Abstract
In recent years the ever so complex field of drug discovery has embraced novel design strategies based on biophysical fragment screening (fragment-based drug design; FBDD) using nuclear magnetic resonance spectroscopy (NMR) and/or structure-guided approaches, most often using X-ray crystallography and computer modeling. Experience from recent years unveiled that these methods are more effective and less prone to artifacts compared to biochemical high-throughput screening (HTS) of large collection of compounds in designing protein inhibitors. Hence these strategies are increasingly becoming the most utilized in the modern pharmaceutical industry. Nonetheless, there is still an impending need to develop innovative and effective strategies to tackle other more challenging targets such as those involving protein-protein interactions (PPIs). While HTS strategies notoriously fail to identify viable hits against such targets, few successful examples of PPIs antagonists derived by FBDD strategies exist. Recently, we reported on a new strategy that combines some of the basic principles of fragment-based screening with combinatorial chemistry and NMR-based screening. The approach, termed HTS by NMR, combines the advantages of combinatorial chemistry and NMR-based screening to rapidly and unambiguously identify bona fide inhibitors of PPIs. This review will reiterate the critical aspects of the approach with examples of possible applications.
Keywords: Drug discovery, fragment-based drug design, FBDD, FBLD, HTS by NMR, NMR, PPIs, protein-protein interactions, positional scanning, POS
1. Introduction
Protein-protein interactions (PPIs) constitute potentially a large number of therapeutic targets. Nonetheless, these targets are generally deemed challenging or undruggable, meaning that small molecule antagonists (MW ≤ 800 Da) cannot be easily found capable of disrupting the interactions with sufficient potency (IC50 < 1 μM) to be effective in cellular assays and subsequently optimized into drug candidates. A possible cause of this challenge is the large surface that is covered by a typical protein-protein interface, which is likely too large and often shallow to bind a small molecule with high affinity.[1] Hence conventional high-throughput screening (HTS) campaigns relying on spectrophotometric, plate-based assays for rapid screening of large collection of compounds is an unlikely approach to identify PPIs antagonists, given the low sensitivity of these methods that can detect only relatively potent hits (IC50 < 10 μM). Weaker binders are often undetected as they are buried in a large number of assay- or compound-based artifacts including non-specific binders and promiscuous aggregators, compounds that can denature or unfold the protein, redox compounds that can interfere with assays components, etc. These go in addition to other possible sources of “noise” that can arise for example from instrumental errors such as from dispending small volumes in the plates, etc.[2] Therefore, given these circumstances, identifying weaker interacting compounds using these HTS approaches is challenging to say the least: any eventual hit compound is either simply not detected by the biochemical assays or buried by a large number of false positives.[2c, 3]
While initially introduced as a way to weed out false positives, biophysical methods such as protein NMR spectroscopy have increasingly played a major role in de novo drug discovery campaigns in the past decade. These approaches have the invaluable property to be able to directly and unambiguously identify and characterize the binding of a test molecule to a given protein or even nuclei acid target, without relying on convoluted biochemical indirect methods.[4] While other biophysical methods have also emerged in recent years, in our opinion and experience, protein-based Nuclear Magnetic Resonance (NMR) spectroscopy remains to date the most reliable method to study ligand binding with low- to medium-throughput capacity (testing several hundred to several thousand compounds in a given typical discovery campaign is attainable). Hence, these methods found fertile ground in guiding the design of PPIs antagonists in recent years.[5]
PPIs can often be recapitulated by short peptide regions from one protein, interacting with a complementary pocket on the surface of its protein binding partner. Hence, a valuable starting point for the design of PPIs antagonists consists in identifying such peptide regions and making subsequently chemical modifications aimed at increasing their affinity along with cell permeability, resistance to proteases, and overall drug likeness. [6]
Most often binding peptides present a set of pharmacophoric groups (the essential side chains or often also backbone atoms) that are accommodated in as many sub-pockets on the surface of the protein partner. Hence, this arrangement of adjacent sub-pockets in PPIs makes this class of targets particularly suitable to fragment-based lead (or drug) discovery (FBLD or FBDD) strategies.[7] A common FBLD strategy consists in first identifying pairs of adjacent binding fragments that are subsequently linked,[8] often guided by structural studies of the ternary complex, leading into more potent bi-dentate compounds.[9] Protein-NMR spectroscopy has been used for the identification, structural characterization and design of such binders, as exemplified in the pioneering SAR by NMR approach.8 Recent examples of the SAR by NMR (Structure Activity Relationships by NMR) include the design of antagonists of Bcl-2 and Bcl-xL (ABT-737) [10] that led to current clinical candidate ABT-199 [11] (one of the first antagonist of PPIs to reach the clinic). Of note is that, as mentioned earlier, HTS approaches against the same targets failed to produce viable hits. [10]
In addition to FBDD approaches, the identification of initial peptides by means of phage display techniques [12] or derived from structural studies of a protein-protein complex, can also enable the design of peptide mimetic with improved pharmacological properties. A recent example is represented by the Genentech compound GDC-0152 that mimics the tetra-peptide of amino-acid sequence AVPI in targeting the anti-apoptotic protein XIAP.[13]
These notable examples suggest that the combination of FBDD approaches guided by NMR spectroscopy and peptide-mimetic design could ideally provide a general and effective mean to obtain viable PPIs antagonists. We have recently proposed a novel approach, termed HTS by NMR,[14] in which the principles of positional scanning combinatorial chemistry [15] and fragment-based drug design are combined with protein-NMR spectroscopy [5] to iteratively identify and optimize PPIs antagonists from collections of >100,000 peptide mimetics. The approach seems also particularly effective in the fragment-hit to lead optimization stages of PPIs, when a positional scanning library is generated from an initial weak binder previously identified from a FBDD campaign, and tested by protein NMR spectroscopy. In next paragraphs we will reiterate the critical aspects of the approach and its applications with several recent examples. Because the use of protein-NMR spectroscopy as biophysical detection methods is critical for the successful implementation of these approaches, we will also briefly reiterate the various possible protein-based NMR strategies for ligand detection and comment of advantages and limitations of these approaches.
2. Protein-based NMR spectroscopy to detect ligand binding
Arguably, the most effective and unambiguous mean to detect ligand binding to a protein target is using protein-NMR spectroscopy techniques. [4–5, 16] In such methods, the NMR spectra of the protein are recorded in presence and absence of test ligands and specific perturbations in the protein spectrum upon ligand titration can be used to determine the dissociation constant and possibly site of binding. In our opinion and based on our experience with a variety of biochemical and other biophysical assays, protein-NMR techniques provide an invaluable support to guide the identification, validation and optimization of ligands, particularly when dealing with targeting PPIs.[5]
Three general strategies can be envisioned when using protein-NMR to detect ligand binding. The first and simplest is measurement of 1D 1H-aliph NMR spectra,[5] that represent the portion of the protein 1D 1H NMR spectrum below 0.7 ppm. This spectral region is typical of several of protein’s methyl groups, and it is almost never populated by signals from small peptides or small molecules. Hence, comparisons of 1D 1H-aliph NMR spectra of the target protein in absence and presence of test ligands allows for detection of ligand binding.[5] This approach is particularly sensitive given the slow relaxation properties of methyl groups, giving sharper and more intense lines in the NMR spectra compared to other protein resonances, allowing the observation of 1D 1H-aliph NMR spectra of most small to medium size proteins (up to 30 kDa) with relatively low protein concentrations (about 1–10 μM) in just a few minutes using a modern high-field NMR instrument. For smaller proteins where the methyl region may be particularly well resolved, ligand titration and 1D 1H-aliph NMR can be used to provide an estimate of the dissociation constant of the complex.[4b]
While in our experience this method is fairly general, it may not be suitable for larger proteins or for proteins that do not possess methyl resonances shifted below 0.7 ppm and that are within the target’s binding site. At the other extreme region of the 1D 1H spectrum of the target protein are usually Trp 1Hε side chain resonances. These usually reside around 10 ppm, hence fall again in a region that is not often populated by NMR signals from small molecules or peptides (although some exceptions do exist). Therefore, if the protein target has Trp residues in its binding side, again observing the signals around 10 ppm in a simple 1D 1H NMR recorded in aqueous buffer can be suitable for detecting and monitoring ligand binding. However, detection of the Trp side chains is far less sensitive than detecting the 3 slow relaxing protons of methyl groups. As mentioned, these simple 1D 1H NMR approaches are most often successfully applicable for smaller or medium size proteins (MW < 30 kDa).
