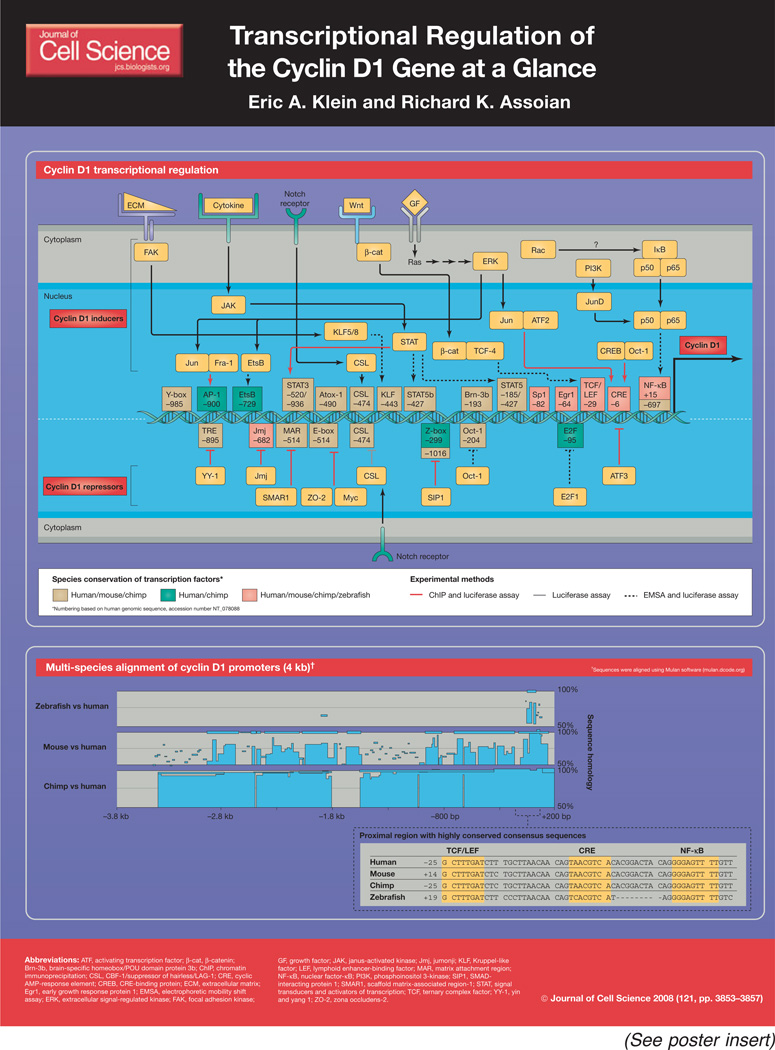
Regulated progression through the cell cycle requires sequential expression of a family of proteins called cyclins. Upon their induction, cyclins form complexes with specific cyclin-dependent kinases (CDKs), creating active holoenzymes that phosphorylate target proteins that are required for cell-cycle progression. Induction of the proto-oncogene cyclin D1, and its binding to CDK4 or CDK6, is a rate-limiting event during cell-cycle progression through G1 phase. Some studies suggest that cyclin D1 also has CDK-independent functions (reviewed by Fu et al., 2004).
In non-transformed cells, the cyclin D1 gene senses the mitogenic potential of the microenvironment during cell-cycle entry from quiescence because its induction requires coordinated signaling from the extracellular matrix (ECM) and soluble growth factors (Assoian and Klein, 2008). These controls can be lost during cellular transformation, and cyclin D1 is correspondingly overexpressed in a number of cancers, including those of the breast, liver, lung and brain (Gillett et al., 1996; Hall and Peters, 1996; Yamamoto et al., 2006; Molenaar et al., 2008; Sanchez-Mora et al., 2008). Conversely, repression of cyclin D1 gene expression is a hallmark of cell differentiation (James et al., 2006; Mejlvang et al., 2007; Takahashi et al., 2007). Since the first descriptions of the cyclin D1 promoter emerged ~15 years ago (Motokura and Arnold, 1993; Herber et al., 1994), many different transcription factors have been identified that directly bind to, or otherwise regulate, the cyclin D1 promoter (reviewed by Wang et al., 2004).
