promiscuity of the specific CoA ligase used. R’” can be many different probes, from acyl chains to fluorescent moieties, and replacement of the thioester by an amide.
Type II ACPs sequester the growing chain within a hydrophobic pocket formed in the core of the helical bundle which expands based on the acyl chain length.[19] Sequestration has been proposed to shape ACP for subsequent protein-protein interactions, define substrate length, and/or protect the labile thioester bond from hydrolytic cleavage.[20,21] Recent work exploiting solvatochromic probes[22] and vibrational probes[23] appended to the PPant arm of ACPs have allowed direct observation of both sequestered and non-sequestered microstates. Type I ACPs are not thought to sequester, as observed by a lack of structural perturbations by NMR of the excised ACP[24] and solvatochromic probes.[22] Other studies have revealed that the structure of the ACP is modulated by its acylation state, including gel shifts observed by conformationally sensitive polyacrylamide gel electrophoresis[25,26] and resonance shifts observed by NMR.[27,28]
Observation of sequestration poses several intriguing questions. Are ACP-partner interactions stochastic, or does ACP communicate its cargo's status? Some studies have suggested cargo-mediated minor changes to the orientation of helix III may be involved[29], but the importance of these small structural changes has not yet been thoroughly validated. How does the ACP translocate the reactive chain to the partner protein? This process was originally named the “switchblade mechanism”,[30] but recently Cronan argued that “chain flipping” (Figure 2a) may be a more accurate description, since no evidence of activated release of the acyl chain has been observed.[31] Do structural changes to the ACP, when sequestering elongated intermediates, signal for TE or AT release of the fatty acid? Evidence has shown that a fully elongated acyl-ACP maintains a hairpin-like structure of the acyl chain[32] that the TE may recognize.
In Nature, the ACP is post-translationally modified from apo- to holo- form by the enzyme phosphopantetheinyl transferase (PPTase). A timely review discussed this class of enzymes in great detail,[33] and only a few key points will be mentioned here. PPTases utilize the high-energy CoA as the substrate for transfer. Three different subfamilies of PPTases have been observed. The AcpS (holo-ACP synthase) type PPTases activate type II FAS ACPs and show very little promiscuity towards non-target ACPs.[34] In E. coli, AcpS is responsible for activation of apo-ACP (Figure 2b). In contrast, Sfp type PPTases are highly promiscuous and often used in conjunction with heterologous synthase expression.[35] Indeed, heterologous expression of homogeneous holo-ACPs can be achieved by co-expressing recombinant Sfp with recombinant ACP[36], whereas without additional Sfp a mixture of apo-ACP and holo-ACP is often expressed. The Sfp family is named for the archetypical B. subtilis surfactin synthase activator. The third family is comprised of PPTases integrated in type I fungal FAS megasynthases.
Generation of homogeneous apo-, holo-, acyl- and (unnatural cargo bearing) crypto-ACPs has been critical to recent studies. Many new strategies have been employed to manipulate the cargo attached to the ACP. Certain fatty acids can be directly ligated onto CoA using an acyl-CoA ligase.[37] Once the acyl-CoA is prepared, addition of Sfp and apo-ACP generates crypto-ACP. The primary limitation of this approach is the requirement of an acyl-CoA ligase with activity for the desired fatty acid. Many studies of ligases are available[37–39], with each ligase demonstrating substrate specificities. It also requires apo-ACP, which has historically been challenging to obtain homogeneously. Finally, this method produces a thioester, which can be beneficial in that it reflects the natural linkage but also suffers from slow hydrolysis in solution. In long experiments, such as multidimensional NMR, such hydrolysis may become detrimental.
Interest in generating CoA analogs without using acyl-CoA ligase led to the development of a “one-pot” chemoenzymatic synthesis.[40,41] Synthetically obtained pantetheine analogs[42,43] can be converted to the corresponding CoA analogs by exploiting CoA biosynthesis through the use of a pantothenate kinase, phosphopantetheine adenylyl transferase and dephosphocoenzyme A kinase (CoaA, CoaD, and CoaE, respectively, in E. coli).[44] Once prepared (isolated or in situ), the CoA analog can be loaded onto apo-ACP by a PPTase, commonly Sfp (Figure 2b). Due to the inherent promiscuity shown by all of the “one-pot” proteins, probes of varying size and functionality, including fluorescent molecules, intermediate analogs and activity-based warhead moieties, can be loaded onto the ACP for visualization, isolation, functional and structural studies. Additionally, pantetheine probes with an amide or oxoester linkage to replace the natural thioester linkage can be loaded onto apo-ACP as a means of preventing slow hydrolysis in solution. The “one-pot” approach greatly broadens the scope of loadable substrates, but its application depends upon access to homogenous apo-ACP.
In E. coli, the ratio of apo- to holo- ACP is thought to offer regulatory control of the FAS.[18] The small protein ACP hydrolase/phosphodiesterase (AcpH) is responsible for cleaving 4’-PPant from holo-ACP to generate apo-ACP.[45] Since its discovery in the 1960s, AcpH has been scarcely used in vitro due to its poor stability characteristics, although it was observed to be very active.[46] In the past, a thiol-sepharose resin had been principally used to separate holo-ACP away from apo-ACP through formation of a disulfide bond, but this method suffered from the loss of large quantities of protein and was only possible with proteins lacking surface cysteine residues.[47] The use of a partially active AcpS mutant in E. coli has also been successfully leveraged for the overexpression of mostly apo-ACP.[18] Recently, the Pseudomonas aeruginosa AcpH has been developed as a much more amenable alternative to generate apo-ACP.[48] Both resin-attached and free protein techniques have been developed, allowing for the preparation of large quantities of apo-ACP and the recycling of high-value, isotopically enriched ACPs for NMR use.[49,50] Reversible tagging has also allowed for quantitative “apo-fication” of holo- and crypto-ACPs, yielding homogenous apo-ACP for further modification (Figure 2b).
Due to their transient nature, ACP-partner protein interactions are difficult to directly observe. In an attempt to trap this interaction, we developed crosslinking probes by leveraging mechanism-based inhibitors incorporated into pantetheine analogs and attaching them to ACP with “one pot” methods. These mechanism-based crosslinkers covalently attach ACP with a partner protein and have been developed for KS,[44,51] DH,[52,53] and TE[54,55] activities, allowing for crystallographic and NMR studies of these complexes. These studies will be discussed in the subsequent sections.
Another ACP cargo-manipulating tool has experienced renewed interest, with applications both in vitro and in vivo. The acyl ACP synthetase (AasS) uses ATP to ligate fatty acids directly onto holo-ACP (Figure 2b). The Vibrio harveyi AasS has been shown to load a range of unique carboxylic acids, including acids with azide, alkyne and halogen functionalities, onto holo-ACP in vitro. AasS allows simple, direct production of acyl- and crypto-ACPs with the natural thioester linkage, although there are some substrate limitations.[56] In E. coli, no cytosolic AasS activity has been observed, and exogenous fatty acids can only be degraded or used as substrate for glycerophospholipid synthesis depending on chain length. By heterologous expression in E. coli with a recombinant AasS, exogenously supplied unique carboxylic acids of varying structures can be imported directly into the FAS pathway and extended. This provides a new avenue to long chain terminally modified carboxylic acids that can be synthetically inaccessible and a way to incorporate unique moieties into FAS and lipid products.[56]
3. Structural Techniques for Studying the Protein-Protein Interactions of the FAS
Both nuclear magnetic resonance (NMR) and X-ray crystallography have been used extensively to study FAS, and the two techniques complement each other. Solution-phase NMR is well equipped to study small, dynamic proteins in a buffered environment, though line broadening inhibits the study of large complexes. Crystallography is well suited for ordered proteins and complexes but dynamic proteins and flexible regions are not ideal.
Computational approaches include Molecular Dynamics (MD), docking, and longer timescale Accelerated Molecular Dynamics (AMD) simulations. These approaches have been exploited significantly in the last few years, as computational power has risen and available structural information has grown. Microscopic techniques will be discussed briefly.
3.1. X-ray Studies and Techniques
X-ray crystallography is a well-established technique for visualization of proteins trapped in a rigid crystal lattice. Crystallographic studies of FASs have yielded many structures of both type I and type II FAS proteins. Due to their connected domain architecture, only type I FAS complexes have yielded crystal structures where most or all of the individual enzymes have been visualized together, while type II FAS enzyme structures have mostly been elucidated individually.
Crystal structures of type I megasynthases include the homodimeric mammalian “gingerbread-man” topology (Figure 1d) (PDB: 2VZ8)[9], the Thermomyces lanuginosus fungal α6β6 heterododecamer “soccer ball”(PDB: 2UVB, 2UVC)[8], and the differently-scaffolded Saccharomyces cerevisiae yeast fungal FAS (PDB: 2UV8)[30]. One critical challenge for crystallographers studying FASs is the highly dynamic ACP. Indeed, in type I structures these dynamic regions are not resolved, with the exception of the stalled ACP in the S. cerevisiae structure.
Crystal structures of individual type II disparate proteins are available, and will be discussed in the following sections. Currently, only one crosslinked protein crystal structure from a native FAS pathway has been solved, demonstrating the ACP trapped during interaction with a partner (Section 8.3).[57] Significant conformational changes of the ACP are well-visualized in the crosslinked structure with FabA (4KEH) in which helix II and helix III are bound by multiple salt bridges and hydrophobic interactions to allow chain translocation, and the hydrophobic pocket has collapsed. As a tool, X-ray crystallography has provided much of the key topological information about FASs, which will be discussed in the subsequent sections (5-10).
3.2. NMR Studies and Techniques
NMR studies enable observation of the dynamics of proteins and can provide structural data on portions of type I FAS that cannot be observed by crystallography. Full structural determination by NMR is data and time intensive, while simpler techniques such as chemical shift perturbation (CSP) analysis offer easily accessible but limited information. CSP analysis allows identification of important residues in protein-protein or protein-ligand interactions by simple experiments.[58,59]
Structures have been solved for a series of acylated ACPs from the spinach type II FAS, revealing that ACP could comfortably sequester ten carbons but no more, and the solvent exposed hairpin formed by longer chains may be a recognition motif for thioesterase activity.[32] Structural calculations and CSPs observed when comparing different reactive intermediates during extension and reduction of C6-ACP to C8-ACP demonstrated very little perturbation of helix II and cargo-mediated changes to helix III.[29] The type I mammalian ACP, when truncated from the megasynthase, did not demonstrate CSPs when comparing acylated and holo-ACP, suggesting type I ACPs do not sequester their cargo.[24]
Dynamics observations are useful for determining which portions of proteins are undergoing motion on a given timescale. Fast timescale dynamics can be accessed by classical relaxation experiments.[60] The application of the Lipari-Szabo Model-free approach[61,62] allows quantification of flexibility residue by residue as a generalized order parameter, revealing significant flexibility of the loop near the post-translationally modified serine.[28]
3.3. Computational Studies and Other Techniques
Computational studies have yielded significant information about protein dynamics and flexibility. Classic MD simulations and docking studies have been used to predict binding and develop inhibitors. In addition to the fast-timescale dynamics mentioned above, slow timescale dynamics are observable using Accelerated Molecular Dynamics (AMD).
MD simulations of acylated ACPs have explored the sequestration preference of the acyl-ACP and the flexibility in the acyl conformations.[63] Simulations of the ACP within the type I fungal FAS have revealed that the ACP's motion is likely stochastic in nature.[17] Optimized by NMR observed Residual Dipolar Couplings[64,65], which encode long timescale motions (μs-ms),[66,67] AMD treatment to compare the long-timescale dynamics of acyl-ACP and crosslinked ACP-DH revealed significant losses in flexibility, suggesting the interaction is well ordered.[57] Docking studies will be explored in the sections below, as part of the analyses of individual domains.
Electron microscopy (EM) is an important tool that has been gaining popularity for the visualization of type I FASs. Early work allowed observation of the mammalian type I FAS and the two multidomain monomers, but in limited resolution.[68] Recent breakthroughs include Grininger's 6 Å EM map of the fungal FAS[69] and mycobacterial type I FAS.[70] EM offers a promising method for study of the FAS in a variety of states and conformation that are expected become more prominent in the future.
3.4. Inhibitors
In the partner domain sections (5-10) inhibitors will be discussed in detail. The search for exploitable drugs has been a primary driving force for inhibitor development in the 20th century. Since FAB is essential for life, and humans and bacteria harbor different machinery, FASs have been a major target of interest. However, given the importance of this target and the sheer number of druggable activities, relatively few FAS-targeting drugs have been developed. More numerous are known inhibitor probes that have been discovered, often by accident, over the course of the last few decades.[71] Mechanism-based “suicide” inhibitors have recently become additionally important in the development of useful probes for elucidation of structure-function relationships in FASs. Noncovalent inhibitors have allowed the study of transient interactions important for enzyme kinetics and structural studies. Protein conformational changes that occur upon inhibitor, cofactor and/or substrate binding can be used together to study important structural, kinetic and thermodynamic properties. We will highlight known inhibitors of each catalytic function in the following sections.
4. FAS Partner Domains
Sections 5-10 will focus on 6 FAS domains (MCAT, KS, KR, DH, ER, and TE/AT). A brief description of the enzyme will precede what is known about its catalytic mechanism, inhibitors with mechanisms of action, and what is currently known about interactions with ACP. Tools for modifying ACP and tools for visualizing protein-protein interactions described earlier will be emphasized as applicable for each domain. This review highlights that while the mechanisms and activities of individual domains are now well understood, individual domain interactions with the ACP remain the next important step for studying the FAS.