Despite the 1D 1H spectrum with unlabeled protein, protein NMR most often relies on detection of 2D spectra by using uniformly or selective 15N and/or 13C labeling. Enriching the protein with these isotopes can follow a variety of approaches, including labeled rich media, minimal media containing 15NH4Cl as sole source of nitrogen and/or 13C-Glucose as sole source of carbon, or incorporation of single or doubly labeled amino acids or amino acid precursors to the culture media. When the protein target is enriched with these NMR observable isotopes, typically 2D [1H, 15N] or 2D [1H, 13C] NMR spectra are collected in absence and presence of test ligand(s) to monitor eventual chemical shift perturbations upon ligand titration, that can be used to directly determine the dissociation constant of the complex. In addition, if the three-dimensional structure of the target and the resonance assignments are available, the chemical shift perturbations induced by a test ligand can be used to roughly locate the site of binding.
Nowadays, 2D sofast-HMQC correlations spectra [17] with 15N labeled protein can be measured with protein sample of as little as 10–50 μM concentration within an hour using an high field instrument equipped with a cryogenic probe, when dealing with small or medium size proteins (MW < 25–30 kDa). The use of deuteration, and TROSY-type correlation spectra,[18] however, can increase the MW limit of the approach. In addition to the backbone 1HN, 15N resonances, other similar [1H, 15N] correlation spectra, using selective excitation and/or a slight modification of the pulse sequences, can be collected to detect the side chains of Trp, Arg, Asn, Gln, and His.[5]
Likewise, 2D [1H, 13C] correlation NMR spectra can be collected for 13C-labeled protein targets in presence and absence of test ligand(s). The most effective and better resolved spectral region in such 2D spectra include the aliphatic (~10–30 ppm in the 13C dimension), and the aromatic regions (~100–130 ppm in the 13C dimension when observing resonances of the side chains of Tyr, Trp, Phe, His). These regions can become fairly crowded for protein of MW > 30kDa. In these cases, amino acid selective labeling is used to simplify the spectra. As mentioned, this can be accomplished by supplementing the bacterial growth medium with the desired labeled amino acids. A common strategy is to add about 100–200 mg of the labeled amino-acid per liter of culture just prior to the induction of protein expression. Some metabolic scrambling can be used to increase the number of labeled amino-acids. For example, introduction of 13C-Thr results also in labeling of the 13Cδ1-Ile. [4a, 19] The simplified 2D [1H, 13C] correlation spectra resulting from these selective labeled protein targets makes it simpler to observe ligand induced perturbations.
More interestingly, if combined with deuteration and measurements of 13C-resolved 2D [1H, 1H] NOESY type of experiments, [4a, 20] such protein samples can be used to detect intermolecular distances (via the NOE) that can turn out to be very useful to guide the docking of ligands. [4a, 20]
In summary, protein-NMR experiments provide methods for detection of ligand binding that are extremely sensitive and unambiguous. Unlike spectrophotometric assays or even other biophysical methods such as SPR (surface plasmon resonance) or ITC (isothermal titration calorimetry), it is highly unlikely for protein-NMR methods to generate false positives or false negatives; hence these methods remain the gold standard for hit validation. Protein aggregators and/or compounds that denature the protein, are easily identified. False positives that interact with assay components in plate based assays, (defined as Pan Assay INterference compoundS or PAINS compounds [2c, 3]), again are easily identified as non-binders in a protein-NMR based assay. In addition, the method is very sensitive to allow detection also of weaker binders, hence can find tremendous utility in the design of PPIs antagonists where again initial low molecular weight binders are not expected to have high affinity. Finally, we want to mention that other popular NMR methods exist that are based on detecting ligand binding by monitoring the indirect effects on the ligand resonances induced upon protein binding. These ligand-based NMR methods [4q] are in principle very useful in fragment screening campaigns; however, similar to the SPR methods, [21] in our experience and opinion these assays are not immune to false positives hence are not as reliable as protein-based NMR experiments to detect and characterize ligand binding.
3. HTS by NMR: a powerful approach for the identification of antagonists of protein-protein interactions
As anticipated earlier, protein NMR spectroscopy found its initial applications in drug design as main screening tool for fragment-based drug discovery. [8] In FBDD campaigns, initial weakly interacting small molecular weight compounds (fragments) are identified using sensitive biophysical methods. Subsequently, these fragments are matured into more potent hits using a variety of approaches, including increasing the complexity of the fragments (fragment growing), as well as linking or merging two fragments occupying adjacent or overlapping binding sites, respectively. Obviously, due to the modular nature of PPIs interfaces, this approach is in principle particularly suitable for the design of antagonists to PPIs. Several biophysical techniques are currently in use in FBDD campaigns, comprising not only ligand- and protein-based NMR, but also SPR or ITC. [21] While these methods seem to reliably identify initial weakly interacting fragments, their optimization into potent hits remains not a trivial task and requires more intensive follow-up approaches including more often structure-based design aided by computational [22] or X-ray studies, [23] fragment growing using medicinal chemistry and SAR, or the fragment linking strategy represented by the original work known as SAR by NMR. [8b] Arguably, all these approaches require some level of structural information on the mode of binding of one or more fragments. This may not always be easily attainable. In addition, weakly binding fragments may adopt an unpredictable orientation following even small modifications in their core structure, making any optimization strategy particularly challenging. As a result, while the identification of initial hits is usually attainable, maturing them into bona fide potent antagonists is a very difficult and often unsuccessful task.
For these reasons, while a few notable exceptions exist, such as the SAR by NMR derived Bcl-2 antagonist, [10, 24] the discovery of PPIs inhibitors remains very challenging. Using small peptides as starting point for the discovery of PPIs antagonists seems therefore a valid alternative. Short peptides have the tremendous advantage over small organic molecules of being most often exquisitely selective and potent against a given target, [25] and rarely produce artifact in assays that plague HTS campaigns.2c, 3 Moreover, peptides can be chemically modified to increase their stability, cell permeability and pharmacokinetics, using a variety of approaches. [25–26]
The discovery of short peptide sequences binding to a given PPI using non-biased screening approaches can be accomplished using phage display techniques,[12a, 12b, 12e, 12g, 12h, 12j, 25a, 27] in which a heterogeneous mixture of such phage clones, each carrying a different peptide of various lengths on their surface is tested. Selection of binding peptides by simple capture techniques and amplification of the selected phage allow the identification of a consensus sequence for those peptides showing higher affinities.[12a, 12b, 12e, 12g, 12h, 12j, 25a, 27–28] A powerful alternative consists in building synthetic combinatorial libraries and use these directly in biochemical assays.[29] These have the intrinsic advantage over the phage display techniques of not being limited to natural amino acids; hence in principle allow the identification of peptide mimetics with improved drug likeness. However, to make the synthesis and testing of these library feasible and practical, mixture of compounds are often utilized. A powerful pooling technique in combinatorial chemistry is the positional scanning (POS) [15] in which compound mixtures are systematically assembled with one element fixed at each given position while the other positions comprise all combinations. For example, a tetra-peptide library composed of the 20 natural amino-acids would require the daunting task of synthesizing and subsequently testing 20 × 20 × 20 × 20 tetra-peptides, hence 160,000 compounds. In the positional scanning mixture approach, mixtures are prepared for each of the 4 positions in which systematically one amino-acid is fixed at one position. For example, a mixture Ala-XXX of 8,000 compounds is composed by all possible tetra-peptides starting with Alanine, while the mixture Tyr-XXX is composed by all possible tetra-peptides with a Tyr a the N-terminal, and so on for all amino-acids and for all positions in the tetra-peptide. Hence, rather than synthesizing and testing individual 160,000 tetra-peptides, this approach entails the synthesis and testing of 20 + 20 + 20 + 20 = 80 mixtures. Similarly, a more drug-like library can be assembled by using non-natural amino-acids, and/or by fragment-like compounds. A practical limitation of the approach lies in the detection methods. Using compounds in mixtures can exacerbate the already artifact prone biochemical assays, reducing even further the signal to noise of a given screening campaign. Hence, only if very potent compounds are present in the library these would be reliably detected using biochemical assays. [15b, 29c, 29e, 30] This often implies that libraries with increased complexity and number of elements in each mixture are necessary. Increasing mixture complexity may in turn render their synthesis and subsequent identification of possible hit problematic and less likely.