Cyclin D1 levels can be regulated transcriptionally and post-transcriptionally (Musgrove, 2006), and here we examine the transcriptional control of the cyclin D1 gene. We have focused this Cell Science at a Glance poster article on the transcription factors and binding sites that are functional in intact cells as determined by luciferase-reporter assays and chromatin immunoprecipitation (ChIP), and refer readers to Wang et al. (Wang et al., 2004) for a review that includes in vitro studies using electrophoretic mobility shift assays (EMSAs) as well as transcription factors, the binding sites for which remain unidentified. The poster that accompanies this text indicates the methods that have been used to implicate individual transcription-factor sequences in the regulation of cyclin D1 transcription. Many of these transcription-factor consensus sequences are regulated by multiple effector pathways, each of which has its own numerous upstream signaling molecules. We have highlighted only a few of the major signaling pathways that regulate each transcription-factor binding site. Wang et al. provide a thorough list of these upstream pathways (Wang et al., 2004). Finally, we have incorporated a bioinformatics-based, multi-species comparison of the cyclin D1 promoter, which allows us to distinguish between highly conserved and specialized regulatory mechanisms that control cyclin D1 gene expression.
Inducers of cyclin D1 gene transcription
Perhaps the best-studied activators of cyclin D1 gene transcription are mitogenic growth factors. The mitogen-activated protein kinases (MAPKs) play a major role in mitogenic signaling, and the canonical Ras-Raf-MEK [MAPK and extracellular signal-regulated kinase (ERK)]-ERK pathway can stimulate expression of AP-1 transcription factors, including members of the Fos, Jun and activating transcription factor (ATF) families (Balmanno and Cook, 1999; Shaulian and Karin, 2001). The human cyclin D1 promoter contains a consensus AP-1 site, at −903 bp†, which is regulated by Fos and Jun (Albanese et al., 1995; Shen et al., 2007). Jun can also form a complex with ATF2 to regulate the cyclic AMP-response element (CRE) (Sabbah et al., 1999; Castro-Rivera et al., 2001). ATF2 forms a complex with CRE-binding protein (CREB) and stimulates cyclin D1 promoter activity (Beier et al., 1999). ATF2-mediated cyclin D1 promoter induction can be stimulated by a number of growth-promoting agents, such as estrogen (Sabbah et al., 1999), hepatocyte growth factor (Recio and Merlino, 2002) and regenerating gene product (Reg) (Takasawa et al., 2006). In addition to regulating AP-1-dependent transcription, growth-factor-dependent Ras activation also promotes Sp1-mediated cyclin D1 gene expression – stimulation of neurons with nerve-growth factor promotes the formation of a transcription complex containing Sp1, p50 [a nuclear factor-κB (NF-κB) family member] and p107 (a pocket protein) (Marampon et al., 2008). The Sp1 consensus site can also bind the transcription factor B-Myb, resulting in cyclin D1 promoter activity (Bartusel et al., 2005).
Mitogen-stimulated Rac activity also induces cyclin D1 transcription, although the mechanism is not fully understood. In certain cellular contexts, Rac is upstream of NF-κB activity (Joyce et al., 1999), whereas in others, Rac and NF-κB signal through parallel pathways (Klein et al., 2007). Despite variances in mechanism, it is well established that both Rac and NF-κB are strong inducers of cyclin D1 gene expression (Guttridge et al., 1999; Hinz et al., 1999; Joyce et al., 1999; Matos and Jordan, 2005; Reddig et al., 2005; Klein et al., 2007; Yang et al., 2008). Furthermore, NF-κB activity can be enhanced by forming a complex with JunD in a phosphoinositide 3-kinase (PI3K)-and phosphoinositide-dependent kinase-1 (PDK1)-dependent manner (Toualbi-Abed et al., 2008).