5. Malonyl-CoA:ACP Transacylase (MCAT)
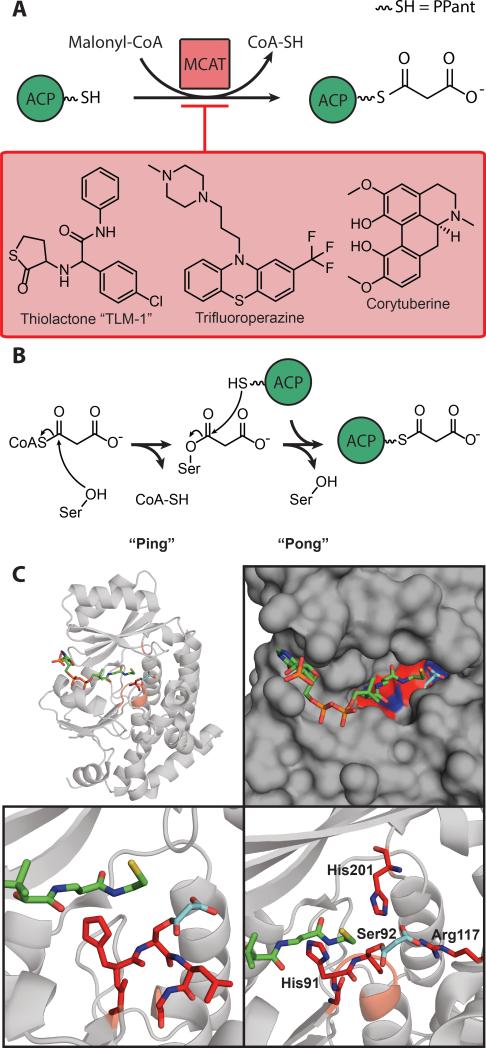
Malonyl-CoA:ACP transacylase (MCAT) catalyzes the transacylation of malonate from malonyl CoA to holo-ACP, which is the first step towards fatty acyl chain elongation (Figure 3a). MCAT is a serine α/β hydrolase, in which the active site serine first becomes malonylated (Ser92 in E. coli) and the enzyme then transfers the malonyl group to ACP. The MCAT domain in E. coli, FabD, is specific for malonyl-CoA and unable to use acetyl-CoA for loading. In vitro, many reaction partners have been observed including CoA, ACP, pantetheine and N-(N-acetyl-β-alanyl)-cysteamine.[72]
Figure 3.
A) Schematic of MCAT chemistry, with several known inhibitors. B) Diagram of MCAT “Ping-pong” mechanism. C) Examination of MCAT's active site. MCAT has been malonylated (cyan) and CoA (green) is present (PDB: 2G2Z). Active-site residues are marked in red. Top-left: global topology of MCAT with critical residues marked in red. Top-right: surface diagram of the active site, demonstrating depth and accessibility. Bottom-left: malonylation of the active-site serine, positioning of CoA. Bottom-right: positioning of catalytic triad.
Deletion of fabD in E. coli is lethal[73] whereas overexpression of fabD in E. coli leads to an increase in cis-vaccenic acid (cis-11-octadecenoic acid) due to an increase in malonyl-ACP concentration and enhanced ketosynthase activity.[74] However, since MCAT does not catalyze a rate-limiting step in FAB, overexpression of fabD in E. coli only leads to an 11% increase in fatty acid content.[75]
5.1. Mechanism
MCAT completes malonyl transfer with the double-displacement or “ping-pong” mechanism common to α/β hydrolases using its His-Ser catalytic domain (Figure 3b).[76,77] Double-displacement is a two-step mechanism: First, the oxyanion hole of MCAT is occupied by the binding of malonyl-CoA, and transacylation of malonate onto to the active site serine forms an acyl-enzyme intermediate. Second, ACP binds to the surface of the MCAT/malonyl complex, which triggers structural movement allowing transacylation to the PPant sulfhydryl to form malonyl-ACP. This transformation has been computationally modeled based upon the EcMCAT/malonyl/CoA complex structure (PDB 2G2Z).[78]
P. falciparum has an apicoplast localized FAS, and sequence alignment of the P. falciparum MCAT (PfMCAT) with template sequences, EcMCAT and HsMCAT, identified conserved motifs and residues. These important motifs include pentapeptides GQGXG and GXSXG, along with four key invariant residues: Gln109, Ser193, Arg218, and His305 in PfMCAT.[79] Glutamine is responsible for stabilization of the oxyanion hole, and the nearby arginine is responsible for the recognition and positioning of the substrate's free carboxyl group via electrostatic interactions.[80] Serine provides the catalytic active site nucleophile, which is stabilized and activated by a histidine.[79] In the S. coelicolor MCAT (ScMCAT) crystal structure an acetate molecule is bound to Gln9 and Arg122 mimicking the carboxyl end of the malonyl group corroborating this mechanistic proposal.[77]
While the ScMCAT, EcMCAT and PfMCAT have a similar catalytic mechanism, the crystal structure of the Mycobacterium tuberculosis MCAT (MtMCAT, PDB: 2QC3) reveals the C-O bond of the catalytic Ser91 turns upwards and results in a different orientation and shape of the active site pocket and subsequently a new catalytic diad.[81]
5.2. Inhibition
MCAT has been considered as an antibacterial target.[82] The binding of ACP to MCAT was found to be competitively inhibited by high CoA concentrations.[72] Some MCAT antimicrobial drugs have been identified, including trifluoroperazine (Figure 3a).[83]
Screening against the MCAT from the apicomplexan parasite Eimeria tenella (EtMCAT) identified an alkaloid natural product, corytuberine, with moderate inhibitory activity against EtMCAT.[84] Virtual screening for inhibitors of PfMCAT was performed, and EDTA, p-toluene sulfonyl fluoride, and a thiolactone derivative, “TLM-1” (Figure 3a), were found to position in the binding pocket and form stable complexes with strong interactions.[79] Crystal structures solved of small molecules (glycerol and malonate) bound to the active site of EcMCAT revealed that the length and amphiphatic character of the substrate define the specificity of the enzyme, but true targeting of MCAT requires further information about which of these structural elements are important to exploit.[80]
5.3. ACP interaction
Four different MCAT-ACP interactions have been studied by modeling and revealed two mechanisms of interaction, one largely electrostatic and one largely hydrophobic.
The interaction between the H. pylori MCAT (HpMCAT) (PDB: 2H1Y) and ACP was studied using computational docking, glutathione S-transferase (GST) fusion protein pull-down, and surface plasmon resonance (SPR). The HpMCAT active site is characterized by Ser92 and His198, which are in a deep gorge between two subdomains (Figure 3c), and part of the GXSXG conserved motif in α/β hydrolases. Helix II of HpACP recognizes a conserved hydrophobic pocket between the two subdomains near the active site entrance of HpMCAT.[76] Additionally, by computation the ACP binding site appears to be adjacent to the GQGXQ motif.[76] Similar to other FAS proteins, the ACP binding site was found to be a positively charged region forming complementary electrostatic interactions.[85,86]
Information on the structural basis of the ping-pong mechanism was obtained computationally by modeling the ScMCAT interaction with the S. coelicolor actinorhodin polyketide synthase (act PKS) apo-ACP.[77] The structure of ScMCAT with active site bound acetate, which mimics the carboxyl of the malonyl (PDB: 1NM2), suggests a ping-pong mechanism where upon substrate binding the oxyanion hole was formed thereby moving the active site from a “closed” to “open” conformation. The second half of the mechanism, which involves ACP, showed a largely hydrophobic interaction between the DSL motif at the prosthetic group attachment site serine of the PKS apo-ScACP and the hydrophobic pocket formed by the helical flap adjacent to the GQGXQ turn of ScMCAT.[77] However, this study did not use the cognate FAS ACP, and since PKS and FAS ACPs are not easily interchangeable, these docking results await experimental validation.[87]
NMR CSP studies and further computational work on the interaction between the ScMCAT and the FAS ScACP shed some light on the situation.[87] In silico macromolecular docking of these proteins returned three different models; the first mimics a model from Keatinge-Clay[77] where the loop region after helix I of FAS ScACP binds the residues near the ScMCAT active site, the second mimics the binding of HpMCAT and HpACP with helix II of FAS ScACP packing against ScMCAT, and the third binds the loop region after helix I of FAS ScACP near the MCAT active site but in a rotated complex.[87] Holo-ScACP (FAS) and ScMCAT NMR titration experiments showed small NMR shifts for negatively charged residues in helix II, suggesting this region interacts with ScMCAT, as shown by other ACP-partner domain interactions studied.[88] However, all NMR shifts were inconclusively small, suggesting ACP is in very fast exchange between its free and bound forms.[87]
The protein-protein interactions of MCAT from Xanthomonas sp. (XoMCAT) and PKS ScACP (PDB 2AF8) were studied using computational docking, revealing that ACP binds to the cleft between the two XoMCAT subdomains, as seen in the HpMCAT-ACP model. A positively charged ACP binding site was observed adjacent to the GQGXQ loop. Gly14 and Ser15 from the GQGXQ loop of XoMCAT hydrogen bond with ACP residues. These residues are near the oxyanion hole, and it is hypothesized this hydrogen bonding could activate the oxyanion hole to initiate malonyl transfer. Further clarification of this interaction will require a co-crystal structure.[78]
6. Ketosynthase (KS)
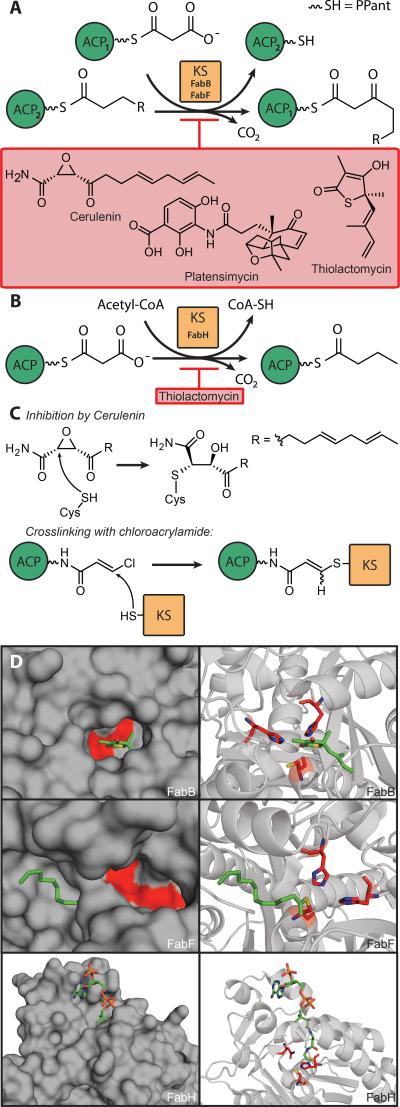
Ketosynthase catalyzes carbon-carbon bond formation via decarboxylative Claisen condensation (Figure 4a, 4b). Type I FASs contains only one KS, whereas type II FASs have two or three KSs. In E. coli, KSI, II and III are encoded for by fabB, fabF and fabH, respectively. The product of these reactions is 3-ketoacyl-ACP by condensation of acyl-ACP with malonyl-ACP, with the exception of FabH (KS III), which condenses malonyl-ACP and acetyl-CoA (Figure 4b). FabH is involved in FAS initiation and FabB and FabF are involved in chain extension. The acyl group is loaded onto an active site cysteine and malonyl-ACP is extended by two carbon units after decarboxylative nucleophilic addition.
Figure 4.
A) Schematic of FabB and FabF chemistry, with known inhibitors. R is an acyl chain, length C1 to C13 (odd values). The two ACP proteins are denoted as ACP1 and ACP2 to clarify the transformation. B) Demonstration of FAS initiation by FabH activity on malonyl-ACP wtih acetyl-CoA, and a known inhibitor. C) Inhibition by Cerulenin, and mechanism-based chloroacrylamide crosslinker based on Cerulenin. D) Comparison of FabB, FabF and FabH demonstrating depth of active site (both FabB and FabF exhibit shallow active sites, while FabH exhibits a deep pocket) and active site geometry. (PDBs: FabB=2VB8; FabF=2GFY; FabH=1HNJ). FabB shows thiolactomycin (green) bound and the catalytic triad (Cys163, His298, His333; red); FabF shows covalently attached C12 fatty acid (green) and the catalytic triad (Cys163, His303, His340; red); and FabH has Malonyl-COA (green) occupying the tunnel and the catalytic triad (Cys112, His244, Asn274; red). The active site geometries of FabB and FabF portray one histidine in position to abstract a proton from the leaving group, and the other in position to provide hydrogen-bond stabilization.