Therefore, we have recently proposed to test mixture based libraries with protein–NMR screening methods, [14] as these approaches present a number of unique advantages over any other assay. First, protein-NMR assays enable the unambiguous identification of hit compounds even in complex mixtures, nearly void of possible false positive or false negatives. This is because, in essence, the protein-NMR spectra intrinsically provide direct information about the protein integrity during the screen. In addition, if resonance assignments, even if limited to a few residues, are available, protein-NMR data can be used to generate initial hypotheses about the binding site and binding mode of hits molecules. Thirds, and perhaps most important, protein-NMR techniques are very sensitive also to weak binding events, hence allowing the identification of initial hits also in mixture libraries of lower complexity. Therefore, these advantages of protein-NMR spectroscopy make it an ideal tool to test positional scanning libraries. In our recent implementations, we combined positional scanning combinatorial libraries of short tri- or tetra-peptides with protein-based NMR screening to identify initial tri- or tetra-peptides or mimetics.
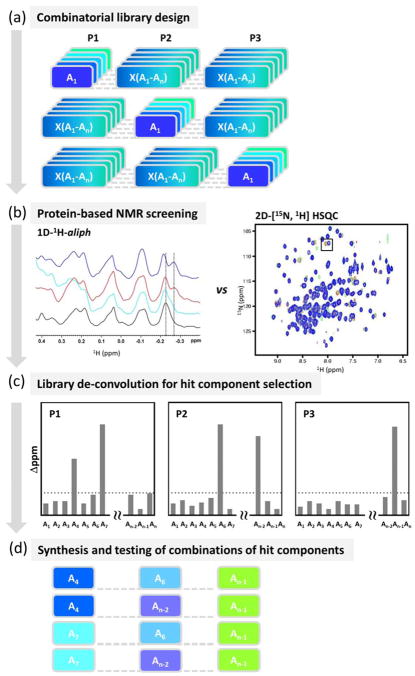
This approach, termed in general HTS by NMR to emphasize the high throughput relative to other fragment screening campaigns, can be used in various implementations for the de novo identification of peptides or peptide mimetics, and for the optimization of known sequences or fragment hits (Figure 1a). The approach is likely more effective in identifying weakly interacting tri-peptoids from the mixtures when using 2D hetero-nuclear NMR experiments with uniformly or selectively labeled target, although the 1D 1H-aliph constitutes a simpler and cost-effective approach, not requiring labeling of the target (Figure 1b).[5] Mixtures causing significant chemical shift perturbations in the spectra of the target are rank ordered for each of the scanned positions, and the ranking is used to prioritize the synthesis of corresponding individual tri-peptoids (Figure 1c). Following the synthesis of each selected compound (usually a few possible molecules; Figure 1d), protein-NMR experiments or other biophysical techniques (such as ITC) are used to verify and characterize the binding of the potential ligands. In our experience with a variety of targets and different libraries, this approach can lead to initial tri- or tetra-peptoids with dissociation constants varying from low micromolar to hundreds of micromolar, again underlying the need of a sensitive and unambiguous screening method as protein-NMR spectroscopy for their initial detection.
Figure 1. Schematic representation of the HTS by NMR approach.
a) A POS library of tri- or tetra-peptoids (or peptides) is designed and synthesized. In the positional scanning (POS) combinatorial library, each sample is a mixtures of compounds in which one position is fixed while other positions consist of all components. In an example of 3-positions POS library, and n-components per position, the library will contain 3 × n mixtures, and each mixture will contain a total n × n compounds. b) The designed library is next screened by collecting either 1D-1H-aliph, or 2D-[15N, 1H], or 2D [13C,1H] correlation spectra of target macromolecule in presence and absence of each mixture. c) The library is deconvoluted by evaluating chemical shit perturbations (Δppm) induced by each mixture. Significant perturbations in the macromolecule spectra are attributable to the fixed component of the given hit mixture. Hence, components at each position are selected as hits based on their ability to induce significant chemical shift perturbations. d) Finally, individual compounds are synthesized with a proper combination of hit components and tested.
Optimization of the hits largely depends on various factors including the nature of the target, availability of structural information and the binding pose of the ligand, desired molecular weight limit of final optimized molecules, availability of analogues of the identified scaffolds, etc. Several hit-to-lead optimizations approaches are envisioned including a) elongation of the polypeptide chain and/or b) modifications at the N- or C-terminal ends, and/or c) traditional SAR in which individual scaffolds are iteratively optimized.
In our recent example, we tested a library consisting of 58 natural and non-natural amino acids linked in tri-peptoids to identify an inhibitor of EphA4–ephrin-B4 PPI,[14] using 1D 1H-aliph as the primary screening method. This PPI is mediated primarily by a long loop region from the ephrin ligand (Figure 2a), however it likely involves additional interactions given that the isolated peptide fails to bind appreciably the EphA4 receptor, while peptides from phage display can be found binding with low micro molar affinities. [31] In this example, a POS library was assembled in 174 pools: 58 + 58 + 58 mixtures each of which contained approximately 3,364 compounds (1 × 58 × 58). The 1D 1H-aliph allows for the screening to be based on relatively low concentrations of protein (10 μM) and mixtures (500 μM – 1 mM; total mixture concentration in each assay tube). Of note is that even if the initial tri-peptoid was of relatively weak affinity (hence likely undetectable by other screening methods) it can be matured into a potent and selective compound using the three general approaches described above (Figure 2a). In this particular case, because the molecule is composed by non-natural amino-acids, it is resistant to proteases present in biological fluids. [14] It is worth mentioning that previous conventional HTS campaigns [32] against this target resulted only in non-specific compounds that are known to produce artifacts in several assays. [2c]
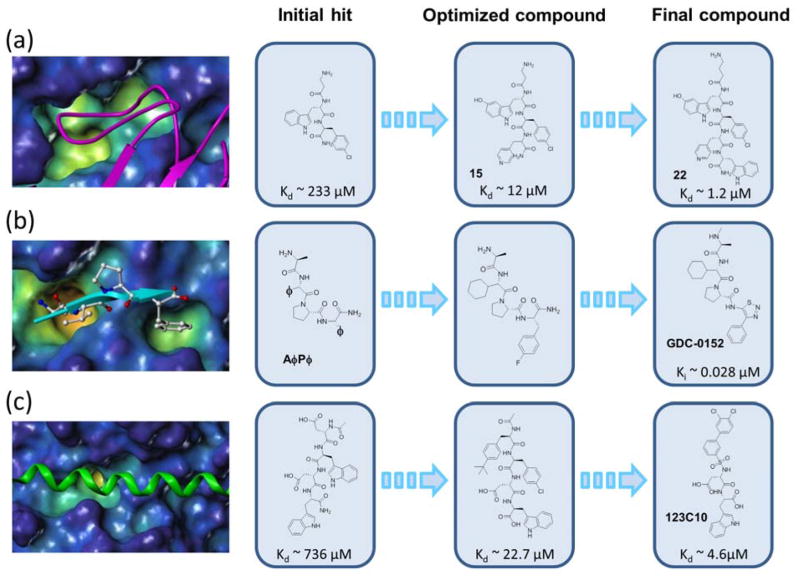
Figure 2. Possible applications of HTS by NMR in targeting PPIs.
a) Example of the discovery of a potent EphA4 inhibitor, targeting a protein-loop interactions. The EphA4 ligand binding domain (surface representation) in complex with ephrin A5 (magenta ribbon) is shown on the left (PDB ID: 4M4R). The evolution into a potent antagonist is illustrated with chemical structures at each step. b) Application of the HTS by NMR to protein-β-strand interactions. The surface representing the BIR3 domain of XIAP in complex with peptide AVPF (cyan ribbon) is shown on the left. (PDB ID: 2OPZ). The HTS by NMR identified the correct essential known consensus AφPφ signature of the peptide. In a second screen using a POS library made up by non-natural aminoacids, a consensus having in particular a cyclohexyl-Glycine in P2 and a 4-F-Phenylalanine in P4 was identified. This consensus motif closely resembles the molecule derived by Genentech, GDC-0152. c) Example of the discovery of compound 121C10 as an inhibitor of Mcl-1, representing an antagonist of protein-α-helix interactions. The surface representing Mcl-1 in complex with Bim BH3 peptide (green ribbon) is shown on the left (PDB ID: 2NL9). The evolution of 121C10 from the initial hits is shown on the right, where the chemical structures and the dissociation constant values (Kd) of selective compound at each step are reported.