Similar to growth factors, cytokines also stimulate cyclin D1 gene expression. Cytokines bind cell-surface receptors and initiate signaling through the JAK-STAT pathway. Janus-activated kinase (JAK) binds to ligand-bound cytokine receptors, and phosphorylates cytoplasmic signal transducers and activators of transcription (STAT) transcription factors, causing their translocation to the nucleus. Cytokines, such as interleukin-3 and interleukin-6, stimulate cyclin D1 promoter activity via STAT3 and STAT5 (Matsumura et al., 1999; Mishra and Das, 2005; Leslie et al., 2006; Wang et al., 2007; Gu et al., 2008).
In addition to these soluble factors, the ECM has a prominent role in regulating cyclin D1 gene expression (Assoian and Klein, 2008; Kothapalli et al., 2008). ECM proteins such as collagen, fibronectin and vitronectin activate focal adhesion kinase (FAK) upon integrin clustering. FAK activity can stimulate ERK signaling, leading to either AP-1- or EtsB-mediated transcription (Renshaw et al., 1999; Zhao et al., 2001). Additionally, FAK can activate the human cyclin D1 promoter by inducing the transcription factor Kruppel-like factor 8 (KLF8) (Zhao et al., 2001; Zhao et al., 2003). ECM proteins also activate Rac and are required to couple Rac-GTP to its effectors (del Pozo et al., 2000). Hyaluronan, a widely distributed, non-proteinaceous component of the ECM, regulates the signaling of ERK and Rac to cyclin D1 through its receptor, CD44 (Kothapalli et al., 2008).
As a consequence of its proliferative potential, cyclin D1 is a crucial regulator of Wnt- and Notch-regulated organism development (Hsu et al., 2001; Pal and Khanna, 2006). When Wnt binds its receptor, Frizzled, β-catenin is released to translocate from the cytoplasm to the nucleus, where it forms a complex with the ternary complex factor (TCF) and/or lymphoid enhancer-binding factor (LEF) transcription factors (Smalley and Dale, 1999) and stimulates cyclin D1 gene transcription (Shtutman et al., 1999; Tetsu and McCormick, 1999). β-catenin–TCF-mediated cyclin D1 gene transcription is further regulated by active Rac signaling (Esufali and Bapat, 2004), phosphorylation by protein kinase A (PKA) (Taurin et al., 2008), overexpression of the androgen receptor (Schweizer et al., 2008) and by forming a complex with AP-1 transcription factors (Toualbi et al., 2006). Proliferation and development are also regulated by Notch signaling through the activation of the CBF-1/suppressor of hairless/LAG-1 (CSL) transcription factor (Stahl et al., 2006). Interestingly, Notch-CSL signaling can also inhibit cyclin D1 induction, as described in the next section. The switch between inducer and repressor might be a result of the context-dependent recruitment of transcriptional co-regulators.
Several transcription factors stimulate cyclin D1 transcription either in a tissue-specific manner or via specialized stimulation. Hepatocyte nuclear factor 6 (HNF6) binds to the cyclin D1 promoter in the mouse liver (Tan et al., 2006). In the presence of Cu(I), the transcription factor Atox1 translocates to the nucleus and stimulates cyclin D1 gene expression (Itoh et al., 2008). Under hypoxic conditions, STAT5b is phosphorylated by JAK2, and expression of cyclin D1 mRNA is upregulated (Joung et al., 2005). Additionally, a number of transcription factors have been identified that induce cyclin D1 gene expression; however, the upstream signaling molecules that activate these pathways remain unknown. These transcription factors include KLF5 (Du et al., 2007), brain-specific homeobox/POU domain protein 3b (Brn-3b) (Budhram-Mahadeo et al., 2007) and DbpA (also called ZONAB) (Sourisseau et al., 2006).