E. coli FabH uses acetyl-CoA as its main substrate, with butyryl-CoA demonstrating much lower activity and hexanoyl-CoA demonstrating no activity.[89] FabH has been shown to be essential to FAB in vivo.[90] Overexpression of fabH leads to a decrease in cis-vaccenate and an increase in myristic acid (C14).[91] FabH is regulated by feedback inhibition by high concentrations of long chain acyl-ACPs.[89]
FabB (KS1) shows activity with C6 to C14 saturated fatty acyl-ACPs, although it is only weakly active on C14-ACP.[92] FabB is unsaturation tolerant and catalyzes the condensation of cis-3-decenoyl-ACP, cis-5-dodecenoyl-ACP and 7-tetradecenoyl-ACP each with malonyl-ACP.[93] fabB deletion leads to auxotrophy for unsaturated fatty acids, and overexpression increases unsaturated fatty acids if fabA is also overexpressed. FabF (KSII) shows activity for C6 to C14 fatty acyl-ACP esters, however C14 is a weak substrate.[92] FabF also shows low activity for C16 and carries out the last step in the unsaturated pathway by elongating cis-9-hexadecenoyl-ACP.[92] Overexpression of fabF is lethal,[94] although interestingly, expression of increased amounts in E. coli did not lead to cell death.[44]
Reconstitution of the cyanobacterium Synechococcus sp. PCC 7002 FAS revealed that FabH was the sole rate limiting step, in contrast to the E. coli FAS with rate limiting FabI (ER) and FabZ (DH) steps.[95]
6.1. Mechanism
KSs adopt a ping-pong mechanism like MCAT. All three E. coli ketosynthases have been crystallized and show invariant hydrophobic residues that line their acyl pocket to facilitate binding of the acyl chain. Each has a catalytic triad that lies at the bottom of the pocket in the structures (Figure 4d), and the acyl chain folds into an extended U-shaped conformation.[96]
With all three KS enzymes, the initial transthioesterification occurs via cysteine attack to form a tetrahedral intermediate, with the oxo group of the bound fatty acid thioester situated in an oxyanion binding site.[96] FabB and FabF both have a Cys-His-His active site triad. The entrance of the FabF active site is blocked by an aromatic “gate keeping” residue, Phe400, and it has been proposed that it must shift in order for the acyl substrate to gain access to the active site and the condensation reaction to occur.[97] A Cys163Gln mutant in EcFabF mimics acyl-KS. In this structure the phenylalanine residue blocking the active site is moved to an open conformation, allowing the second half of the “ping-pong” mechanism.[98] Of the two nearby histidine side chains, one is proposed to abstract a proton from the carboxylic acid leaving group of the malonyl residue, while the other histidine accepts a hydrogen bond and then makes contact with the thioester oxo group of the malonyl residue inducing a partial negative charge on that group. These two processes prompt the decarboxylation reaction, which is followed by condensation.[96,97]
FabH on the other hand, has a different proposed decarboxylating Claisen mechanism. The FabH catalytic triad consists of Cys-His-Asn. The histidine and asparagine make an oxyanion binding site, which binds the malonyl thioester oxo group and promotes decarboxylation.[99]
6.2. Inhibition
The fungal polyketide cerulenin targets KSs of FAS and PKS pathways Figure 4a).[100] Cerulenin is a covalent inhibitor that selectively targets FabB and FabF through modification of the active-site cysteine sulfhydryl (Figure 4c). A crystal structure with FabF reveals that cerulenin binds in a hydrophobic pocket at the dimer interface and covalently attaches to the active site cysteine, mimicking the condensation transition state. Phe400 and Ile108 completely rotate for this to occur, as seen with the Cys163Gln EcFabF acyl-KS mimic, providing access to the active site cysteine and opening a hydrophobic picket for the tail of the inhibitor.[101]
Thiolactomycin (TLM) is a natural product that inhibits all FAS condensing enzymes, including FabH.[102] A FabB-TLM crystal structure shows that TLM mimics malonyl-ACP in the active site, and both active site histidines aid the protein-antibiotic interaction. This explains the greater inhibition of FabB than FabH with TLM, since FabH harbors different catalytic machinery.[103] Additionally TLM inhibits two ketosynthases from Mycobacterium tuberculosis, KasA and KasB, which are involved in chain extension like FabB and FabF.[104] Neither cerulenin nor thiolactomycin are used in the clinic. Cerulenin also inhibits the KS function of the mammalian FAS, while the total synthesis of thiolactomycin is problematic due to its inherent instability.[105,106]
Platensimycin, a natural product isolated from Streptomyces platensis, is a selective FabB/F inhibitor with potent antibiotic properties. The formation of the acyl-enzyme complex is essential for platensimycin binding. The compound was found to be 200 fold more potent than cerulenin and 50 times more efficient than thiolactomycin in inhibiting FabF.[98]
6.3. Interactions with ACP
All three KSs from E. coli have been crystallized, but only the interactions of FabH and FabF with ACP have been shown by modeling studies.[55,85] No co-crystal structure of an ACP bound to a type II FAS KS has been published.[13]
ACP-protein interactions were investigated using computational analysis to dock the NMR structure of EcACP with the crystal structure of EcFabH, and the generated model was experimentally validated by in vitro characterization of FabH mutants.[85] The EcFabH catalytic triad sits at the bottom of a tunnel, and the entrance to the active site is flanked by four conserved basic residues (Arg36, Arg151, Lys214, Arg249) and two conserved aromatic residues (Trp32 and Phe213), thereby giving it hydrophobic and electropositive character.[99] EcACP docked to the area adjacent to the active site of EcFabH, and a conserved arginine (Arg249) in this patch was required for docking, as seen by an ACP-dependent assay and direct binding studies monitored by surface plasmon resonance. This patch also included alanine residues, which were proposed to be important for the close approach of the two proteins. Zhang et. al. analyzed the crystal structures of FabA, FabD, FabI, FabF, and LpxA and noted a conserved arginine/lysine residue in an electropositive/hydrophobic patch adjacent to their active sites (within 10 to 14 Å). ACPs from a variety of species have a highly conserved region from residues 32 to 50, which encompasses helix II. Glu41 predicted to interact with Arg249 of FabH, is conserved in all 49 ACP sequences known at the time. The Ser36 of ACP, the point of PPant attachment, is oriented correctly in the docked model, but a 10 Å movement would be required for the chain to reach the FabH active site. The “chain-flipping” mechanism is proposed, in which dissociation of the acyl group from ACP upon binding with FabH promotes this proposed conformational change in ACP and subsequent 10 Å movement.[85]
Wothington et al. designed an E-chloroacrylamide probe as an active site crosslinking reagent to covalently crosslink KSs and ACPs. The warhead portion of the probe is a Michael acceptor that, when attacked by the KS active site cysteine residue, undergoes tandem 1,4-conjugate addition and β-chloro-elimination to yield a stable covalent linkage between the probe and active site (Figure 4c). The chloroacrylamide compound was used to crosslink EcACP with EcFabB and EcFabF. Since EcFabH did not readily crosslink, but does bind ACP subsequent to priming by acetyl-CoA, it is proposed that EcFabH goes through a conformational change upon substrate loading before ACP can bind.[44]
Further studies utilizing the chloroacrylamide probe revealed that select ACPs from polyketide synthases, the frenolicin synthase carrier protein (FrenACP) from Streptomyces roseofulvus and the oxytetracycline synthase carrier protein (OtcACP) from Streptomyces rimosus, were able to crosslink with EcFabB. EcFabB was unable to crosslink with nonribosomal peptide synthase carrier proteins.[51] Additionally, Chlamydomonas reinhardtii chloroplastic cACP crosslinked with EcFabF.[55] Crosslinking was used as a measure of interaction strength between ACP and KS since it is dependent on the ability of KS to recognize the carrier protein via protein-protein interactions and react with the substrate analog. Crosslinking showed that EcFabF-ACP has tighter binding than EcFabB-ACP as seen by crosslinking efficiency. EcFabF also acts more efficiently than EcFabB on longer substrate mimics, with a 4-fold larger preference, in accordance with its native substrate specificity. The two enzymes have equal preference for shorter substrate mimics.[107]
7. Ketoreductase (KR)
3-Ketoacyl-ACP is reduced to (R)-3-hydroxyacyl-ACP by an NAD(P)H dependent KR, a member of the short-chain dehydrogenases/reductases (SDR) family (Figure 5a). FabG, the KR from E. coli, was first purified and characterized in 1966[108] and found to be active over a wide range of different 3-ketoacyl-ACPs.[109] FabG can function on acyl-CoAs but with much lower efficiency.[108]
Figure 5.
A) Schematic of KR chemistry, with several known inhibitors. R is an acyl chain, length C1 to C15 (odd values). B) Diagram of NAD(P)H based reduction mechanism. C) Demonstration of active site restructuring due to NAD+ binding. Top left: KR topology with NADP+ highlighted in red; middle left: surface filled close up on NADP+ binding and remote Arg129, Arg172 patch for ACP binding (PDB: 1Q7B). For comparison, Top right: KR topology without NADP+; middle right: surface close up demonstrating different topology, and highlighted arginine patch Arg129, Arg172 (PDB: 1I01). At bottom, demonstration of distance between cofactor binding site and ACP binding site, requiring chain translocation for activity (PDB: 1Q7B).
FabG is ubiquitously expressed in all bacteria, conserved across all species and the only known enzyme to catalyze the ketoreduction step in fatty acid biosynthesis.[71] Experimental evidence for gene essentiality has been reported for E. coli, Salmonella enterica, Mycobacterium tuberculosis and Pseudomonas aeruginosa.[110–113] Overexpression of fabG in E. coli increases the content of C16 acid two-fold and C18 acid three-fold.[114]
7.1. Mechanism
During the catalytic reaction, the β-keto group is reduced to a β-hydroxy group by the pro-4S hydride ion from the nicotinamide cofactor and a proton donated by the hydroxyl of an adjacent tyrosine (Figure 5b).[115–117] FabG is a tetramer and exhibits negative cooperativity, as seen by the binding of NADPH to one active site increasing the affinity at that site for ACP and decreasing the affinity for the cofactor elsewhere.[118] KRs exhibit an ordered kinetic mechanism, with the NADPH cofactor binding first and the NADP+ being the last product to leave.[116]
The structure of E. coli FabG in binary complex with NADPH reveals mechanistically important conformational changes that result from cofactor binding (Figure 5c). The cofactor binds in a two-stage mechanism, and when bound the cofactor is in the syn conformation and the nicotinamide ribose is stabilized in the active site via hydrogen bonds from Tyr151 and Lys155. In the structure of the Y151F EcFabG mutant, the nicotinamide ribose is disordered and not bound to the active site. Once NADPH was bound to wildtype FabG, there was a rearrangement of active site triad residues Ser138, Tyr151 and Lys155 and the associated β5-α5 loop, as well as a shift of the β4-α4 loop and dimer interface. These conformational changes were functionally important because they primed the active site for catalysis; allowed the substrate access to the active site; and created a hydrogen-bonded network of ribose hydroxyls, the catalytic triad and four water molecules to relay a proton to tyrosine after it donates a proton to the substrate during catalysis.[116] Finally, the B. napus FabG and NADP+ cofactor binary complex suggests that the α6/α7 subdomain rotates to enclose the active site and create a well-defined tunnel as the final step in ternary complex formation.[119]
7.2. Inhibition
Studies suggest that FabG could be susceptible to inhibition at its cofactor binding site.[120] Inhibition studies for FabG commonly follow the consumption of NADPH by absorbance as FabG acts on a substrate, acetoacetyl-coenzyme A, for example.[111]
Epigallocatechin gallate, the major component of green tea extracts, and other related plant polyphenols were found to inhibit FAS II by targeting FabG and FabI. The galloyl moiety of the catechins is essential for inhibition, disrupting cofactor binding by associating with the cofactor free FabG.[121] Isoflavonoids and flavonoid derivatives inhibit FabG from H. pylori (HpFabG) in a noncompetitive manner with respect to acetoacetyl-CoA and NADPH.[122] The first inhibitor of microbial origin, macrolactin S from Bacillus sp. AT28, showed dose dependent selective inhibition of S. aureus FabG (SaFabG), but did not inhibit SaFabI.[123]
The crystal structure of FabG from P. falciparum (PfFabG) was solved, and four structural analogues of triclosan were found to have 75% inhibition in vitro against PfFabG: hexachlorophene, bithionol, di-resocinol sulphide and bromochlorophen. The inhibitors displayed non-linear competitive inhibition with NADPH, but further mechanistic studies performed by varying substrate concentrations were inconclusive.[122–125] The above potential inhibitors, however, are largely natural product extracts facing drug development challenges and have not reached clinical usage.[111]
Recently an allosteric inhibitor-binding site at the inter-subunit interface in FabG from Pseudomonas aeruginosa (PaFabG) was identified. Two hydrophobic cavities at the “dimer-dimer” interface formed from residues in helices α4 and α5, not seen in apo-PaFabG, were induced upon ligand binding. The size of the cavity was dependent on the inhibitor size, demonstrating induced fit. These conformational changes were shown to propagate to the catalytic triad (Ser141, Tyr154, Lys158), resulting in an active site conformation that was incompatible with NADPH binding. All ligands tested adopted almost planar conformations and contained key hydrogen bond donors and acceptors. Absence of this cavity in the non-inhibited enzyme reveals a limitation of most present-day, docking-based virtual screening methods for drug discovery.[111]
7.3. Interactions with ACP
KRs remain poorly investigated, but NMR and in silico studies of the ACP binding site have been performed. No co-crystal structure of an ACP with a KR has been solved.