As mentioned earlier, linear peptides sequences can be more directly converted into drug candidates, as demonstrated by the recent example of the orally active XIAP antagonist GDC-015213 that closely mimics the structure of the natural antagonist tetra-peptide motif Ala-Val-Pro-Phe/Ile. Indeed the HTS by NMR was successful in identifying the known consensus motif AφPφ (where φ represents hydrophobic residues) for this target used for proof of concept (Figure 2b). [14] Interestingly, when the HTS by NMR was subsequently applied using a POS library made up by non-natural aminoacids against the same protein, a consensus motif Ala-(cyclohexyl)-Gly-Pro-(4F)-Phe was identified (middle panel of Figure 2b) that closely resembles the structure of the clinical candidate GDC-0152.
While the two examples listed above tackled PPIs that are mediated by a loop or by a peptide that binds in an extended conformation, we sought to verify if the approach would also lead to linear peptides when targeting PPIs that are mediated by an α-helix. As an example we targeted the anti-apoptotic protein Mcl-1, a member of the Bcl-2 family of proteins. Binding of Mcl-1 with its partners is mediated by an α-helical motif called BH3 (Figure 2c) and attempts to use directly these helices as therapeutics have been recently reported.[33]
Hence, we tested a library of natural tetra-peptides arranged in 76 positional scanning mixtures (19 + 19 + 19 + 19, comprised by all natural L- amino-acids excluding Cys) against 15N-labeled mouse Mcl-1(152–308)ΔTM Interestingly, the consensus motif Ac-Asp-Trp-Asp-Trp-NH2 emerged with a dissociation constant of 736 μM by NMR titration (Figure 2c). As mentioned above, even when the initial affinity is relatively weak, these initial peptides can be optimized into more potent and selective molecules as illustrated above for the case of EphA4. However, unlike the EphA4 ligand binding domain, Mcl-1 is intracellular hence, rather than elongating the molecule, optimizations strategies that keep MW as low as possible are more suitable to obtain potentially a cell permeable peptide-mimetic. These efforts led to an initial compound that possesses a dissociation constant of 4.6 μM by NMR and ITC (Figure 2c), hence amenable for further optimizations.
These examples demonstrate the general applicability of the approach in the identification of short peptide and peptide-mimetic sequences that can be used as platform for lead optimizations into possible therapeutics.
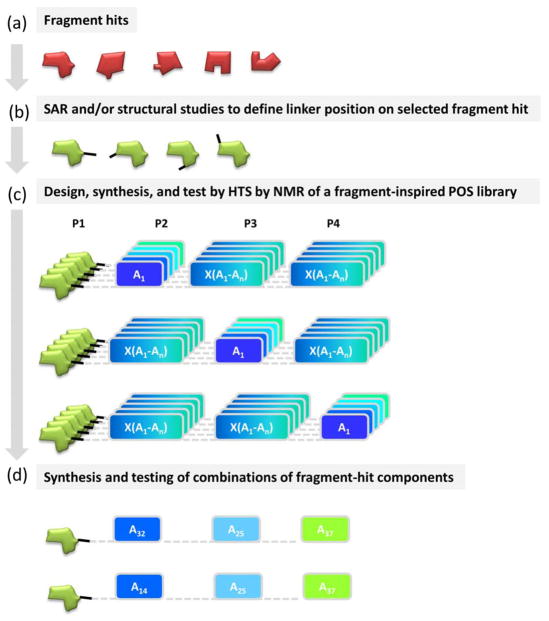
As mentioned above, in FBDD, while the identification of initial hit molecules is relatively straightforward, their optimization into more potent compounds is often quite difficult and unsuccessful. One of the most effective approaches is the fragment growing, in which the initial fragment hit is further iteratively derivatized into a more potent hit molecule, often guided by structural information of the binding mode of the compound. Here we propose to deploy the HTS by NMR approach for hit optimizations by generating a positional scanning library in which one position is fixed and occupied by the given fragment hit (Figure 3a). When possible, structural information should be used to identify a suitable way to link the fragment to the rest of the library, however, in absence of such structural studies, SAR data on the fragment hit can be conducted to hypothesize a possible site of derivatization (Figure 3b).
Figure 3. Schematic representation of the Fragment-Inspired HTS by NMR approach.
a) Fragment hits are identified by screening methods and/or by de-fragmentation of known inhibitors or of natural substrates (such as an ATP mimetic, a metal chelating group, a phospho-Tyrosine mimetic, etc.). b) SAR studies, including possibly structural information, are performed with a selected fragment hit to provide suggestions on a suitable position to place a linker to be used for synthesis of the POS library. c) The optimized fragment is introduced into the POS combinatorial library to be tested using the HTS by NMR strategy.
This approach, in which libraries of compounds are produced with one fixed anchoring fragment while other positions are randomized, can be very effective in the hit optimization process.
Likewise, one can envision assembling target specific positional scanning combinatorial libraries, where the anchoring fragment is common to several members of a given protein family. For example, positional scanning libraries of the type pTyr-XXX could be used to identify and optimize peptide-mimetics binding to proteins that recognize phosphorylated Tyrosine. A similar approach can be envisioned for metallo-proteins or protein kinases, using a metal chelating moiety or a weak ATP mimetic as baits, respectively, for example. These examples clearly suggest that the method is potentially of general applicability for the identification of inhibitors not only of PPIs but also of other targets.
With the resurgence of peptide-mimetics as therapeutics, we envision that the HTS by NMR in its various implementations, also given the relatively ease of synthesis of the POS libraries, [15c, 15d, 34] may provide viable hit compounds for more immediate hit-to-lead optimizations.
Acknowledgments
Financial support to MP was obtained in part by NIH grants CA168517 and CA081534.
List of Abbreviations
- FBDD
Fragment Based Drug Discovery
- FBLD
Fragment Based Ligand Design
- HMQC
Heteronuclear Multiple Quantum Correlation
- HSQC
Heteronuclear Single Quantum Correlation
- HTS
High-Throughput Screening
- ITC
Isothermal Titration Calorimetry
- NMR
Nuclear Magnetic Resonance
- NOESY
Nuclear Overhauser Effect SpectroscopY
- PAINS
Pan Assay INterference compoundS
- POS
POsitional Scanning
- PPIs
Protein-Protein Interactions
- SAR
Structure Activity Relationships
- SPR
Surface Plasmon Resonance
- TROSY
Transverse Relaxation Optimized SpectroscopY
Footnotes
The authors declare no conflict of interest.