Misregulation of cyclin D1 gene expression and increased proliferation are hallmarks of a number of proliferative diseases, including cancer and atherosclerosis. Activating mutations in of any of these transcription factors or their upstream signaling pathways could conceivably cause constitutive cyclin D1 expression and contribute to disease progression. Additionally, several transcription factors have been reported to selectively stimulate cyclin D1 gene transcription in cancer. Bombesin is a 14-amino-acid peptide that has been used as a marker for gastric cancer and neuroblastoma. Treatment of prostate cells with bombesin activates the Ras-MAPK pathway, which in this case leads to an early growth response protein 1 (Egr1)-dependent (rather than AP-1-dependent; see above) stimulation of cyclin D1 gene transcription (Xiao et al., 2005). Serine/threonine-protein kinase 11 (LKB1) normally functions as a tumor-suppressor by upregulating p21 in a p53-dependent manner, but LKB1 sporadically acquires a gain-of-function mutation in some cancers, which allows it to bind to the cyclin D1 promoter and stimulate gene transcription (Scott et al., 2007). Finally, the Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP1) is an oncogene that regulates the nuclear translocation of epidermal growth factor receptor (EGFR). Nuclear EGFR binds the cyclin D1 promoter and promotes gene transcription (Tao et al., 2005).
The numerous transcription-factor binding sites in the cyclin D1 promoter, and the extensive list of signaling pathways that regulate these binding sites, allows for a wide variety of regulatory mechanisms that induce cyclin D1 gene expression under an array of cellular conditions. However, the regulation of cyclin D1 transcription is not limited to transcriptional activation – a number of repressor proteins bind to the cyclin D1 promoter to inhibit its transcription.
Repressors of cyclin D1 gene transcription
Repression of cyclin D1 is a hallmark of cell differentiation for certain cell lineages. Cell-type-specific transcriptional repressors have been identified that inhibit the cyclin D1 promoter. These include jumonji (Jmj; also known as JARID2), SMAD-interacting protein 1 (SIP1) and POU domain, class 2, transcription factor 1 (Oct-1). Neurogenesis in the mouse hindbrain requires cyclin D1 repression by Jmj, as mice that lack this protein have abnormal mitotic clusters in the mantle zone (Takahashi et al., 2007). Cardiac myocyte proliferation is also repressed by Jmj, which suggests a role for this protein in controlling cardiac morphogenesis (Toyoda et al., 2003).
Epithelial cells can undergo an epithelial-mesenchymal transition (EMT) via the expression of a number of transcription factors, including SIP1. In A431 epidermoid cells, SIP1 induces an EMT that displays the characteristic invasive phenotype, while also downregulating cyclin D1 mRNA expression (Mejlvang et al., 2007). Inhibition of cyclin D1 mRNA expression blocks S-phase entry in these cells. Although the overexpression of cyclin D1 rescues S-phase entry in SIP1-treated cells, it not does not reduce cellular invasiveness (Mejlvang et al., 2007).
Oct-1 can function both as a cyclin D1 transcriptional repressor and inducer. Oct-1 is thought to repress cyclin D1 transcription because stimulation of mammary epithelial cells with prolactin induces cyclin D1 promoter activity by removing Oct-1 from the cyclin D1 promoter (Brockman and Schuler, 2005). Conversely, Oct-1 can form a complex with CREB to activate CRE-mediated cyclin D1 gene transcription (Boulon et al., 2002). In this context, Oct-1 does not bind directly to the cyclin D1 promoter, but acts as a transcriptional co-activator for CREB.
In addition to regulating cell differentiation, transcriptional repression of the cyclin D1 gene is crucial for maintaining cellular quiescence and preventing unwanted cell proliferation. One mechanism of maintaining quiescence in mammary epithelial cells is through yin and yang 1 (YY-1; also known as TYY1), which represses cyclin D1 by recruiting histone deactylase 1 (HDAC1) to a TRE/Oct-1-binding site on the cyclin D1 promoter. Treatment of human breast cancer cells with estrogen causes a Jun-Fos-estrogen receptor complex to displace YY-1 and stimulate cyclin D1 gene transcription (Cicatiello et al., 2004).