Two surface residues, Arg129 and Arg172, located in a positive hydrophobic patch adjacent to the EcFabG active site (Figure 5c), were mutated and showed a decrease in ability to use ACP thioester substrates but were fully active on a non-ACP substrate analog, β-ketobutyryl-CoA.[86] These two residues are conserved in PfFabG as well.[124] NMR chemical shift perturbations (CSPs) revealed that EcFabG-ACP interactions occur along ACP helix II and extend into the adjacent loop 2.[86] The acylated prosthetic groups are seen to reside in the hydrophobic cavity between helices II and III,[126] and therefore the acyl chain of the prosthetic group would interact with residues in loop 2. NMR titration studies showing a shift in Ile54 from loop 2 of ACP suggest that FabG interactions alter the acyl chain binding site in ACP to facilitate the release of the prosthetic group for modification.[86] These studies from 2003 were validated by the X-ray structures of hexanoyl-, heptanoyl-, and decanoyl-ACP solved in 2007 that showed the tethered substrate interacting with loop 2.[19]
Based on computational docking of EcACP to EcFabG-NADPH, the FabG ACP binding site is found closest to the active site of an adjacent monomer in the tetramer (Figure 5c). Substrate delivery via the PPant arm takes place across the dimer interface. Arg172 of the ACP binding site moves towards Ile136, which is at the base of the active site pocket, by 1.3 Å when the cofactor binds, showing the conformational change preparing for ACP interaction. The H-bonding of Glu168 located between the two sites also changes upon cofactor binding and is hypothesized to facilitate ACP interaction and substrate delivery. Additionally in this region of EcFabG is the β5-α5 loop, which becomes ordered upon cofactor binding and contains the conserved Asn145 residue, suggesting this may bind the pantetheine moiety.[116,119]
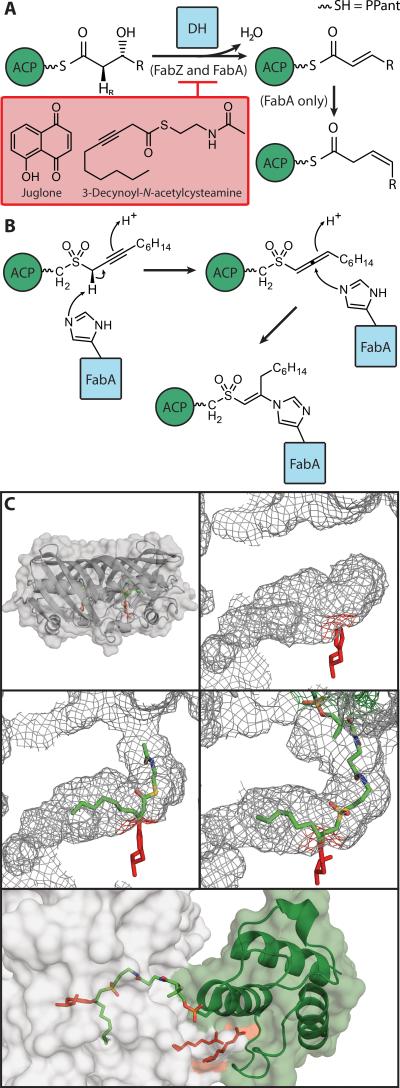
8. Dehydratase (DH)
The dehydration of (R)-3-hydroxyacyl-ACP to the enoyl moiety is performed by β-hydroxyl-ACP dehydratase via the elimination of water. The DH catalyzes syn elimination of the pro-2S hydrogen and (3R)-hydroxy group producing a trans product.[127] In E. coli there are two DHs encoded by fabA and fabZ. Besides dehydration, FabA isomerizes trans-2-decenoyl-ACP into cis-3-decenoyl-ACP, which is the first reaction towards the synthesis of unsaturated fatty acids (Figure 6a).[128] Bacteria and anaerobes employ a dual DH system to achieve unsaturation, as seen with FabA in E. coli, whereas other organisms utilize desaturases to achieve unsaturation.[129] The DH domain is a homodimer comprised of monomers with double-hotdog topology in which α helixes are wrapped by β sheets.
Figure 6.
A) Schematic of DH chemistry, with known inhibitors. R is an acyl chain, length C1 to C15 (odd values). FabA isomerization is depicted, but observed only when R is C5-C9 with preference for C7. The anti-periplanar pro-2R hydrogen is highlighted for elimination. B) Diagram of mechanism-based crosslinking activity described in text. C) Demonstration of EcFabA topology and a comparison of conformational changes induced by substrates. The catalytic His70 is shown in gray at the bottom of the active site. Top left: overall topology of the DH, FabA; top right: wireframe topology of empty active site (PDB: 1MKB). Middle left: wireframe topology of occupied active site, covalently attached to 3-Decynoyl-N-acetylcysteamine (marked in red) (PDB: 1MKA). Middle right: wireframe topology of occupied active site when crosslinked to ACP using the mechanism-based crosslinker described in B; Bottom, cartoon of ACP when crosslinked to FabA with overlayed electrostatic surface, demonstrating unfolding of helix III by interaction with arginine (gray sticks) from the FabA (PDB: 4KEH).
FabA and FabZ exhibit broad overlapping chain length specificities. The substrate preference for FabA is intermediate chain length acyl-ACPs centered on 10 carbons. FabZ demonstrates preference for shorter chains yet also has low activity for 16-carbon substrates. FabA does not participate in elongating unsaturated acyl-ACPs, so only FabZ is part of the unsaturated fatty acid arm of the pathway after FabA performs the initial isomerization.[130] Insight into why these two enzymes have different substrate specificities has yet to be elucidated.
E. coli overexpressing both fabA and fabB produced an enhanced amount of unsaturated fatty acids.[131] Deletion and overproduction of fabA leads to a decrease in unsaturated fatty acid content of phospholipids and an increase in saturated fatty acids.[130] Deletion of fabZ leads to reduced (3R)-hydroxymyristoyl-ACP activity suggesting an enhanced amount of 3-hydroxymyristoyl-ACP could be found in vivo.[132] Heterologous overexpression of fabZ leads to a 2-fold increase in palmitic acid and stearic acid.[114]
8.1. Mechanism
Crystal structures of E. coli FabA (EcFabA), P. aeruginosa FabZ (PaFabZ) and P. falciparum hexameric FabZ (PfFabZ) show active site tunnels formed between two monomers that accommodate the growing acyl chain. A catalytic diad, a conserved histidine from loop B and conserved glutamate (FabZ) or aspartate (FabA) from the N-terminal region of the central helix, each from a different subunit are responsible for dehydrating the acyl substrate[133–135]
3-hydroxydecanoyl-N-acetylcysteamine (3-hydroxydecanoyl-NAC) bound in the pantetheine tunnel in the PaFabA crystal structure yields insight into the catalytic mechanism.[136] The NAC molecule hydrogen-bonds with loop C as well as with a water molecule bridging the other subunit. The NAC moiety binds closely to catalytic residues Asp84 and His70 (the same residues found in EcFabA). His70Asn and Asp84Asn mutants show a complete loss in PaFabA activity. His70 abstracts the proton from C2 while Asp84 hydrogen bonds the C3 hydroxyl along with a water molecule, and only the R configuration puts the hydroxyl group in proximity of Asp84. The mechanism for the protonation of the hydroxyl has yet to be elucidated.[136]
FabAs have a conserved α1-β1 loop that is larger than in FabZ sequences. PaFabA and EcFabA structures shows this loop contributes extensively to the alkyl-binding tunnel, whereas the α3-β3 loop is the major contributor to the alkyl-binding tunnel in FabZ.[136]
8.2. Inhibition
Studies of FabZ as a drug target exploit crotonyl-CoA as a substrate, monitoring the reverse reaction by decrease in absorbance at 260 nm. Crotonyl-CoA studies give a lower KM than β-hydroxybutyryl-CoA.[137]
Helicobacter pylori FabZ (HpFabZ) exists as a hexamer in its native state, with its active site located in a dimer interface. The substrate binding tunnel is long, narrow and hydrophobic, with the catalytic histidines and glutamate residues half way down the tunnel.[138] Isoflavonoids, flavonoid derivatives and synthetic inhibitors have been reported to inhibit HpFabZ as competitive inhibitors of the substrate crotonyl-CoA, blocking the active site and extending to the groove around the pantetheine-binding tunnel.[122,125,139] The natural product juglone (Figure 6a) was found to potently inhibit HpFabZ in a competitive fashion (it also targets HpFabD as an uncompetitive inhibitor). The HpFabZ-juglone crystal structure revealed that juglone experienced π-π interactions with tyrosine and proline residues, was stabilized by H-bonds with water and inserted deeply into the active site pocket.[140] The discovery of binding fragments that occupy various positions in the PaFabA active site may help future inhibitor design. Two fragments were found to block the pantetheine-binding tunnel and partly mimic the flavonoids and synthetic inhibitors reported for HpFabZ.[136]
The catalytic site of PfFabZ is also located in a deep narrow pocket formed at the dimer interface. PfFabZ experiences multimeric state regulation by pH control. An inactive form of PfFabZ was discovered existing as a dimer rather than the active hexamer. As the pH decreases, the dimeric form prevails, and the two active site histidines have electrostatic repulsion, resulting in their expulsion from the active site. This was proposed to help aid in drug design by designing inhibitors to stabilize the inactive dimeric state.[141]
There are currently no published inhibitors for FabA, although it is an interesting drug target due to its essential isomerase activity and absence from type I systems. Enterococcus faecalis was found to have two FabZ homolog dehydratases, one with isomerase activity (FabN) and one without (FabZ). Lu et al. constructed EfFabZ/N chimeric proteins in an attempt to transform the FabZ into an isomerase. Single point mutations of FabZ were not sufficient, but the addition of the β3-β4 region of EfFabN to the EfFabZ showed a gain of isomerase activity, 38% of the original EfFabN. Isomerase activity likely depends on the shape of the substrate binding tunnel, controlled by the positioning of the β-strands surrounding the long central helix, and not the catalytic machinery of the active site. Disruptions to the active site shape eliminate isomerase activity.[142]
8.3. Interaction with ACP
A mechanism-based crosslinker was used to trap the transient interaction of AcpP and EcFabA to obtain a crystal structure. The crosslinked AcpP-EcFabA complex shows AcpP in two different conformations, one presumably a snapshot of the AcpP in transition and one when docking was complete (Figure 6c).[57]
The dehydratase-specific mechanistic crosslinking probe was designed and synthesized to covalently link FabA and ACP. The probe utilizes a 3-alkynyl sulfone warhead, where the sulfone mimics the natural thioester, yet is safe from hydrolysis. The warhead gets deprotonated and forms a reactive allene intermediate, which is attacked by the FabA active site His70, forming a covalent linkage (Figure 6b). Subsequently, when loaded onto EcACP the 3-alkynyl sulfone probe and was demonstrated to be fully sequestered by ACP, indicating that it mimics the hydrophobicity of the natural DH substrate.[52,53]
The FabA-AcpP complex crosslinked crystal structure, along with chemical shift perturbations from NMR titration experiments between either holo-ACP or octanoyl-ACP and FabA, were used to elucidate a four step mechanism by which FabA prompts the release of the sequestered acyl chain from AcpP for dehydration. First, a “positive patch” of EcFabA interacts with the PPant attached to Ser36 of AcpP, which is consistent with previous studies,[30,143–145] that show these charge complementary surfaces dictate protein-protein interactions. Secondly, Arg132 and Lys161 of EcFabA form salt bridges with Glu41 and Glu47 of AcpP to anchor the complex. Thirdly, Arg136 and Arg137 of EcFabA interact with Ala59 and Glu60 of AcpP to pry away helix III. Finally, Leu138 and Val134 of EcFabA have hydrophobic interactions with Leu37 and Val40 from AcpP helix II to further stabilize the complex.[57] Interestingly, one FabA pries apart the AcpP, facilitating acyl chain translocation into the cleft between the FabA monomers, but the other monomer presents the active-site histidine for crosslinking.
These four events together stabilize AcpP in its open conformation so the acyl substrate can be released into the binding pocket of EcFabA. As this occurs, five hydrophobic pocket residues between helices II and III of AcpP move inward and collapse the substrate pocket. This provides experimental evidence for the previously proposed switchblade (or chain-flipping) mechanism.[30,31]
Additionally, the gate-keeping residues Phe165 and Phe171 at the entrance to the active site were seen to be completely rotated or displaced in the crosslinked structure as compared to apo-EcFabA. This active site entrance control is also seen in HpFabZ, where the top of the active site is controlled by an aromatic residue, Tyr100. It switches between open and closed conformations helping to control substrate acceptance and product exit.[146]
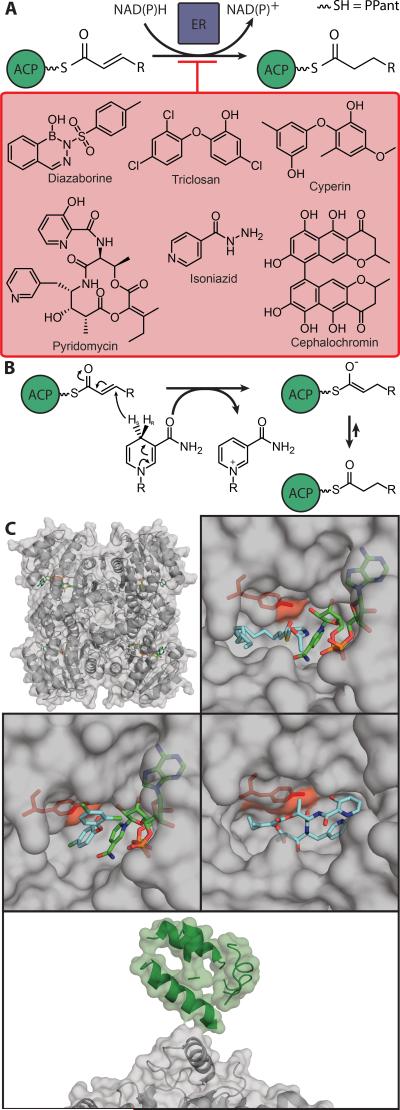
9. Enoyl Reductase (ER)
Enoyl-ACP reductase (ER) is a member of the short-chain dehydrogenase (SDR) family and performs the last step in each fatty acid biosynthetic cycle by reducing 2-enoyl-ACP to fatty acyl-ACP (Figure 7a). In E. coli there is a single enoyl-ACP reductase, FabI, which is essential.[147] Since the KR and DH activities that precede the ER are both reversible, FabI plays a determinant role in completing each elongation cycle and is thought to be the rate-limiting step.[148] Overexpression of fabI in E. coli did not result in growth defect but also did not influence fatty acid accumulation.[114] Three other ER isozymes have been reported in bacteria: FabL and FabV in the SDR family and FabK, a TIM barrel flavoprotein unrelated to the SDR family.[149–151]
Figure 7.
A) Schematic of ER chemistry and several known inhibitors. B) Diagram of ER mechanism with enolate intermediate. C) Comparison of binding of substrates and inhibitors for InhA, the MtFabI. Top left: overall topology with NAD+ (green) bound, with transparent overlay of electrostatic surface; top right: active site occupation by NAD+ (green) and the fatty acid trans-2-hexadecenoyl-(N-acetyl-cysteamine)-thioester (cyan) with the hydrogen bond positioning tyrosine (Tyr158, red) (PDB: 1BVR). Middle-left: binding of triclosan (cyan) and NAD+ (green), with positioning tyrosine (Tyr158, red), forming a tight-binding but noncovalent complex (PDB: 2B35). Middle-right: binding of pyridomycin (cyan), with Tyr158 (red) in the absence of NAD+ (PDB: 4BII). Transparent surface over cartoon of the EcACP interface with EcFabI, with residues not resolved (PDB: 2FHS).