References
- 1.(a) Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–9. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]; (b) Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nature reviews Drug discovery. 2004;3(4):301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 2.(a) Bocker A, Bonneau PR, Edwards PJ. HTS promiscuity analyses for accelerating decision making. Journal of biomolecular screening. 2011;16(7):765–74. doi: 10.1177/1087057111407763. [DOI] [PubMed] [Google Scholar]; (b) Coan KE, Maltby DA, Burlingame AL, Shoichet BK. Promiscuous aggregate-based inhibitors promote enzyme unfolding. Journal of medicinal chemistry. 2009;52(7):2067–75. doi: 10.1021/jm801605r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of medicinal chemistry. 2010;53(7):2719–40. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]; (d) Rishton GM. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov Today. 2003;8(2):86–96. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]; (e) Hann MM, Oprea TI. Pursuing the leadlikeness concept in pharmaceutical research. Curr Opin Chem Biol. 2004;8(3):255–63. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]; (f) Coan KE, Shoichet BK. Stability and equilibria of promiscuous aggregates in high protein milieus. Mol Biosyst. 2007;3(3):208–13. doi: 10.1039/b616314a. [DOI] [PubMed] [Google Scholar]; (g) Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat Chem Biol. 2005;1(3):146–8. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]; (h) Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat Protoc. 2006;1(2):550–3. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Feng BY, Shoichet BK. Synergy and antagonism of promiscuous inhibition in multiple-compound mixtures. J Med Chem. 2006;49(7):2151–4. doi: 10.1021/jm060029z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50(10):2385–90. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]; (k) McGovern SL, Helfand BT, Feng B, Shoichet BK. A specific mechanism of nonspecific inhibition. J Med Chem. 2003;46(20):4265–72. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]; (l) Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J Med Chem. 2003;46(21):4477–86. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]; (m) Shoichet BK. Interpreting steep dose-response curves in early inhibitor discovery. J Med Chem. 2006;49(25):7274–7. doi: 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- 3.Baell J, Walters MA. Chemistry: Chemical con artists foil drug discovery. Nature. 2014;513(7519):481–3. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 4.(a) Pellecchia M, Meininger D, Dong Q, Chang E, Jack R, Sem DS. NMR-based structural characterization of large protein-ligand interactions. J Biomol NMR. 2002;22(2):165–73. doi: 10.1023/a:1014256707875. [DOI] [PubMed] [Google Scholar]; (b) Pellecchia M. Solution nuclear magnetic resonance spectroscopy techniques for probing intermolecular interactions. Chemistry & biology. 2005;12(9):961–71. doi: 10.1016/j.chembiol.2005.08.013. [DOI] [PubMed] [Google Scholar]; (c) Stockman BJ, Dalvit C. NMR screening techniques in drug discovery and drug design. Progress in Nuclear Magnetic Resonance Spectroscopy. 2002;41:187–231. [Google Scholar]; (d) Lepre CA, Peng J, Fejzo J, Abdul-Manan N, Pocas J, Jacobs M, Xie X, Moore JM. Applications of SHAPES screening in drug discovery. Comb Chem High Throughput Screen. 2002;5(8):583–90. doi: 10.2174/1386207023329950. [DOI] [PubMed] [Google Scholar]; (e) Wyss DF, McCoy MA, Senior MM. NMR-based approaches for lead discovery. Curr Opin Drug Discov Devel. 2002;5(4):630–47. [PubMed] [Google Scholar]; (f) Jahnke W, Widmer H. Protein NMR in biomedical research. Cell Mol Life Sci. 2004;61(5):580–99. doi: 10.1007/s00018-003-3382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Huth JR, Sun C, Sauer DR, Hajduk PJ. Utilization of NMR-derived fragment leads in drug design. Methods Enzymol. 2005;394:549–71. doi: 10.1016/S0076-6879(05)94023-8. [DOI] [PubMed] [Google Scholar]; (h) Kessler H, Klages J. Lead Search and Optimization: NMR in Drug Discovery. In: Triggle DJ, Taylor JB, editors. Comprehensive Medicinal Chemistry II. Vol. 3. Elsevier; Oxford: 2006. pp. 901–920. [Google Scholar]; (i) Coles M, Heller M, Kessler H. NMR-based screening technologies. Drug Discov Today. 2003;8(17):803–10. doi: 10.1016/s1359-6446(03)02796-x. [DOI] [PubMed] [Google Scholar]; (j) Luy B, Frank A, Kessler H. Conformational analysis of drugs by NMR. In: Mannhold R, editor. Molecular Drug Properties: Measurement and Prediction. Wiley-VCH; Weinheim: 2008. pp. 207–254. [Google Scholar]; (k) Takeuchi K, Wagner G. NMR studies of protein interactions. Curr Opin Struct Biol. 2006;16(1):109–17. doi: 10.1016/j.sbi.2006.01.006. [DOI] [PubMed] [Google Scholar]; (l) Zartler ER, Shapiro MJ. Protein NMR-based screening in drug discovery. Curr Pharm Des. 2006;12(31):3963–72. doi: 10.2174/138161206778743619. [DOI] [PubMed] [Google Scholar]; (m) Fry DC, Emerson SD. Applications of biomolecular NMR to drug discovery. Drug Des Discov. 2000;17(1):13–33. [PubMed] [Google Scholar]; (n) Salvatella X, Giralt E. NMR-based methods and strategies for drug discovery. Chem Soc Rev. 2003;32(6):365–72. doi: 10.1039/b210047a. [DOI] [PubMed] [Google Scholar]; (o) Betz M, Saxena K, Schwalbe H. Biomolecular NMR: a chaperone to drug discovery. Curr Opin Chem Biol. 2006;10(3):219–25. doi: 10.1016/j.cbpa.2006.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (p) Klages J, Coles M, Kessler H. NMR-based screening: a powerful tool in fragment-based drug discovery. In: Bartlett PA, Etzeroth M, editors. NMR-Based Screening in Exploiting Chemical Diversity for Drug Discovery. Vol. 12. RSC Publishing; 2006. pp. 263–290. [Google Scholar]; (q) Pellecchia M, Bertini I, Cowburn D, Dalvit C, Giralt E, Jahnke W, James TL, Homans SW, Kessler H, Luchinat C, Meyer B, Oschkinat H, Peng J, Schwalbe H, Siegal G. Perspectives on NMR in drug discovery: a technique comes of age. Nature reviews Drug discovery. 2008;7(9):738–45. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barile E, Pellecchia M. NMR-based approaches for the identification and optimization of inhibitors of protein-protein interactions. Chemical reviews. 2014;114(9):4749–63. doi: 10.1021/cr500043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henchey LK, Jochim AL, Arora PS. Contemporary strategies for the stabilization of peptides in the alpha-helical conformation. Current opinion in chemical biology. 2008;12(6):692–7. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Valkov E, Sharpe T, Marsh M, Greive S, Hyvonen M. Targeting protein-protein interactions and fragment-based drug discovery. Topics in current chemistry. 2012;317:145–79. doi: 10.1007/128_2011_265. [DOI] [PubMed] [Google Scholar]; (b) Zartler ER, Shapiro MJ. Fragonomics: fragment-based drug discovery. Current opinion in chemical biology. 2005;9(4):366–70. doi: 10.1016/j.cbpa.2005.05.002. [DOI] [PubMed] [Google Scholar]; (c) Talamas FX, Ao-Ieong G, Brameld KA, Chin E, de Vicente J, Dunn JP, Ghate M, Giannetti AM, Harris SF, Labadie SS, Leveque V, Li J, Lui AS, McCaleb KL, Najera I, Schoenfeld RC, Wang B, Wong A. De novo fragment design: a medicinal chemistry approach to fragment-based lead generation. Journal of medicinal chemistry. 2013;56(7):3115–9. doi: 10.1021/jm4002605. [DOI] [PubMed] [Google Scholar]; (d) Fischer M, Hubbard RE. Fragment-based ligand discovery. Molecular interventions. 2009;9(1):22–30. doi: 10.1124/mi.9.1.7. [DOI] [PubMed] [Google Scholar]; (e) Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nature reviews Drug discovery. 2004;3(8):660–72. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]; (f) Jahnke W, Erlanson DA. Fragment-based Approaches in Drug Discovery. Wiley-VCH; 2006. [Google Scholar]