A number of transcription factors have been characterized as tumor suppressors on the basis of their ability to inhibit cyclin D1 gene transcription and cell-cycle progression. These include CSL, ATF3, scaffold matrix-associated region-1 (SMAR1; also known as BANP), zona occludens 2 (ZO-2) and p53. In contrast to its role as a transcriptional activator, as described above, the expression of CSL downstream of Notch signaling also confers protection against cutaneous squamous-cell carcinoma in a mouse model of skin cancer (Proweller et al., 2006). ATF3 is a stress-inducible member of the AP-1 family. ATF3 binds to the cyclin D1 promoter and represses its transcription, whereas knocking out ATF3 increases the growth rate and ability of mouse embryonic fibroblasts (MEFs) to form colonies in soft agar (Lu et al., 2006). SMAR1 inhibits cyclin D1 gene transcription by recruiting a repressor complex, containing the HDAC-complex component Sin3, HDAC1, and the pocket proteins p107 and p130, to the cyclin D1 promoter (Rampalli et al., 2005). The proto-oncogene myc can form a complex with the tight-junction protein ZO-2. This complex inhibits cyclin D1 transcription via the E-box of the promoter (Huerta et al., 2007). Finally, p19ARF represses cyclin D1 gene expression in human mammary epithelial cells by recruiting p53 to a cis-acting element in the cyclin D1 promoter (D’Amico et al., 2004).
Knowledge of the diverse set of regulatory elements that are described in this Cell Science at a Glance poster article was generated using a large number of cell lines from various cell lineages and species. As a consequence, it remains unclear which transcription factors are global regulators of proliferation and differentiation and which are species specific. The acquisition of whole-genome sequences from a number of mammalian species allows us to use bioinformatic analysis to study the conservation of consensus regulatory sites on the cyclin D1 promoter.
Multi-species bioinformatic analysis of the cyclin D1 promoter
Eto reported a multi-sequence analysis of the cyclin D1 promoter in 2000 (Eto, 2000). However, the recent accumulation of genomic sequence data combined with advances in bioinformatics tools for promoter analysis prompted us to update the multi-species alignment of the cyclin D1 promoter to identify conserved and species-specific regulatory elements. We performed an alignment of the human, chimp, mouse and zebrafish cyclin D1 promoters (accession numbers NT_078088, NW_001222300, NT_039437 and NW_001879279, respectively) using 3.8 kb of sequence upstream and 200 bp downstream of the transcription start sites as annotated in GenBank. The sequences were aligned using Mulan software (mulan.dcode.org) and the results were submitted to MultiTF (multitf.dcode.org) to identify conserved transcription-factor consensus sequences.
Evolutionarily conserved regions (ECRs) were defined as regions that were at least 70 bp long with a minimum of 70% homology. Similar to Eto (Eto, 2000), we found that mammalian promoters share a large number of ECRs. However, the zebrafish cyclin D1 promoter contains only one ECR immediately upstream of the transcription start site. Interestingly, this highly conserved region contains the TCF, CRE and NF-κB consensus sequences. The conservation of these binding sites suggests that they have a crucial and fundamental role in cyclin D1 induction and cell proliferation.
Multi-species alignment also allowed us to determine which transcription factors might function in a species-specific manner. As shown in the accompanying poster, most promoter elements are conserved from mice to humans, yet there are notable exceptions. The human and chimp cyclin D1 promoters contain an E2F consensus site (TTTGGCGCCCG) at positions −95 and −94, respectively, and E2F1 can repress cyclin D1 gene expression in assays using human luciferase-promoter constructs (Watanabe et al., 1998). However, no homologous sequence exists in the mouse.