9.1. Mechanism
In E. coli, NADPH reduction of the double bond proceeds by conjugate syn-addition of the pro-4S hydrogen from NAD(P)H to carbon 3 of the trans-2-acyl group, forming an enolate anion intermediate which is stabilized by a lysine residue in the ER active site. Protonation of C2 to form the final product acyl-ACP is performed by an active site tyrosine residue (Figure 7b).[152,153] In InhA, the ER from M. tuberculosis, the stabilization of the enolate anion intermediate is presumed to be by a tyrosine rather than a lysine, because in the X-ray structure of InhA in complex with NAD+ and a C16 fatty acyl substrate (PDB 1BVR), Tyr158 is seen to make a direct hydrogen bond with the fatty acyl substrate thioester carbonyl oxygen (Figure 7c).[154]
9.2. Inhibition
FabI is inhibited by low concentrations of palmitoyl-CoA.[147] Additionally, there are currently three well-known small molecule inhibitors of ER: triclosan, isoniazid and diazoborines (Figure 7a), which bind to the active site of ER and form a tight complex with the nicotinamide cofactor.[120] Isoniazid, the primary anti-tuberculosis drug, is a prodrug that must first undergo activation to a free radical by KatG prior to acylation of the NAD+ radical, forming a covalent complex with the cofactor nicotinamide ring.[71] This species has been found to inhibit InhA from M. tuberculosis as well as ER domains from other mycobacteria. Acyl-ACP substrates can prevent isoniazid inhibition of InhA, suggesting the activated isoniazid interacts with the substrate-binding region.[154]
Triclosan was added to consumer products, such as antiseptic soaps, toothpaste and cosmetics, prior to understanding its target. In 1999, the EcFabI-NAD+-triclosan complex was reported.[155] Triclosan was found to non-covalently interact with the 2’-hydroxyl of ribose through hydrogen bonds while also hydrogen bonding to the catalytic tyrosine residue and experiencing π-π stacking interactions with the pyridine ring of the charged cofactor.[155,156] In the same year, a conformational change of the Ile192-Ser198 loop of EcFabI was shown to close the active site and allow this region to form van der Waals interactions with the inhibitor and hydrogen bond the bound NAD+. This appears critical for the enhanced binding activity of triclosan.[157] In addition, triclosan has been found to inhibit FabI from the malarial causative apicomplexan P. falciparum, but the validity of FAS II as a drug target against P. falciparum is currently under debate after the findings that the microorganism can scavenge fatty acids during the blood stage, leaving no indispensible role to FAS II.[158]
Diazoborines are a class of heterocyclic boron-containing compounds that inhibit FabI by covalent attachment of the boron atom with the 2’-hydroxyl of the NAD+ ribose.[156] Additionally, diazoborines π stack with the nicotinamide ring, and the boron and hydroxyl group occupy the space of the enolate intermediate. Diazoborines are not clinically used due to their undesirable inhibition of RNA processing.[159] Isoniazid and triclosan are used clinically, however a single point mutation in the M. smegmatis and M. bovis FabIs confers resistance to isoniazid.[154] As of 2009, 30% of clinical isolates of M. tuberculosis were isoniazid resistant.[150] Also, mutants of the EcFabI active site, G93C for example, acquire resistance to triclosan by interfering with the formation of a stable FabI-NAD+-triclosan ternary complex.[155]
More recently, cyperin, a natural compound of fungal origin, was found to inhibit ER from Arabidopsis thaliana, mimicking the binding of triclosan.[160] Cephalochromin, a fungal secondary metabolite, inhibits FabI but the mechanism has not been elucidated.[161] Pyridomycin, a natural product with potent anti-tuberculosis activity, inhibits InhA with a new mechanism not yet seen. Pyridomycin is a competitive inhibitor of NADH binding in InhA and blocks both the NADH cofactor and fatty acid substrate binding pockets. This inhibitor's unique mechanism can be a starting point for future inhibitor design (Figure 7a).[162]
9.3. Interaction with ACP
EcFabI is a tetramer, and a combination of X-ray crystallography and molecular dynamics (MD) simulation show that two ACPs loaded with a 2-dodecenoyl acyl group react with one EcFabI. This is unusual since each EcFabI has 4 active sites. The FabI-acyl ACP complex of E. coli is stabilized by largely electrostatic interactions between the acidic residues in the ACP helix II (Asp35, Asp38, Glu41, Glu48) and a patch of basic residues (Lys201, Arg204, Lys205) adjacent to the EcFabI substrate-binding loop, consistent with the structural and modeling studies of ACP with AcpS, FabA, FabH, and FabG. The ACP PPant is proposed to deliver the enoyl substrate to the minor portal between EcFabI helix α8 and the mobile loop of residues 152-156. However, the absence of electron density for the ACP pantetheine hindered the ability to predict how the ACP delivers the substrate into the EcFabI active site (Figure 7c).[163]
When the substrate binds, EcFabI does not experience an overall conformational change, but rather the substrate-binding loop (residues 191-200) goes through a major conformational change upon complexation with ACP. In the triclosan-EcFabI crystal structure, triclosan was shielded from the solvent by the substrate-binding loop, but in the EcFabI-acyl ACP complex crystal structure the loop adopts an open lid conformation, presumably because of the interaction with ACP in this area. Details of this interaction could not be elucidated from the crystal structure due to missing side chain density. In regards to the catalytic triad (Tyr146, Tyr156, and Lys163 in EcFabI and Phe149, Tyr158 and Lys165 in M. tuberculosis InhA); Tyr156 does not play a significant role in the reduction and the substrate carbonyl group is pointing away from it in the crystal structure. Tyr146 is directly involved in catalysis and hydrogen bonds with the substrate carbonyl. This differs from than the corresponding Phe149 in InhA, which is proposed to position the cofactor for hydride transfer. Finally, the function of Lys165 is not yet established.[163]
InhA, the enoyl reductase from M. tuberculosis, prefers long chain fatty acyl substrates, C16 and longer. The InhA crystal structure with NAD+ and a C16 acyl substrate, trans-2-hexadecenoyl-(N-acetylcysteamine)-thioester, revealed that the substrate binds in a “U-shaped” conformation, with the trans double bond positioned adjacent to the nicotinamide ring of NAD+. Hydrophobic residues from the substrate-binding loop (residues 196-219) engulf the fatty acyl chain portion of the substrate. The substrate-binding loop is longer than EcFabI, creating a deeper binding pocket to accommodate longer chain acyl substrates.[154] The interaction of InhA with ACP has not yet been elucidated.
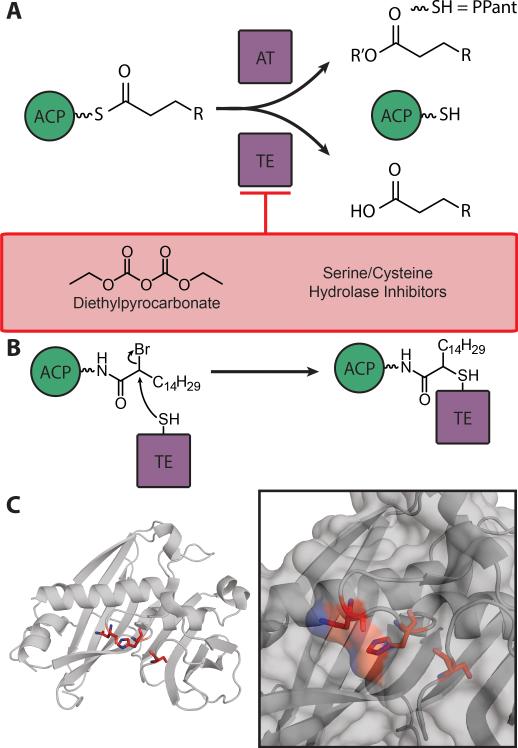
10. Thioesterase (TE)/Acyl Transferase (AT)
After the iterative chain elongation cycle, FAB is terminated either by offloading the fatty acid from the ACP by an acyl-ACP thioesterase releasing a free fatty acid or by transesterification onto a lipid by an acyltransferase (Figure 8a). Most often, prokaryotes utilize ATs and eukaryotes utilize TEs.
Figure 8.
A) Schematic of AT and TE chemistry, with inhibitors of TE. R’ = substrate of AT activity, including sugars, proteins and small molecules. B) Diagram of TE crosslinker mechanism C) Demonstration of catalytic triad of a Bacteroides thetaiotaomicron TE. At left: overall topology; at right: close-up on triad orientation and nearness to protein surface (PDB: 2ESS). The catalytic triad (Asn172, His174, Ser209) is denoted in red.
In E. coli, the acyl group is directly transferred from ACP to glycerol-3-phosphate by a glycerol-3-phosphate AT, PlsB (a membrane protein). Free fatty acids are not found as intermediates in bacterial lipid biosynthesis.[164] A plsB mutant in E. coli leads to the cessation of phospholipid biosynthesis, an accumulation of long chain acyl-ACP and the inhibition of de novo fatty acid biosynthesis.[165] A conserved domain containing histidine and aspartic acid separated by four residues (HX4D) was identified for ATs from bacterial, plant and animal kingdoms. The catalytic histidine is thought to function as a base to deprotonate the hydroxyl moiety of the acyl acceptor. The PlsB[His36Ala] mutant lacked AT activity. The PlsB[Asp311Ala] mutant also lacked activity and did not assemble into the membrane, suggesting that proper folding and membrane insertion is also dictated by the active site aspartic acid.[166] Additionally, a conserved glycine is involved in catalysis by playing a steric role in the active site, and an invariant arginine is critical for glycerol 3-phosphate binding, as seen by a 13-fold higher KM for EcPlsB mutant Arg354Cys.[167] This acyl transferase is one of the proteins that controls chain length in E. coli.[164] However, in spite of this mechanistic insight, very little is known about PlsB protein-protein interactions, and little has been done with this enzyme in terms of metabolic engineering. TEs from plants, algae and bacteria have been used extensively in metabolic engineering of FAB and will be the focus of the remainder of this section.
As of 2010, there were 23 families of TEs grouped by their substrate and tertiary structure, and these were almost completely unrelated by primary structure. Of these 23 families, only 6 break bonds between acyl groups and ACP (the other TEs act on CoA, glutathione and other proteins). Two of the groups (TE14 and TE15) contain TEs found in plants, algae, and bacteria and have a typical hotdog fold, whereas the remaining groups are found in fungi and mammals and are TEs with an α/β hydrolase fold (TE16-TE19). The most characterized TEs that have garnered interest for metabolic engineering fall into the TE14 family.[168] The N-terminal hot dog domain has been shown by chimeric enzyme construction to control chain length specificity.[169] E. coli has seven hotdog TEs, but none are believed to be dedicated acyl-ACP TEs and are therefore not directly involved in fatty acid or lipid biosynthesis.[170] TesA and TesB do work on acyl-ACP and are more active on longer chain fatty acids. However, TesA and TesB activity for palmitoyl-ACP is 0.6% and 0.1% the rate they work on palmitoyl-CoA, respectively.[171] When tesA is overexpressed in E. coli, without targeting periplasmic sequence, the amount of free fatty acids is increased, producing titers as high as 0.32 g/L.[172,173]
In plants the TEs can be the sole factor in determining fatty acid chain length. There are two distinct TE gene classes in higher plants that are dedicated acyl-ACP TEs, fatA and fatB. fatA encodes a TE with preference for 18:1-ACP and a minor preference for 18:0 and 16:0-ACP. fatB encodes a TE with a preference for short/medium chain saturated fatty acids ranging from 8 to 16 carbons.[174,175] When plant TEs are overexpressed in E. coli, the fatty acid profile is altered by the specificity of the TE.[172] When the C12:0-ACP selective TE from Umbellularia californica (UcFatB1) was overexpressed in E. coli, a 500-fold increase in C12:0 fatty acid production was seen.[176,177]
10.1. Mechanism
The α/β hydrolase TEs have a conserved catalytic triad: a nucleophile-histidine-acid triad, with the nucleophile commonly being a serine residue. The acid stabilizes the basic histidine, which accepts a proton from the nucleophile. The nucleophilic serine forms a tetrahedral intermediate with the substrate before it is attacked by water.[178] The active site residues in TEs are on the surface of the protein, rather than buried in an acyl-binding pocket as seen with other FAS domains (Figure 8c). The hotdog-fold TEs lack defined non-solvated binding pockets and conserved catalytic residues, and therefore a variety of catalytic residues and mechanisms exist.[170] Plant TEs likely have a papain-like catalytic triad, as they are inhibited by thiol inhibitors and carry a conserved histidine required for catalysis .[179] A bioinformatics-guided site-directed mutagenesis study proposed the catalytic triad for the A. thaliana TE (FatB) to be Cys264, His229, and Asn227, but there is currently no structural verification.[180,181]
10.2. Inhibition
Plant TEs have a cysteine catalytic residue and therefore are generally inhibited by compounds that are known to react with thiols. Animal and bacterial TEs have a serine catalytic residue and are inhibited by serine reactive reagents.[179]
When the Umbellularia californica plant TE (UcTE) was pre-incubated with diethylpyrocarbonate, a 97% loss in activity was seen (Figure 8a).[182] Diethylpyrocarbonate acts by covalently modifying histidine at the N-Ω-2 nitrogen of the imidazole ring, showing the evidence for a catalytic histidine residue. UcTE was also completely inhibited by N-ethylmaleimide suggesting an active-cysteine mechanism, since it acts as a Michael acceptor for soft nucleophiles.[183]
10.3. Interaction with ACP
There has been considerable interest in the metabolic engineering of plant TEs, but thus far it has been met with limited success due to a lack of structural information.