- 8.(a) Hajduk PJ, Sheppard G, Nettesheim DG, Olejniczak ET, Shuker SB, Meadows RP, Steinman DH, Carrera GM, Marcotte PA, Severin J, Walter K, Smith H, Gubbins E, Simmer R, Holzman TF, Morgan DW, Davidsen SK, Summers JB, Fesik SW. Discovery of potent nonpeptide inhibitors of stromelysin using SAR by NMR. Journal of the American Chemical Society. 1997;119:5818–5827. [Google Scholar]; (b) Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274(5292):1531–4. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 9.Pellecchia M. Antagonists of protein-protein interactions made easy? Journal of medicinal chemistry. 2013;56(1):13–4. doi: 10.1021/jm301837n. [DOI] [PubMed] [Google Scholar]
- 10.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O’Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435(7042):677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 11.(a) Vandenberg CJ, Cory S. ABT-199, a new Bcl-2-specific BH3 mimetic, has in vivo efficacy against aggressive Myc-driven mouse lymphomas without provoking thrombocytopenia. Blood. 2013;121(12):2285–8. doi: 10.1182/blood-2013-01-475855. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DC, Hymowitz SG, Jin S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC, Mason KD, Mitten MJ, Nimmer PM, Oleksijew A, Park CH, Park CM, Phillips DC, Roberts AW, Sampath D, Seymour JF, Smith ML, Sullivan GM, Tahir SK, Tse C, Wendt MD, Xiao Y, Xue JC, Zhang H, Humerickhouse RA, Rosenberg SH, Elmore SW. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nature medicine. 2013;19(2):202–8. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 12.(a) Hamzeh-Mivehroud M, Alizadeh AA, Morris MB, Church WB, Dastmalchi S. Phage display as a technology delivering on the promise of peptide drug discovery. Drug discovery today. 2013;18(23–24):1144–57. doi: 10.1016/j.drudis.2013.09.001. [DOI] [PubMed] [Google Scholar]; (b) Tan WS, Ho KL. Phage display creates innovative applications to combat hepatitis B virus. World journal of gastroenterology: WJG. 2014;20(33):11650–11670. doi: 10.3748/wjg.v20.i33.11650. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnology advances. 2010;28(6):849–58. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]; (d) Deutscher SL. Phage display in molecular imaging and diagnosis of cancer. Chemical reviews. 2010;110(5):3196–211. doi: 10.1021/cr900317f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Current opinion in structural biology. 2007;17(4):481–7. doi: 10.1016/j.sbi.2007.08.007. [DOI] [PubMed] [Google Scholar]; (f) Brissette R, Prendergast JK, Goldstein NI. Identification of cancer targets and therapeutics using phage display. Current opinion in drug discovery & development. 2006;9(3):363–9. [PubMed] [Google Scholar]; (g) Samoylova TI, Morrison NE, Globa LP, Cox NR. Peptide phage display: opportunities for development of personalized anti-cancer strategies. Anti-cancer agents in medicinal chemistry. 2006;6(1):9–17. doi: 10.2174/187152006774755492. [DOI] [PubMed] [Google Scholar]; (h) Mullen LM, Nair SP, Ward JM, Rycroft AN, Henderson B. Phage display in the study of infectious diseases. Trends in microbiology. 2006;14(3):141–7. doi: 10.1016/j.tim.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Paschke M. Phage display systems and their applications. Applied microbiology and biotechnology. 2006;70(1):2–11. doi: 10.1007/s00253-005-0270-9. [DOI] [PubMed] [Google Scholar]; (j) Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chemical reviews. 2005;105(11):4056–72. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 13.Flygare JA, Beresini M, Budha N, Chan H, Chan IT, Cheeti S, Cohen F, Deshayes K, Doerner K, Eckhardt SG, Elliott LO, Feng B, Franklin MC, Reisner SF, Gazzard L, Halladay J, Hymowitz SG, La H, LoRusso P, Maurer B, Murray L, Plise E, Quan C, Stephan JP, Young SG, Tom J, Tsui V, Um J, Varfolomeev E, Vucic D, Wagner AJ, Wallweber HJ, Wang L, Ware J, Wen Z, Wong H, Wong JM, Wong M, Wong S, Yu R, Zobel K, Fairbrother WJ. Discovery of a potent small-molecule antagonist of inhibitor of apoptosis (IAP) proteins and clinical candidate for the treatment of cancer (GDC-0152) Journal of medicinal chemistry. 2012;55(9):4101–13. doi: 10.1021/jm300060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu B, Zhang Z, Noberini R, Barile E, Giulianotti M, Pinilla C, Houghten RA, Pasquale EB, Pellecchia M. HTS by NMR of combinatorial libraries: a fragment-based approach to ligand discovery. Chemistry & biology. 2013;20(1):19–33. doi: 10.1016/j.chembiol.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Dooley CT, Houghten RA. Synthesis and screening of positional scanning combinatorial libraries. Methods in molecular biology. 1998;87:13–24. doi: 10.1385/0-89603-392-9:13. [DOI] [PubMed] [Google Scholar]; (b) Dooley CT, Houghten RA. The use of positional scanning synthetic peptide combinatorial libraries for the rapid determination of opioid receptor ligands. Life sciences. 1993;52(18):1509–17. doi: 10.1016/0024-3205(93)90113-h. [DOI] [PubMed] [Google Scholar]; (c) Houghten RA, Ostresh JM, Pratt SM. Modified solid-phase methods for the rapid synthesis of opioid peptides. NIDA research monograph. 1991;112:239–55. [PubMed] [Google Scholar]; (d) Pinilla C, Appel JR, Blanc P, Houghten RA. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. BioTechniques. 1992;13(6):901–5. [PubMed] [Google Scholar]
- 16.Klages J, Coles M, Kessler H. NMR-based Screening. Vol. 12. RSC Publishing; 2006. pp. 263–290. [Google Scholar]
- 17.(a) Gal M, Schanda P, Brutscher B, Frydman L. UltraSOFAST HMQC NMR and the repetitive acquisition of 2D protein spectra at Hz rates. Journal of the American Chemical Society. 2007;129(5):1372–7. doi: 10.1021/ja066915g. [DOI] [PubMed] [Google Scholar]; (b) Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. Journal of biomolecular NMR. 2005;33(4):199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 18.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(23):12366–71. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Religa TL, Kay LE. Optimal methyl labeling for studies of supra-molecular systems. Journal of biomolecular NMR. 2010;47(3):163–9. doi: 10.1007/s10858-010-9419-7. [DOI] [PubMed] [Google Scholar]; (b) Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nature protocols. 2006;1(2):749–54. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]; (c) Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1 methyl-protonated 15N-, 13C-, 2H-labeled proteins. Journal of biomolecular NMR. 1999;13(4):369–74. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]; (d) Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE. Selective methyl group protonation of perdeuterated proteins. Journal of molecular biology. 1996;263(5):627–36. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 20.Pellecchia M, Sem DS, Wuthrich K. NMR in drug discovery. Nat Rev Drug Discov. 2002;1(3):211–9. doi: 10.1038/nrd748. [DOI] [PubMed] [Google Scholar]