Multi-species promoter alignment can be a valuable tool for resolving mechanisms by which transcription factors affect promoter activity. For example, many studies have reported that AP-1 transcription factors regulate cyclin D1 gene expression, and the human and chimp cyclin D1 promoters contain a consensus AP-1 site (see above). However, our analysis of the human and mouse cyclin D1 promoters demonstrates that this site (TGAGTCA at position −903 in the human promoter) is not conserved in the mouse. In fact, the first conserved AP-1 site that we can identify (TGAGTCA) is very far upstream of the transcription start site (−24 kb in the mouse promoter and −53 kb in the human promoter). Eto used bioinformatics to identify a potential AP-1 site in the mouse promoter (position −795; TGTCTCA) on the basis of its similarity to the consensus AP-1 site (Eto, 2000). However, this sequence does not register as an AP-1 site when analyzed by JASPAR (Vlieghe et al., 2006) or Transfac (Wingender, 1994). Additionally, the third base in AP-1 sites is almost always an adenine, but rarely a thymine (Pollock and Treisman, 1990).
As ERK and AP-1 stimulate cyclin D1 gene transcription in mouse as well as human cells (Brown et al., 1998; Burch et al., 2004; Shen et al., 2006; Villanueva et al., 2007), there may be alternative mechanisms by which ERK regulates cyclin D1 promoter activity. In addition to a possible distant AP-1 site, ERK regulates Ets-family transcription factors, and AP-1 family members have the potential to activate cyclin D1 transcription at other consensus sites, such as the conserved CRE, which is known to bind Jun and ATF2 (Beier et al., 1999; Sabbah et al., 1999). It is important to emphasize, however, that the likelihood of alternative mechanisms for ERK-dependent cyclin D1 gene induction in the mouse does not lessen the probable importance of the −903 AP-1 site in the primate cyclin D1 promoters.
Perspectives
Research that spans 15 years has revealed the extremely complex regulation of cyclin D1 transcription, presumably to maintain proper physiological quiescence and differentiation, and to allow for cellular proliferation upon appropriate extracellular stimulation. Modern molecular-biology tools combined with bioinformatic approaches using whole-genome sequences can help advance our understanding of the transcriptional regulation of the cyclin D1 gene in these physiological and pathological contexts. The high reliance of modern biomedical research on the mouse emphasizes the importance of recognizing these interspecies promoter differences during data interpretation and the need to use species-matched reagents (i.e. species-specific promoter constructs and cells) for studying non-conserved elements.
Acknowledgments
Our research is supported by grants from the National Institutes of Health.
Abbreviations
- ATF
activating transcription factor
- β-cat
β-catenin
- Brn-3b
brain-specific homeobox/POU domain protein 3b
- ChIP
chromatin immunoprecipitation
- CSL
CBF-1/suppressor of hairless/LAG-1
- CRE
cyclic AMP-response element
- CREB
CRE-binding protein
- ECM
extracellular matrix
- Egr1
early growth response protein 1
- EMSA
electrophoretic mobility shift assay
- ERK
extracellular signal-regulated kinase
- FAK
focal adhesion kinase
- GF
growth factor
- JAK
janus-activated kinase
- Jmj
jumonji
- KLF
Kruppel-like factor
- LEF
lymphoid enhancer-binding factor
- MAR
matrix attachment region
- NF-κB
nuclear factor-κB
- PI3K
phosphoinositol 3-kinase
- SIP1
SMAD-interacting protein 1
- SMAR1
scaffold matrix-associated region-1
- STAT
signal transducers and activators of transcription
- TCF
ternary complex factor
- YY-1
yin and yang 1
- ZO-2
zona occludens-2
Footnotes
All numbering is based on the human genomic sequence (accession number NT_078088).