In one recent study, the Jatropha curcas FatA and FatB were docked with 4’-PPant substrates 4’-PPT-16:0, 4’-PPT-18:0 and 4’-PPT-18:1, and the top models bound to the active catalytic triad. These models were then used for protein-protein docking between spinach ACP and JcFatA or JcFatB. For 5 JcFatB models and 9 JcFatA models, ACP docked near the catalytic triad, and the serine of ACP was oriented towards the TE active site.[184]
Unlike plants, green algae only have one TE, Fat1. Some plant TEs have been engineered into E. coli and other various plants, and this led to changes in fatty acid quantity and profile observed.[55] However, engineering plant TEs into algae has been unpredictable, likely due to mismatched protein-protein interactions. Cr-cACP and CrTE from Chlamydomonas reinhardtii were docked, identifying a protein-protein recognition surface. The fatty-acyl thioester attached to Cr-cACP docked closely to the active site cysteine of CrTE.
Additionally, an α-bromopalmitic pantetheine activity-based probe was utilized to investigate ACP-TE interactions. This probe is very electrophilic at the α carbon and allows for nucleophilic attack by the active site TE nucleophile, leading to covalent attachment of the probe (Figure 8b). When the probe is chemoenzymatically loaded onto ACP and a partner TE is added, a covalent crosslinked complex is achieved. CrTE, UcTE and ChTE orient in a similar conformation to Cr-cACP in computational docking studies. However, only Cr-cACP and CrTE successfully formed a crosslinked complex, showing that vascular plant TEs do not interact with CrACP in vitro. Far less complex formation of CrcACP-CrTE was seen with a α-bromohexanoic pantetheine probe than with the C16 analog, showing that protein-protein interactions, along with chain length, govern the hydrolysis by TE.[54]
11. Conclusions and Outlook
This review has established that the mechanistic, structural and inhibition understanding of type II FAS is substantial. However, drug resistance, a lack of clinical viability and unachieved FAS engineering prospects highlight the need for a new generation of studies focusing on protein-protein interactions with ACP. Future investigations will delve deeper into the fundamentals of the FAS to future inhibitor design and metabolic engineering efforts.
It is evident that protein-protein interactions are necessary for substrate modification attached to ACP in the FAS.[13] Each interaction between ACP and its partners must be specific enough to allow precise chain flipping and substrate presentation, but weak enough to allow rapid turnover.[85] In 2000, the ACP-AcpS crystal structure allowed the first visualization of domain-ACP interactions and served as a starting point for future work. However, AcpS acts only on serine, not the PPant arm of ACP, raising questions about the generality of the observed interactions. In 2001, Zhang et. al. anticipated that ACP conformational changes facilitate the presentation of cargo and the release post-modification, but could not demonstrate how.[85] It was not until 2006 that a crystal structure of a partner domain (FabI) and ACP was solved, but the lack of resolution of the PPant and nearby side chain density precluded insight into substrate delivery (Section 9.3).[185] It was not until 2014 that the crosslinked crystal structure of ACP and FabA was elucidated, allowing a snapshot of the AcpP in transition and when fully interacting with a native partner (Section 8.3). Using new technologies, including a functional AcpH resin, a “one-pot” chemoenzymatic reaction for crypto-ACP preparation and mechanistic crosslinking probes (Section 2), along with protein crystallography and NMR experiments (Section 3), among others we have been able to directly observe substrate delivery.
Some directions for future work could include the design, synthesis and application of mechanistic crosslinking probes for cofactor dependent ER and KR domains, since these domains are essential and their protein-protein interactions could prove to be an ideal target for inhibitor design. Additionally, cryo-electron microscopy has been used recently to study large type I FAS complexes in motion.[70,186] The application of mechanistic crosslinking probes to stabilize type I FAS complexes for better resolution can enhance future cryo-EM studies. To build upon current in vitro studies, AasS can be utilized as a means to study the FAS and subsequent products in vivo by loading unique visualization and activity based acids onto ACP in a fast and efficient manner.[56]
The outlook for FAS research is bright. New tools are available, allowing visualization of previously unobservable states. Our understanding of the ACP interaction with partner proteins allows future studies of processivity, sequestration and new cargo-dependent conformational changes. Specifically, studies on the ACP interaction with the TE will yield crucial information for engineering attempts to modify fatty acid profiles, in the same manner that unsaturation may be controlled by careful engineering of dehydratases. Finally, while the chemistry of FAB is conserved between type I and type II FAS systems, sequestration and ACP-partner interactions are different, providing an opportunity for directed development of antibiotics.
Acknowledgements
We thank J. Beld for critical advice. This work was supported by NIH R01GM094924 and R01GM086225.
Biography
 Kara Finzel is originally from Long Island, New York. She graduated from the University of Richmond in 2011 where she conducted research under the tutelage of Dr. John Gupton and received a B.S in Chemistry. She has been in the PhD program at the University of California-San Diego in the Department of Chemistry and Biochemistry under the guidance of Prof. Michael Burkart since 2011. Her current work is focused on the development of new tools for in vitro and in vivo carrier protein labeling to study protein-protein interactions in fatty acid biosynthesis.
Kara Finzel is originally from Long Island, New York. She graduated from the University of Richmond in 2011 where she conducted research under the tutelage of Dr. John Gupton and received a B.S in Chemistry. She has been in the PhD program at the University of California-San Diego in the Department of Chemistry and Biochemistry under the guidance of Prof. Michael Burkart since 2011. Her current work is focused on the development of new tools for in vitro and in vivo carrier protein labeling to study protein-protein interactions in fatty acid biosynthesis.
 Originally from San Diego, D. John Lee traveled to University of California, Santa Cruz to earn BS degrees in Chemistry and Biology. While there, he studied under Prof. Bakthan Singaram advancing boronic asymmetric reducing agents. He returned to San Diego in 2010, and joined Prof. Michael Burkart's lab as a doctoral student. He currently studies protein-protein interactions of FAS and PKS enzymes by NMR, exploiting chemical tools developed in the Burkart laboratory.
Originally from San Diego, D. John Lee traveled to University of California, Santa Cruz to earn BS degrees in Chemistry and Biology. While there, he studied under Prof. Bakthan Singaram advancing boronic asymmetric reducing agents. He returned to San Diego in 2010, and joined Prof. Michael Burkart's lab as a doctoral student. He currently studies protein-protein interactions of FAS and PKS enzymes by NMR, exploiting chemical tools developed in the Burkart laboratory.
 A native Texan, Michael Burkart received a BA in chemistry from Rice University in 1994. He received a PhD in Organic Chemistry from the Scripps Research Institute in 1999 under the mentorship of Prof. Chi-Huey Wong, after which time he pursued postdoctoral studies at Harvard Medical School with Prof. Christopher Walsh. Since 2002 he has taught and performed research at the University of California, San Diego, where he is a full Professor of Chemistry and Biochemistry and Associate Director of the California Center for Algae Biotechnology. His research interests include natural product synthesis and biosynthesis, enzyme mechanism, and metabolic engineering. Prof. Burkart has been a fellow of the Alfred P. Sloan Foundation and Ellison Medical Foundation and was the RSC Organic and Biomolecular Chemistry Lecturer for 2010.
A native Texan, Michael Burkart received a BA in chemistry from Rice University in 1994. He received a PhD in Organic Chemistry from the Scripps Research Institute in 1999 under the mentorship of Prof. Chi-Huey Wong, after which time he pursued postdoctoral studies at Harvard Medical School with Prof. Christopher Walsh. Since 2002 he has taught and performed research at the University of California, San Diego, where he is a full Professor of Chemistry and Biochemistry and Associate Director of the California Center for Algae Biotechnology. His research interests include natural product synthesis and biosynthesis, enzyme mechanism, and metabolic engineering. Prof. Burkart has been a fellow of the Alfred P. Sloan Foundation and Ellison Medical Foundation and was the RSC Organic and Biomolecular Chemistry Lecturer for 2010.
References
- 1.Dunn BJ, Khosla C, Soc JR. Interface. 2013;10:20130297. doi: 10.1098/rsif.2013.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong FT, Khosla C. Curr. Opin. Chem. Biol. 2012;16:117–123. doi: 10.1016/j.cbpa.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das A, Khosla C. Acc. Chem. Res. 2009;42:631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khosla C. J. Org. Chem. 2009;74:6416–6420. doi: 10.1021/jo9012089. [DOI] [PubMed] [Google Scholar]
- 5.Khosla C, Kapur S, Cane DE. Curr. Opin. Chem. Biol. 2009;13:135–143. doi: 10.1016/j.cbpa.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keatinge-Clay AT. Nat. Prod. Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 7.Piel J. Nat. Prod. Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 8.Jenni S, Leibundgut M, Boehringer D, Frick C, Mikolásek B, Ban N. Science. 2007;316:254–261. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- 9.Maier T, Leibundgut M, Ban N. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 10.Rock CO, Cronan J. Methods Enzymol. 1981;71:341–351. doi: 10.1016/0076-6879(81)71043-7. [DOI] [PubMed] [Google Scholar]
- 11.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Nat. Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 12.Butland G, Peregrín-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 13.Crosby J, Crump MP. Nat. Prod. Rep. 2012;29:1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- 14.Falb M, Müller K, Königsmaier L, Oberwinkler T, Horn P, von Gronau S, Gonzalez O, Pfeiffer F, Bornberg-Bauer E, Oesterhelt D. Extremophiles. 2008;12:177–196. doi: 10.1007/s00792-008-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombard J, López-García P, Moreira D. Mol. Biol. Evol. 2012;29:3261–3265. doi: 10.1093/molbev/mss160. [DOI] [PubMed] [Google Scholar]
- 16.V Dibrova D, Galperin MY, Mulkidjanian AY. Environ. Microbiol. 2014;16:907–918. doi: 10.1111/1462-2920.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anselmi C, Grininger M, Gipson P, Faraldo-Gómez JD. J. Am. Chem. Soc. 2010;132:12357–12364. doi: 10.1021/ja103354w. [DOI] [PubMed] [Google Scholar]
- 18.Keating DH, Carey MR, Cronan JE. J. Biol. Chem. 1995;270:22229–22235. doi: 10.1074/jbc.270.38.22229. [DOI] [PubMed] [Google Scholar]
- 19.Roujeinikova A, Simon WJ, Gilroy J, Rice DW, Rafferty JB, Slabas AR. J. Mol. Biol. 2007;365:135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 20.Evans SE, Williams C, Arthur CJ, Burston SG, Simpson TJ, Crosby J, Crump MP. ChemBioChem. 2008;9:2424–2432. doi: 10.1002/cbic.200800180. [DOI] [PubMed] [Google Scholar]
- 21.Zornetzer GA, Tanem J, Fox BG, Markley JL. Biochemistry. 2009;49:470–477. doi: 10.1021/bi9014659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beld J, Cang H, Burkart MD. Angew. Chemie. 2014 doi: 10.1002/anie.201408576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson MNR, Londergan CH, Charkoudian LK. J. Am. Chem. Soc. 2014;136:11240–11243. doi: 10.1021/ja505442h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Płoskoń E, Arthur CJ, Evans SE, Williams C, Crosby J, Simpson TJ, Crump MP. J. Biol. Chem. 2008;283:518–528. doi: 10.1074/jbc.M703454200. [DOI] [PubMed] [Google Scholar]