- 21.(a) Giannetti AM. From experimental design to validated hits a comprehensive walk-through of fragment lead identification using surface plasmon resonance. Methods in enzymology. 2011;493:169–218. doi: 10.1016/B978-0-12-381274-2.00008-X. [DOI] [PubMed] [Google Scholar]; (b) Frostell A, Vinterback L, Sjobom H. Protein-Ligand Interactions Using SPR Systems. Methods in molecular biology. 2013;1008:139–65. doi: 10.1007/978-1-62703-398-5_6. [DOI] [PubMed] [Google Scholar]; (c) Elinder M, Geitmann M, Gossas T, Kallblad P, Winquist J, Nordstrom H, Hamalainen M, Danielson UH. Experimental validation of a fragment library for lead discovery using SPR biosensor technology. Journal of biomolecular screening. 2011;16(1):15–25. doi: 10.1177/1087057110389038. [DOI] [PubMed] [Google Scholar]; (d) Danielson UH. Fragment library screening and lead characterization using SPR biosensors. Current topics in medicinal chemistry. 2009;9(18):1725–35. doi: 10.2174/156802609790102392. [DOI] [PubMed] [Google Scholar]; (e) Torres FE, Recht MI, Coyle JE, Bruce RH, Williams G. Higher throughput calorimetry: opportunities, approaches and challenges. Curr Opin Struct Biol. 2010;20(5):598–605. doi: 10.1016/j.sbi.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Chen Y, Shoichet BK. Molecular docking and ligand specificity in fragment-based inhibitor discovery. Nature chemical biology. 2009;5(5):358–64. doi: 10.1038/nchembio.155. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Clark M, Meshkat S, Talbot GT, Carnevali P, Wiseman JS. Fragment-based computation of binding free energies by systematic sampling. Journal of chemical information and modeling. 2009;49(8):1901–13. doi: 10.1021/ci900132r. [DOI] [PubMed] [Google Scholar]
- 23.(a) Recht MI, Sridhar V, Badger J, Hernandez L, Chie-Leon B, Nienaber V, Torres FE. Fragment-based screening for inhibitors of PDE4A using enthalpy arrays and X-ray crystallography. Journal of biomolecular screening. 2012;17(4):469–80. doi: 10.1177/1087057111430987. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hennig M, Ruf A, Huber W. Combining biophysical screening and X-ray crystallography for fragment-based drug discovery. Topics in current chemistry. 2012;317:115–43. doi: 10.1007/128_2011_225. [DOI] [PubMed] [Google Scholar]; (c) Wyss DF, Wang YS, Eaton HL, Strickland C, Voigt JH, Zhu Z, Stamford AW. Combining NMR and X-ray crystallography in fragment-based drug discovery: discovery of highly potent and selective BACE-1 inhibitors. Topics in current chemistry. 2012;317:83–114. doi: 10.1007/128_2011_183. [DOI] [PubMed] [Google Scholar]; (d) Wang YS, Strickland C, Voigt JH, Kennedy ME, Beyer BM, Senior MM, Smith EM, Nechuta TL, Madison VS, Czarniecki M, McKittrick BA, Stamford AW, Parker EM, Hunter JC, Greenlee WJ, Wyss DF. Application of fragment-based NMR screening, X-ray crystallography, structure-based design, and focused chemical library design to identify novel microM leads for the development of nM BACE-1 (beta-site APP cleaving enzyme 1) inhibitors. Journal of medicinal chemistry. 2010;53(3):942–50. doi: 10.1021/jm901472u. [DOI] [PubMed] [Google Scholar]; (e) Jhoti H, Cleasby A, Verdonk M, Williams G. Fragment-based screening using X-ray crystallography and NMR spectroscopy. Current opinion in chemical biology. 2007;11(5):485–93. doi: 10.1016/j.cbpa.2007.07.010. [DOI] [PubMed] [Google Scholar]; (f) Gill A, Cleasby A, Jhoti H. The discovery of novel protein kinase inhibitors by using fragment-based high-throughput x-ray crystallography. Chembiochem: a European journal of chemical biology. 2005;6(3):506–12. doi: 10.1002/cbic.200400188. [DOI] [PubMed] [Google Scholar]; (g) Hartshorn MJ, Murray CW, Cleasby A, Frederickson M, Tickle IJ, Jhoti H. Fragment-based lead discovery using X-ray crystallography. Journal of medicinal chemistry. 2005;48(2):403–13. doi: 10.1021/jm0495778. [DOI] [PubMed] [Google Scholar]; (h) Lesuisse D, Lange G, Deprez P, Benard D, Schoot B, Delettre G, Marquette JP, Broto P, Jean-Baptiste V, Bichet P, Sarubbi E, Mandine E. SAR and X-ray. A new approach combining fragment-based screening and rational drug design: application to the discovery of nanomolar inhibitors of Src SH2. Journal of medicinal chemistry. 2002;45(12):2379–87. doi: 10.1021/jm010927p. [DOI] [PubMed] [Google Scholar]
- 24.Petros AM, Dinges J, Augeri DJ, Baumeister SA, Betebenner DA, Bures MG, Elmore SW, Hajduk PJ, Joseph MK, Landis SK, Nettesheim DG, Rosenberg SH, Shen W, Thomas S, Wang X, Zanze I, Zhang H, Fesik SW. Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J Med Chem. 2006;49(2):656–63. doi: 10.1021/jm0507532. [DOI] [PubMed] [Google Scholar]
- 25.(a) Mok WW, Li Y. Therapeutic peptides: new arsenal against drug resistant pathogens. Curr Pharm Des. 2014;20(5):771–92. doi: 10.2174/13816128113199990011. [DOI] [PubMed] [Google Scholar]; (b) Farkhani SM, Valizadeh A, Karami H, Mohammadi S, Sohrabi N, Badrzadeh F. Cell penetrating peptides: efficient vectors for delivery of nanoparticles, nanocarriers, therapeutic and diagnostic molecules. Peptides. 2014;57:78–94. doi: 10.1016/j.peptides.2014.04.015. [DOI] [PubMed] [Google Scholar]; (c) Vasconcelos L, Parn K, Langel U. Therapeutic potential of cell-penetrating peptides. Therapeutic delivery. 2013;4(5):573–91. doi: 10.4155/tde.13.22. [DOI] [PubMed] [Google Scholar]; (d) Joo SH. Cyclic peptides as therapeutic agents and biochemical tools. Biomolecules & therapeutics. 2012;20(1):19–26. doi: 10.4062/biomolther.2012.20.1.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Boohaker RJ, Lee MW, Vishnubhotla P, Perez JM, Khaled AR. The use of therapeutic peptides to target and to kill cancer cells. Current medicinal chemistry. 2012;19(22):3794–804. doi: 10.2174/092986712801661004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lindberg S, Copolovici DM, Langel U. Therapeutic delivery opportunities, obstacles and applications for cell-penetrating peptides. Therapeutic delivery. 2011;2(1):71–82. doi: 10.4155/tde.10.78. [DOI] [PubMed] [Google Scholar]; (g) Johnson RM, Harrison SD, Maclean D. Therapeutic applications of cell-penetrating peptides. Methods in molecular biology. 2011;683:535–51. doi: 10.1007/978-1-60761-919-2_38. [DOI] [PubMed] [Google Scholar]; (h) Mason JM. Design and development of peptides and peptide mimetics as antagonists for therapeutic intervention. Future medicinal chemistry. 2010;2(12):1813–22. doi: 10.4155/fmc.10.259. [DOI] [PubMed] [Google Scholar]; (i) Juliano RL, Alam R, Dixit V, Kang HM. Cell-targeting and cell-penetrating peptides for delivery of therapeutic and imaging agents. Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology. 2009;1(3):324–35. doi: 10.1002/wnan.4. [DOI] [PubMed] [Google Scholar]; (j) Bidwell GL, 3rd, Raucher D. Therapeutic peptides for cancer therapy. Part I - peptide inhibitors of signal transduction cascades. Expert opinion on drug delivery. 2009;6(10):1033–47. doi: 10.1517/17425240903143745. [DOI] [PubMed] [Google Scholar]
- 26.(a) Lee S, Braun CR, Bird GH, Walensky LD. Photoreactive stapled peptides to identify and characterize BCL-2 family interaction sites by mass spectrometry. Methods in enzymology. 2014;544:25–48. doi: 10.1016/B978-0-12-417158-9.00002-9. [DOI] [PubMed] [Google Scholar]; (b) Walensky LD, Bird GH. Hydrocarbon-stapled peptides: principles, practice, and progress. Journal of medicinal chemistry. 2014;57(15):6275–88. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bird GH, Gavathiotis E, LaBelle JL, Katz SG, Walensky LD. Distinct BimBH3 (BimSAHB) stapled peptides for structural and cellular studies. ACS chemical biology. 2014;9(3):831–7. doi: 10.1021/cb4003305. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Nomura W, Aikawa H, Ohashi N, Urano E, Metifiot M, Fujino M, Maddali K, Ozaki T, Nozue A, Narumi T, Hashimoto C, Tanaka T, Pommier Y, Yamamoto N, Komano JA, Murakami T, Tamamura H. Cell-permeable stapled peptides based on HIV-1 integrase inhibitors derived from HIV-1 gene products. ACS chemical biology. 2013;8(10):2235–44. doi: 10.1021/cb400495h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Demizu Y, Nagoya S, Shirakawa M, Kawamura M, Yamagata N, Sato Y, Doi M, Kurihara M. Development of stapled short helical peptides capable of inhibiting vitamin D receptor (VDR)-coactivator interactions. Bioorganic & medicinal chemistry letters. 2013;23(15):4292–6. doi: 10.1016/j.bmcl.2013.06.002. [DOI] [PubMed] [Google Scholar]; (f) Verdine GL, Hilinski GJ. Stapled peptides for intracellular drug targets. Methods in enzymology. 2012;503:3–33. doi: 10.1016/B978-0-12-396962-0.00001-X. [DOI] [PubMed] [Google Scholar]