References
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- Assoian RK, Klein EA. Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol. 2008;18:347–352. doi: 10.1016/j.tcb.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmanno K, Cook SJ. Sustained MAP kinase activation is required for the expression of cyclin D1, p21Cip1 and a subset of AP-1 proteins in CCL39 cells. Oncogene. 1999;18:3085–3097. doi: 10.1038/sj.onc.1202647. [DOI] [PubMed] [Google Scholar]
- Bartusel T, Schubert S, Klempnauer K-H. Regulation of the cyclin D1 and cyclin A1 promoters by B-Myb is mediated by Sp1 binding sites. Gene. 2005;351:171–180. doi: 10.1016/j.gene.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Beier F, Lee RJ, Taylor AC, Pestell RG, LuValle P. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc. Natl. Acad. Sci. USA. 1999;96:1433–1438. doi: 10.1073/pnas.96.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Dantonel J-C, Binet V, Vie A, Blanchard J-M, Hipskind RA, Philips A. Oct-1 potentiates CREB-driven Cyclin D1 promoter activation via a phospho-CREB- and CREB binding protein-independent mechanism. Mol. Cell. Biol. 2002;22:7769–7779. doi: 10.1128/MCB.22.22.7769-7779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman JL, Schuler LA. Prolactin signals via Stat5 and Oct-1 to the proximal cyclin D1 promoter. Mol. Cell. Endocrinol. 2005;239:45–53. doi: 10.1016/j.mce.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Nigh E, Lee RJ, Ye H, Thompson MA, Saudou F, Pestell RG, Greenberg ME. Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol. 1998;18:5609–5619. doi: 10.1128/mcb.18.9.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhram-Mahadeo VS, Irshad S, Bowen S, Lee SA, Samady L, Tonini GP, Latchman DS. Proliferation-associated Brn-3b transcription factor can activate cyclin D1 expression in neuroblastoma and breast cancer cells. Oncogene. 2007;27:145–154. doi: 10.1038/sj.onc.1210621. [DOI] [PubMed] [Google Scholar]
- Burch PM, Yuan Z, Loonen A, Heintz NH. An extracellular signal-regulated kinase 1- and 2-dependent program of chromatin trafficking of c-Fos and Fra-1 is required for Cyclin D1 expression during cell cycle reentry. Mol. Cell. Biol. 2004;24:4696–4709. doi: 10.1128/MCB.24.11.4696-4709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Rivera E, Samudio I, Safe S. Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J. Biol. Chem. 2001;276:30853–30861. doi: 10.1074/jbc.M103339200. [DOI] [PubMed] [Google Scholar]
- Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, Cancemi M, Caporali S, Caristi S, Scafoglio C, et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/Estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of Cyclin D1 to its own gene promoter. Mol. Cell. Biol. 2004;24:7260–7274. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico M, Wu K, Fu M, Rao M, Albanese C, Russell RG, Lian H, Bregman D, White MA, Pestell RG. The inhibitor of Cyclin-dependent kinase 4a/Alternative Reading Frame (INK4a/ARF) locus encoded proteins p16INK4a and p19ARF repress Cyclin D1 transcription through distinct cis elements. Cancer Res. 2004;64:4122–4130. doi: 10.1158/0008-5472.CAN-03-2519. [DOI] [PubMed] [Google Scholar]
- del Pozo MA, Price LS, Alderson NB, Ren XD, Schwartz MA. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 2000;19:2008–2014. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du JX, Yun CC, Bialkowska A, Yang VW. Protein Inhibitor of activated STAT1 interacts with and up-regulates activities of the pro-proliferative transcription factor kruppel-like factor 5. J. Biol. Chem. 2007;282:4782–4793. doi: 10.1074/jbc.M603413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esufali S, Bapat B. Cross-talk between Rac1 GTPase and dysregulated Wnt signaling pathway leads to cellular redistribution of [beta]-catenin and TCF//LEF-mediated transcriptional activation. Oncogene. 2004;23:8260–8271. doi: 10.1038/sj.onc.1208007. [DOI] [PubMed] [Google Scholar]