- 25.Rock CO, Cronan JE, Armitage IM. J. Biol. Chem. 1981;256:2669–2674. [PubMed] [Google Scholar]
- 26.Post-Beittenmiller D, Jaworski JG, Ohlrogge JB. J. Biol. Chem. 1991;266:1858–1865. [PubMed] [Google Scholar]
- 27.Jones PJ, Holak TA, Prestegard JH. Biochemistry. 1987;26:3493–3500. doi: 10.1021/bi00386a037. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y, Kovrigin EL, Eletr Z. Biochem. Biophys. Res. Commun. 2006;341:776–783. doi: 10.1016/j.bbrc.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 29.Płoskoń E, Arthur CJ, Kanari ALP, Wattana-amorn P, Williams C, Crosby J, Simpson TJ, Willis CL, Crump MP. Chem. Biol. 2010;17:776–785. doi: 10.1016/j.chembiol.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 30.Leibundgut M, Jenni S, Frick C, Ban N. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 31.Cronan JE. Biochem. J. 2014;460:157–163. doi: 10.1042/BJ20140239. [DOI] [PubMed] [Google Scholar]
- 32.Zornetzer GA, Fox BG, Markley JL. Biochemistry. 2006;45:5217–5227. doi: 10.1021/bi052062d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beld J, Sonnenschein EC, Vickery CR, Noel JP, Burkart MD. Nat. Prod. Rep. 2014;31:61–108. doi: 10.1039/c3np70054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gehring AM, Lambalot RH, Vogel KW, Drueckhammer DG, Walsh CT. Chem. Biol. 1997;4:17–24. doi: 10.1016/s1074-5521(97)90233-7. [DOI] [PubMed] [Google Scholar]
- 35.Wenzel SC, Müller R. Curr. Opin. Biotechnol. 2005;16:594–606. doi: 10.1016/j.copbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Weissman KJ, Hong H, Oliynyk M, Siskos AP, Leadlay PF. ChemBioChem. 2004;5:116–125. doi: 10.1002/cbic.200300775. [DOI] [PubMed] [Google Scholar]
- 37.Fernández-Valverde M, Reglero A, Martinez-Blanco H, Luengo JM. Appl. Environ. Microbiol. 1993;59:1149–1154. doi: 10.1128/aem.59.4.1149-1154.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Go MK, Chow JY, Cheung VWN, Lim YP, Yew WS. Biochemistry. 2012;51:4568–79. doi: 10.1021/bi300425j. [DOI] [PubMed] [Google Scholar]
- 39.Holliday GL, Rahman SA, Furnham N, Thornton JM. J. Mol. Biol. 2014;426:2098–111. doi: 10.1016/j.jmb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarke KM, Mercer AC, La Clair JJ, Burkart MD. J. Am. Chem. Soc. 2005;127:11234–11235. doi: 10.1021/ja052911k. [DOI] [PubMed] [Google Scholar]
- 41.Worthington AS, Burkart MD. Org. Biomol. Chem. 2006;4:44–46. doi: 10.1039/b512735a. [DOI] [PubMed] [Google Scholar]
- 42.Mandel AL, La Clair JJ, Burkart MD. Org. Lett. 2004;6:4801–4803. doi: 10.1021/ol048853+. [DOI] [PubMed] [Google Scholar]
- 43.Meier JL, Mercer AC, Rivera H, Burkart MD. J. Am. Chem. Soc. 2006;128:12174–12184. doi: 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- 44.Worthington AS, Rivera H, Torpey JW, Alexander MD, Burkart MD. ACS Chem. Biol. 2006;1:687–691. doi: 10.1021/cb6003965. [DOI] [PubMed] [Google Scholar]
- 45.Vagelos P, Larrabee A. J. Biol. Chem. 1967;242:1776–1781. [PubMed] [Google Scholar]
- 46.Thomas J, Cronan JE. J. Biol. Chem. 2005;280:34675–34683. doi: 10.1074/jbc.M505736200. [DOI] [PubMed] [Google Scholar]
- 47.Flugel RS, Hwangbo Y, Lambalot RH, Cronan JE, Walsh CT. J. Biol. Chem. 2000;275:959–968. doi: 10.1074/jbc.275.2.959. [DOI] [PubMed] [Google Scholar]
- 48.Murugan E, Kong R, Sun H, Rao F, Liang Z-X. Protein Expr. Purif. 2010;71:132–138. doi: 10.1016/j.pep.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Kosa NM, Haushalter RW, Smith AR, Burkart MD. Nat. Methods. 2012;9:981–984. doi: 10.1038/nmeth.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rothmann M, Kosa NM, Burkart MD. RSC Adv. 2014;4:9092–9097. doi: 10.1039/C3RA47847E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Worthington AS, Porter DF, Burkart MD. Org. Biomol. Chem. 2010;8:1769–1772. doi: 10.1039/b925966j. [DOI] [PubMed] [Google Scholar]
- 52.Ishikawa F, Haushalter RW, Burkart MD. J. Am. Chem. Soc. 2012;134:769–72. doi: 10.1021/ja2082334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa F, Haushalter RW, Lee DJ, Finzel K, Burkart MD. J. Am. Chem. Soc. 2013;135:8846–8849. doi: 10.1021/ja4042059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blatti JL, Beld J, Behnke CA, Mendez M, Mayfield SP, Burkart MD. PLoS One. 2012;7:e42949. doi: 10.1371/journal.pone.0042949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beld J, Blatti J, Behnke C, Mendez M, Burkart M. J. Appl. Phycol. 2013;26:1619–1629. doi: 10.1007/s10811-013-0203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beld J, Finzel K, Burkart MD. Chem. Biol. 2014;21:1257–1259. doi: 10.1016/j.chembiol.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen C, Haushalter RW, Lee DJ, Markwick PRL, Bruegger J, Caldara-Festin G, Finzel K, Jackson DR, Ishikawa F, O'Dowd B, McCammon JA, Opella SJ, Tsai S-C, Burkart MD. Nature. 2014;505:427–31. doi: 10.1038/nature12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williamson MP. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Bonvin AMJJ, Boelens R, Kaptein R. Curr. Opin. Chem. Biol. 2005;9:501–508. doi: 10.1016/j.cbpa.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 60.Jarymowycz VA, Stone MJ. Chem. Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- 61.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 62.Lipari G, Szabo A. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 63.Chan DI, Stockner T, Tieleman DP, Vogel HJ. J. Biol. Chem. 2008;283:33620–33629. doi: 10.1074/jbc.M805323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gasper PM, Fuglestad B, Komives EA, Markwick PRL, McCammon JA. Proc. Natl. Acad. Sci. U. S. A. 2012;109:21216–21222. doi: 10.1073/pnas.1218414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Markwick PRL, McCammon JA. Phys. Chem. Chem. Phys. 2011;13:20053–20065. doi: 10.1039/c1cp22100k. [DOI] [PubMed] [Google Scholar]
- 67.Blackledge M. Prog. Nucl. Magn. Reson. Spectrosc. 2005;46:23–61. [Google Scholar]
- 68.Asturias FJ, Chadick JZ, Cheung IK, Stark H, Witkowski A, Joshi AK, Smith S. Nat Struct Mol Biol. 2005;12:225–232. doi: 10.1038/nsmb899. [DOI] [PubMed] [Google Scholar]
- 69.Gipson P, Mills DJ, Wouts R, Grininger M, Vonck J, Kühlbrandt W. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9164–9169. doi: 10.1073/pnas.0913547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boehringer D, Ban N, Leibundgut M. J. Mol. Biol. 2013;425:841–849. doi: 10.1016/j.jmb.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 71.Heath R, White S, Rock C. Appl. Microbiol. Biotechnol. 2002;58:695–703. doi: 10.1007/s00253-001-0918-z. [DOI] [PubMed] [Google Scholar]
- 72.Joshi VC, Wakil SJ. Arch. Biochem. Biophys. 1971;143:493–505. doi: 10.1016/0003-9861(71)90234-7. [DOI] [PubMed] [Google Scholar]
- 73.Verwoert IIGS, Verhagen EF, van der Linden KH, Verbree EC, Nijkamp HJJ, Stuitje AR. FEBS Lett. 1994;348:311–316. doi: 10.1016/0014-5793(94)00630-x. [DOI] [PubMed] [Google Scholar]
- 74.Garwin JL, Klages AL, Cronan JE. J. Biol. Chem. 1980;255:3263–3265. [PubMed] [Google Scholar]
- 75.Jeon E, Lee S, Won J-I, Han SO, Kim J, Lee J. Enzyme Microb. Technol. 2011;49:44–51. doi: 10.1016/j.enzmictec.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 76.Zhang L, Liu W, Xiao J, Hu T, Chen J, Chen K, Jiang H, Shen X. Protein Sci. 2007;16:1184–1192. doi: 10.1110/ps.072757307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keatinge-Clay AT, Shelat AA, Savage DF, Tsai S-C, Miercke LJW, O'Connell JD, III, Khosla C, Stroud RM. Structure. 2003;11:147–154. doi: 10.1016/s0969-2126(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 78.Natarajan S, Kim J-K, Jung T-K, Doan T, Ngo H-P-T, Hong M-K, Kim S, Tan V, Ahn S, Lee S, Han Y, Ahn Y-J, Kang L-W. Mol. Cells. 2012;33:19–25. doi: 10.1007/s10059-012-2155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sreshty MAL, Surolia A, Sastry GN, Murty US. Mol. Inform. 2012;31:281–299. doi: 10.1002/minf.201100051. [DOI] [PubMed] [Google Scholar]
- 80.Oefner C, Schulz H, D'Arcy A, Dale GE. Acta Crystallogr. Sect. D. 2006;62:613–618. doi: 10.1107/S0907444906009474. [DOI] [PubMed] [Google Scholar]
- 81.Li Z, Huang Y, Ge J, Fan H, Zhou X, Li S, Bartlam M, Wang H, Rao Z. J. Mol. Biol. 2007;371:1075–1083. doi: 10.1016/j.jmb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 82.Campbell JW, Cronan JE. Annu. Rev. Microbiol. 2001;55:305–332. doi: 10.1146/annurev.micro.55.1.305. [DOI] [PubMed] [Google Scholar]
- 83.Sinha I, Dick T. J. Antimicrob. Chemother. 2004;53:1072–1075. doi: 10.1093/jac/dkh241. [DOI] [PubMed] [Google Scholar]
- 84.Sun M, Zhu G, Qin Z, Wu C, Lv M, Liao S, Qi N, Xie M, Cai J. Mol. Biochem. Parasitol. 2012;184:20–28. doi: 10.1016/j.molbiopara.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 85.Zhang Y-M, Rao MS, Heath RJ, Price AC, Olson AJ, Rock CO, White SW. J. Biol. Chem. 2001;276:8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y-M, Wu B, Zheng J, Rock CO. J. Biol. Chem. 2003;278:52935–52943. doi: 10.1074/jbc.M309874200. [DOI] [PubMed] [Google Scholar]
- 87.Arthur CJ, Williams C, Pottage K, Płoskoń E, Findlow SC, Burston SG, Simpson TJ, Crump MP, Crosby J. ACS Chem. Biol. 2009;4:625–636. doi: 10.1021/cb900099e. [DOI] [PubMed] [Google Scholar]
- 88.Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Structure. 2000;8:883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 89.Heath RJ, Rock CO. J. Biol. Chem. 1996;271:10996–11000. doi: 10.1074/jbc.271.18.10996. [DOI] [PubMed] [Google Scholar]
- 90.Lai C-Y, Cronan JE. J. Biol. Chem. 2003;278:51494–503. doi: 10.1074/jbc.M308638200. [DOI] [PubMed] [Google Scholar]
- 91.Tsay JT, Oh W, Larson TJ, Jackowski S, Rock CO. J. Biol. Chem. 1992;267:6807–6814. [PubMed] [Google Scholar]
- 92.Edwards P, Sabo Nelsen J, Metz JG, Dehesh K. FEBS Lett. 1997;402:62–66. doi: 10.1016/s0014-5793(96)01437-8. [DOI] [PubMed] [Google Scholar]
- 93.Feng Y, Cronan JE. J. Biol. Chem. 2009;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Subrahmanyam S, Cronan JE. J. Bacteriol. 1998;180:4596–4602. doi: 10.1128/jb.180.17.4596-4602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuo J, Khosla C. Metab. Eng. 2014;22:53–59. doi: 10.1016/j.ymben.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Olsen JG, Kadziola A, von Wettstein-Knowles P, Siggaard-Andersen M, Larsen S. Structure. 2001;9:233–243. doi: 10.1016/s0969-2126(01)00583-4. [DOI] [PubMed] [Google Scholar]
- 97.Huang Jia Edwards WJ, Dehesh Patricia, Schneider Katayoon, Lindqvist Gunter, Ylva EMBO J. 1998;17:1183–1191. doi: 10.1093/emboj/17.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio Á, et al. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 99.Davies C, Heath RJ, White SW, Rock CO. Structure. 2000;8:185–195. doi: 10.1016/s0969-2126(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 100.Torella JP, Ford TJ, Kim SN, Chen AM, Way JC, Silver PA. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11290–11295. doi: 10.1073/pnas.1307129110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moche M, Schneider G, Edwards P, Dehesh K, Lindqvist Y. J. Biol. Chem. 1999;274:6031–6034. doi: 10.1074/jbc.274.10.6031. [DOI] [PubMed] [Google Scholar]
- 102.Jackowski S, Murphy CM, Cronan JE, Rock CO. J. Biol. Chem. 1989;264:7624–7629. [PubMed] [Google Scholar]
- 103.Price AC, Choi K-H, Heath RJ, Li Z, White SW, Rock CO. J. Biol. Chem. 2001;276:6551–6559. doi: 10.1074/jbc.M007101200. [DOI] [PubMed] [Google Scholar]
- 104.Kremer L, Douglas JD, Baulard AR, Morehouse C, Guy MR, Alland D, Dover LG, Lakey JH, Jacobs WR, Brennan PJ, Minnikin DE, Besra GS. J. Biol. Chem. 2000;275:16857–16864. doi: 10.1074/jbc.M000569200. [DOI] [PubMed] [Google Scholar]
- 105.Young K, Jayasuriya H, Ondeyka JG, Herath K, Zhang C, Kodali S, Galgoci A, Painter R, Brown-Driver V, Yamamoto R, Silver LL, Zheng Y, Ventura JI, Sigmund J, Ha S, Basilio A, Vicente F, Tormo JR, Pelaez F, Youngman P, et al. Antimicrob. Agents Chemother. 2006;50:519–526. doi: 10.1128/AAC.50.2.519-526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heath RJ, White SW, Rock CO. Prog. Lipid Res. 2001;40:467–497. doi: 10.1016/s0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- 107.Worthington AS, Hur GH, Meier JL, Cheng Q, Moore BS, Burkart MD. ChemBioChem. 2008;9:2096–2103. doi: 10.1002/cbic.200800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Toomey RE, Wakil SJ. Biochim. Biophys. Acta - Lipids Lipid Metab. 1966;116:189–197. doi: 10.1016/0005-2760(66)90001-4. [DOI] [PubMed] [Google Scholar]