- 27.(a) Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annual review of immunology. 1994;12:433–55. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]; (b) Smith GP, Petrenko VA. Phage Display. Chemical reviews. 1997;97(2):391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 28.(a) Heinis C, Rutherford T, Freund S, Winter G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nature chemical biology. 2009;5(7):502–7. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]; (b) Landon LA, Deutscher SL. Combinatorial discovery of tumor targeting peptides using phage display. Journal of cellular biochemistry. 2003;90(3):509–17. doi: 10.1002/jcb.10634. [DOI] [PubMed] [Google Scholar]
- 29.(a) Nefzi A, Ostresh JM, Yu Y, Houghten RA. Combinatorial chemistry: libraries from libraries, the art of the diversity-oriented transformation of resin-bound peptides and chiral polyamides to low molecular weight acyclic and heterocyclic compounds. The Journal of organic chemistry. 2004;69(11):3603–9. doi: 10.1021/jo040114j. [DOI] [PubMed] [Google Scholar]; (b) Hoesl CE, Nefzi A, Ostresh JM, Yu Y, Houghten RA. Mixture-based combinatorial libraries: from peptides and peptidomimetics to small molecule acyclic and heterocyclic compounds. Methods in enzymology. 2003;369:496–517. doi: 10.1016/S0076-6879(03)69025-7. [DOI] [PubMed] [Google Scholar]; (c) Dooley CT, Houghten RA. New opioid peptides, peptidomimetics, and heterocyclic compounds from combinatorial libraries. Biopolymers. 1999;51(6):379–90. doi: 10.1002/(SICI)1097-0282(1999)51:6<379::AID-BIP2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]; (d) Nefzi A, Dooley C, Ostresh JM, Houghten RA. Combinatorial chemistry: from peptides and peptidomimetics to small organic and heterocyclic compounds. Bioorganic & medicinal chemistry letters. 1998;8(17):2273–8. doi: 10.1016/s0960-894x(98)00412-0. [DOI] [PubMed] [Google Scholar]; (e) Blondelle SE, Takahashi E, Weber PA, Houghten RA. Identification of antimicrobial peptides by using combinatorial libraries made up of unnatural amino acids. Antimicrobial agents and chemotherapy. 1994;38(10):2280–6. doi: 10.1128/aac.38.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Houghten RA, Appel JR, Blondelle SE, Cuervo JH, Dooley CT, Pinilla C. The use of synthetic peptide combinatorial libraries for the identification of bioactive peptides. BioTechniques. 1992;13(3):412–21. [PubMed] [Google Scholar]; (g) Wang J, Anania VG, Knott J, Rush J, Lill JR, Bourne PE, Bandeira N. Combinatorial approach for large-scale identification of linked peptides from tandem mass spectrometry spectra. Molecular & cellular proteomics: MCP. 2014;13(4):1128–36. doi: 10.1074/mcp.M113.035758. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Gray BP, Brown KC. Combinatorial peptide libraries: mining for cell-binding peptides. Chemical reviews. 2014;114(2):1020–81. doi: 10.1021/cr400166n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Bohrer BC, Li YF, Reilly JP, Clemmer DE, DiMarchi RD, Radivojac P, Tang H, Arnold RJ. Combinatorial libraries of synthetic peptides as a model for shotgun proteomics. Analytical chemistry. 2010;82(15):6559–68. doi: 10.1021/ac100910a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Molecular pharmaceutics. 2007;4(5):631–51. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]; (k) Bersanetti PA, Andrade MC, Casarini DE, Juliano MA, Nchinda AT, Sturrock ED, Juliano L, Carmona AK. Positional-scanning combinatorial libraries of fluorescence resonance energy transfer peptides for defining substrate specificity of the angiotensin I-converting enzyme and development of selective C-domain substrates. Biochemistry. 2004;43(50):15729–36. doi: 10.1021/bi048423r. [DOI] [PubMed] [Google Scholar]; (l) Real E, Rain JC, Battaglia V, Jallet C, Perrin P, Tordo N, Chrisment P, D’Alayer J, Legrain P, Jacob Y. Antiviral drug discovery strategy using combinatorial libraries of structurally constrained peptides. Journal of virology. 2004;78(14):7410–7. doi: 10.1128/JVI.78.14.7410-7417.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Boon CL, Frost D, Chakrabartty A. Identification of stable helical bundles from a combinatorial library of amphipathic peptides. Biopolymers. 2004;76(3):244–57. doi: 10.1002/bip.20074. [DOI] [PubMed] [Google Scholar]; (n) Cowell SM, Gu X, Vagner J, Hruby VJ. Intelligent design in combinatorial chemistry: use of designed peptide libraries to explore secondary and tertiary structures in peptides and proteins. Methods in enzymology. 2003;369:288–97. doi: 10.1016/S0076-6879(03)69016-6. [DOI] [PubMed] [Google Scholar]; (o) Nefzi A, Ostresh JM, Houghten RA. Combinatorial chemistry: mixture-based combinatorial libraries of acyclic and heterocyclic compounds from amino acids and short peptides. EXS. 2003;93:109–23. doi: 10.1007/978-3-0348-7997-2_6. [DOI] [PubMed] [Google Scholar]; (p) Horton DA, Bourne GT, Smythe ML. Exploring privileged structures: the combinatorial synthesis of cyclic peptides. Molecular diversity. 2002;5(4):289–304. doi: 10.1023/a:1021365402751. [DOI] [PubMed] [Google Scholar]
- 30.Dooley CT, Kaplan RA, Chung NN, Schiller PW, Bidlack JM, Houghten RA. Six highly active mu-selective opioid peptides identified from two synthetic combinatorial libraries. Peptide research. 1995;8(3):124–37. [PubMed] [Google Scholar]
- 31.(a) Lamberto I, Qin H, Noberini R, Premkumar L, Bourgin C, Riedl SJ, Song J, Pasquale EB. Distinctive binding of three antagonistic peptides to the ephrin-binding pocket of the EphA4 receptor. The Biochemical journal. 2012;445(1):47–56. doi: 10.1042/BJ20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lamberto I, Lechtenberg BC, Olson EJ, Mace PD, Dawson PE, Riedl SJ, Pasquale EB. Development and Structural Analysis of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor. ACS chemical biology. 2014 doi: 10.1021/cb500677x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noberini R, Koolpe M, Peddibhotla S, Dahl R, Su Y, Cosford ND, Roth GP, Pasquale EB. Small molecules can selectively inhibit ephrin binding to the EphA4 and EphA2 receptors. The Journal of biological chemistry. 2008;283(43):29461–72. doi: 10.1074/jbc.M804103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph TL, Lane DP, Verma CS. Stapled BH3 peptides against MCL-1: mechanism and design using atomistic simulations. PloS one. 2012;7(8):e43985. doi: 10.1371/journal.pone.0043985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.(a) Houghten RA, Pinilla C, Appel JR, Blondelle SE, Dooley CT, Eichler J, Nefzi A, Ostresh JM. Mixture-based synthetic combinatorial libraries. Journal of medicinal chemistry. 1999;42(19):3743–78. doi: 10.1021/jm990174v. [DOI] [PubMed] [Google Scholar]; (b) Pinilla C, Appel J, Blondelle S, Dooley C, Dorner B, Eichler J, Ostresh J, Houghten RA. A review of the utility of soluble peptide combinatorial libraries. Biopolymers. 1995;37(3):221–40. doi: 10.1002/bip.360370306. [DOI] [PubMed] [Google Scholar]