- 109.Nomura CT, Taguchi K, Gan Z, Kuwabara K, Tanaka T, Takase K, Doi Y. Appl. Environ. Microbiol. 2005;71:4297–4306. doi: 10.1128/AEM.71.8.4297-4306.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Y, Cronan JE. J. Bacteriol. 1998;180:3295–3303. doi: 10.1128/jb.180.13.3295-3303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cukier CD, Hope AG, Elamin AA, Moynie L, Schnell R, Schach S, Kneuper H, Singh M, Naismith JH, Lindqvist Y, Gray DW, Schneider G. ACS Chem. Biol. 2013;8:2518–2527. doi: 10.1021/cb4005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lai C-Y, Cronan JE. J. Bacteriol. 2004;186:1869–1878. doi: 10.1128/JB.186.6.1869-1878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parish T, Roberts G, Laval F, Schaeffer M, Daffé M, Duncan K. J. Bacteriol. 2007;189:3721–3728. doi: 10.1128/JB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jeon E, Lee S, Lee S, Han SO, Yoon YJ, Lee J. J. Microbiol. Biotechnol. 2012;22:990–999. doi: 10.4014/jmb.1112.12057. [DOI] [PubMed] [Google Scholar]
- 115.Oppermann U, Filling C, Hult M, Shafqat N, Wu X, Lindh M, Shafqat J, Nordling E, Kallberg Y, Persson B, Jörnvall H. Chem. Biol. Interact. 2003:143–144. 247–253. doi: 10.1016/s0009-2797(02)00164-3. [DOI] [PubMed] [Google Scholar]
- 116.Price AC, Zhang Y-M, Rock CO, White SW. Structure. 2004;12:417–428. doi: 10.1016/j.str.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 117.Nambiar KP, Stauffer DM, Kolodziej PA, Benner SA. J. Am. Chem. Soc. 1983;105:5886–5890. [Google Scholar]
- 118.Price AC, Zhang Y-M, Rock CO, White SW. Biochemistry. 2001;40:12772–12781. doi: 10.1021/bi010737g. [DOI] [PubMed] [Google Scholar]
- 119.Fisher M, Kroon JTM, Martindale W, Stuitje AR, Slabas AR, Rafferty JB. Structure. 2014;8:339–347. doi: 10.1016/s0969-2126(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 120.Wright HT. Structure. 2014;12:358–359. doi: 10.1016/j.str.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 121.Zhang Y-M, Rock CO. J. Biol. Chem. 2004;279:30994–31001. doi: 10.1074/jbc.M403697200. [DOI] [PubMed] [Google Scholar]
- 122.Tasdemir D, Lack G, Brun R, Rüedi P, Scapozza L, Perozzo R. J. Med. Chem. 2006;49:3345–3353. doi: 10.1021/jm0600545. [DOI] [PubMed] [Google Scholar]
- 123.Sohn M-J, Zheng C-J, Kim W-G. J. Antibiot. (Tokyo) 2008;61:687–691. doi: 10.1038/ja.2008.98. [DOI] [PubMed] [Google Scholar]
- 124.Wickramasinghe SR, Inglis KA, Urch JE, Müller S, van Aalten DMF, Fairlamb AH. Biochem. J. 2006;393:447–457. doi: 10.1042/BJ20050832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang L, Kong Y, Wu D, Zhang H, Wu J, Chen J, Ding J, Hu L, Jiang H, Shen X. Protein Sci. 2008;17:1971–1978. doi: 10.1110/ps.036186.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu B-N, Zhang Y-M, Rock CO, Zheng JJ. Protein Sci. 2009;18:240–246. doi: 10.1002/pro.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sedgwick B, Morris C, French SJ. J. Chem. Soc. Chem. Commun. 1978:193–194. [Google Scholar]
- 128.Kass LR, Bloch K. Proc. Natl. Acad. Sci. U. S. A. 1967;58:1168–1173. doi: 10.1073/pnas.58.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marrakchi H, Zhang Y-M, Rock CO. Biochem. Soc. Trans. 2002;30:1050–1055. doi: 10.1042/bst0301050. [DOI] [PubMed] [Google Scholar]
- 130.Heath RJ, Rock CO. J. Biol. Chem. 1996;271:27795–27801. doi: 10.1074/jbc.271.44.27795. [DOI] [PubMed] [Google Scholar]
- 131.Cao Y, Yang J, Xian M, Xu X, Liu W. Appl. Microbiol. Biotechnol. 2010;87:271–280. doi: 10.1007/s00253-009-2377-x. [DOI] [PubMed] [Google Scholar]
- 132.Mohan S, Kelly TM, Eveland SS, Raetz CR, Anderson MS. J. Biol. Chem. 1994;269:32896–32903. [PubMed] [Google Scholar]
- 133.Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. Structure. 2014;4:253–264. doi: 10.1016/s0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 134.Kimber MS, Martin F, Lu Y, Houston S, Vedadi M, Dharamsi A, Fiebig KM, Schmid M, Rock CO. J. Biol. Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]
- 135.Kostrewa D, Winkler FK, Folkers G, Scapozza L, Perozzo R. Protein Sci. 2005;14:1570–1580. doi: 10.1110/ps.051373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moynié L, Leckie SM, McMahon SA, Duthie FG, Koehnke A, Taylor JW, Alphey MS, Brenk R, Smith AD, Naismith JH. J. Mol. Biol. 2013;425:365–377. doi: 10.1016/j.jmb.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 137.Sharma SK, Kapoor M, Ramya TNC, Kumar S, Kumar G, Modak R, Sharma S, Surolia N, Surolia A. J. Biol. Chem. 2003;278:45661–45671. doi: 10.1074/jbc.M304283200. [DOI] [PubMed] [Google Scholar]
- 138.Liu W, Luo C, Han C, Peng S, Yang Y, Yue J, Shen X, Jiang H. Biochem. Biophys. Res. Commun. 2005;333:1078–1086. doi: 10.1016/j.bbrc.2005.05.197. [DOI] [PubMed] [Google Scholar]
- 139.He L, Zhang L, Liu X, Li X, Zheng M, Li H, Yu K, Chen K, Shen X, Jiang H, Liu H. J. Med. Chem. 2009;52:2465–2481. doi: 10.1021/jm8015602. [DOI] [PubMed] [Google Scholar]
- 140.Kong Y, Zhang L, Yang Z, Han C, Hu L, Jiang H, Shen X. Acta Pharmacol. Sin. 2008;29:870–876. doi: 10.1111/j.1745-7254.2008.00808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Swarnamukhi PL, Sharma SK, Bajaj P, Surolia N, Surolia A, Suguna K. FEBS Lett. 2006;580:2653–2660. doi: 10.1016/j.febslet.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 142.Lu Y-J, White SW, Rock CO. J. Biol. Chem. 2005;280:30342–30348. doi: 10.1074/jbc.M504637200. [DOI] [PubMed] [Google Scholar]
- 143.Agarwal V, Lin S, Lukk T, Nair SK, Cronan JE. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17406–17411. doi: 10.1073/pnas.1207028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cryle MJ, Schlichting I. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Babu M, Greenblatt JF, Emili A, Strynadka NCJ, Reithmeier RAF, Moraes TF. Structure. 2010;18:1450–1462. doi: 10.1016/j.str.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 146.Zhang L, Liu W, Hu T, Du L, Luo C, Chen K, Shen X, Jiang H. J. Biol. Chem. 2007;283:5370–5379. doi: 10.1074/jbc.M705566200. [DOI] [PubMed] [Google Scholar]
- 147.Bergler H, Fuchsbichler S, Högenauer G, Turnowsky F. Eur. J. Biochem. 1996;242:689–694. doi: 10.1111/j.1432-1033.1996.0689r.x. [DOI] [PubMed] [Google Scholar]
- 148.Heath RJ, Rock CO. J. Biol. Chem. 1995;270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- 149.Heath RJ, Su N, Murphy CK, Rock CO. J. Biol. Chem. 2000;275:40128–40133. doi: 10.1074/jbc.M005611200. [DOI] [PubMed] [Google Scholar]
- 150.Massengo-Tiassé RP, Cronan JE. Cell. Mol. Life Sci. 2009;66:1507–1517. doi: 10.1007/s00018-009-8704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Marrakchi H, Dewolf WE, Quinn C, West J, Polizzi BJ, So CY, Holmes DJ, Reed SL, Heath RJ, Payne DJ, Rock CO, Wallis NG. Biochem. J. 2003;370:1055–1062. doi: 10.1042/BJ20021699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Quemard A, Sacchettini JC, Dessen A, Vilcheze C, Bittman R, Jacobs WR, Blanchard JS. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 153.Saito K, Kawaguchi A, Seyama Y, Yamakawa T, Okuda S. Eur. J. Biochem. 1981;116:581–586. doi: 10.1111/j.1432-1033.1981.tb05375.x. [DOI] [PubMed] [Google Scholar]
- 154.Rozwarski DA, Vilcheze C, Sugantino M, Bittman R, Sacchettini JC. J. Biol. Chem. 1999;274:15582–15589. doi: 10.1074/jbc.274.22.15582. [DOI] [PubMed] [Google Scholar]
- 155.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. J. Biol. Chem. 1999;274:11110–11114. doi: 10.1074/jbc.274.16.11110. [DOI] [PubMed] [Google Scholar]
- 156.Levy CW, Roujeinikova A, Sedelnikova S, Baker PJ, Stuitje AR, Slabas AR, Rice DW, Rafferty JB. Nature. 1999;398:383–384. doi: 10.1038/18803. [DOI] [PubMed] [Google Scholar]
- 157.Qiu X, Abdel-Meguid SS, Janson CA, Court RI, Smyth MG, Payne DJ. Protein Sci. 1999;8:2529–2532. doi: 10.1110/ps.8.11.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Botté CY, Dubar F, McFadden GI, Maréchal E, Biot C. Chem. Rev. 2011;112:1269–1283. doi: 10.1021/cr200258w. [DOI] [PubMed] [Google Scholar]
- 159.Pertschy B, Zisser G, Schein H, Köffel R, Rauch G, Grillitsch K, Morgenstern C, Durchschlag M, Högenauer G, Bergler H. Mol. Cell. Biol. 2004;24:6476–6487. doi: 10.1128/MCB.24.14.6476-6487.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dayan FE, Ferreira D, Wang Y-H, a Khan I, a McInroy J, Pan Z. Plant Physiol. 2008;147:1062–1071. doi: 10.1104/pp.108.118372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zheng CJ, Sohn M-J, Lee S, Hong Y-S, Kwak J-H, Kim W-G. Biochem. Biophys. Res. Commun. 2007;362:1107–1112. doi: 10.1016/j.bbrc.2007.08.144. [DOI] [PubMed] [Google Scholar]
- 162.Hartkoorn RC, Pojer F, Read JA, Gingell H, Neres J, Horlacher OP, Altmann K-H, Cole ST. Nat. Chem. Biol. 2014;10:96–98. doi: 10.1038/nchembio.1405. [DOI] [PubMed] [Google Scholar]
- 163.Rafi S, Novichenok P, Kolappan S, Zhang X, Stratton CF, Rawat R, Kisker C, Simmerling C, Tonge PJ. J. Biol. Chem. 2006;281:39285–39293. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Magnuson K, Jackowski S, Rock CO, Cronan JE. Microbiol. Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heath RJ, Rock CO. J. Biol. Chem. 1995;270:15531–15538. doi: 10.1074/jbc.270.26.15531. [DOI] [PubMed] [Google Scholar]
- 166.Heath RJ, Rock CO. J. Bacteriol. 1998;180:1425–1430. doi: 10.1128/jb.180.6.1425-1430.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lewin TM, Wang P, Coleman RA. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 168.Cantu DC, Chen Y, Reilly PJ. Protein Sci. 2010;19:1281–1295. doi: 10.1002/pro.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yuan L, Voelker TA, Hawkins DJ. Proc. Natl. Acad. Sci. U. S. A. 1995;92:10639–10643. doi: 10.1073/pnas.92.23.10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhuang Z, Song F, Zhao H, Li L, Cao J, Eisenstein E, Herzberg O, Dunaway-Mariano D. Biochemistry. 2008;47:2789–2796. doi: 10.1021/bi702334h. [DOI] [PubMed] [Google Scholar]
- 171.Spencer AK, Greenspan AD, Cronan JE. J. Biol. Chem. 1978;253:5922–5926. [PubMed] [Google Scholar]
- 172.Lu X, Vora H, Khosla C. Metab. Eng. 2008;10:333–339. doi: 10.1016/j.ymben.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 173.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 174.Jones A, Davies HM, Voelker TA. Plant Cell. 1995;7:359–371. doi: 10.1105/tpc.7.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Voelker T. In: Genet. Eng. SE - 8. Setlow J, editor. Springer; US: 1996. pp. 111–133. [DOI] [PubMed] [Google Scholar]
- 176.Voelker TA, Worrell AC, Anderson L, Bleibaum J, Fan C, Hawkins DJ, Radke SE, Davies HM. Science. 1992;257:72–74. doi: 10.1126/science.1621095. [DOI] [PubMed] [Google Scholar]
- 177.Voelker TA, Davies HM. J. Bacteriol. 1994;176:7320–7327. doi: 10.1128/jb.176.23.7320-7327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 179.Yuan L, Nelson BA, Caryl G. J. Biol. Chem. 1996;271:3417–3419. doi: 10.1074/jbc.271.7.3417. [DOI] [PubMed] [Google Scholar]
- 180.Mayer KM, Shanklin J. J. Biol. Chem. 2005;280:3621–3627. doi: 10.1074/jbc.M411351200. [DOI] [PubMed] [Google Scholar]
- 181.Mayer KM, Shanklin J. BMC Plant Biol. 2007;7:1. doi: 10.1186/1471-2229-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Davies HM, Anderson L, Fan C, Hawkins DJ. Arch. Biochem. Biophys. 1991;290:37–45. doi: 10.1016/0003-9861(91)90588-a. [DOI] [PubMed] [Google Scholar]
- 183.Pollard MR, Anderson L, Fan C, Hawkins DJ, Davies HM. Arch. Biochem. Biophys. 1991;284:306–312. doi: 10.1016/0003-9861(91)90300-8. [DOI] [PubMed] [Google Scholar]
- 184.Srikanta Dani KG, Hatti KS, Ravikumar P, Kush A. Plant Biol. 2011;13:453–461. doi: 10.1111/j.1438-8677.2010.00410.x. [DOI] [PubMed] [Google Scholar]
- 185.Rafi S, Novichenok P, Kolappan S, Zhang X, Stratton CF, Rawat R, Kisker C, Simmerling C, Tonge PJ. J. Biol. Chem. 2006;281:39285–39293. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kumari P, Vonck J, Schröder RR, Kühlbrandt W. Microsc. Microanal. 2007;13:158–159. [Google Scholar]