- Eto I. Molecular cloning and sequence analysis of the promoter region of mouse cyclin D1 gene: implication in phorbol ester-induced tumour promotion. Cell Prolif. 2000;33:167–187. doi: 10.1046/j.1365-2184.2000.00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- Gillett C, Smith P, Gregory W, Richards M, Millis R, Peters G, Barnes D. Cyclin D1 and prognosis in human breast cancer. Int. J. Cancer. 1996;69:92–99. doi: 10.1002/(SICI)1097-0215(19960422)69:2<92::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Gu J, Li G, Sun T, Su Y, Zhang X, Shen J, Tian Z, Zhang J. Blockage of the STAT3 signaling pathway with a decoy oligonucleotide suppresses growth of human malignant glioma cells. J. Neurooncol. 2008;16:16. doi: 10.1007/s11060-008-9590-9. [DOI] [PubMed] [Google Scholar]
- Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS., Jr NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv. Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- Herber B, Truss M, Beato M, Muller R. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:1295–1304. [PubMed] [Google Scholar]
- Hinz M, Krappmann D, Eichten A, Heder A, Scheidereit C, Strauss M. NF-kappaB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol. Cell. Biol. 1999;19:2690–2698. doi: 10.1128/mcb.19.4.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu W, Shakya R, Costantini F. Impaired mammary gland and lymphoid development caused by inducible expression of Axin in transgenic mice. J. Cell Biol. 2001;155:1055–1064. doi: 10.1083/jcb.200107066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta M, Munoz R, Tapia R, Soto-Reyes E, Ramirez L, Recillas-Targa F, Gonzalez-Mariscal L, Lopez-Bayghen E. Cyclin D1 is transcriptionally down-regulated by ZO-2 via an E Box and the transcription factor c-Myc. Mol. Biol. Cell. 2007;18:4826–4836. doi: 10.1091/mbc.E07-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S, Kim HW, Nakagawa O, Ozumi K, Lessner SM, Aoki H, Akram K, McKinney RD, Ushio-Fukai M, Fukai T. Novel role of Antioxidant-1 (Atox1) as a copper-dependent transcription factor involved in cell proliferation. J. Biol. Chem. 2008;283:9157–9167. doi: 10.1074/jbc.M709463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CG, Woods A, Underhill TM, Beier F. The transcription factor ATF3 is upregulated during chondrocyte differentiation and represses cyclin D1 and A gene transcription. BMC Mol. Biol. 2006;7:30. doi: 10.1186/1471-2199-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung YH, Lim EJ, Lee MY, Park JH, Ye SK, Park EU, Kim SY, Zhang Z, Lee KJ, Park DK, et al. Hypoxia activates the cyclin D1 promoter via the Jak2/STAT5b pathway in breast cancer cells. Exp. Mol. Med. 2005;37:353–364. doi: 10.1038/emm.2005.45. [DOI] [PubMed] [Google Scholar]
- Joyce D, Bouzahzah B, Fu M, Albanese C, D’Amico M, Steer J, Klein JU, Lee RJ, Segall JE, Westwick JK, et al. Integration of Rac-dependent regulation of cyclin D1 transcription through a nuclear factor-kappaB-dependent pathway. J. Biol. Chem. 1999;274:25245–25249. doi: 10.1074/jbc.274.36.25245. [DOI] [PubMed] [Google Scholar]
- Klein EA, Yang C, Kazanietz MG, Assoian RK. NFkappaB-independent signaling to the cyclin D1 gene by Rac. Cell Cycle. 2007;6:1115–1121. doi: 10.4161/cc.6.9.4147. [DOI] [PubMed] [Google Scholar]
- Kothapalli D, Flowers J, Xu T, Pure E, Assoian RK. Differential activation of ERK and Rac mediates the proliferative and anti-proliferative effects of hyaluronan and CD44. J. Biol. Chem. 2008;19:19. doi: 10.1074/jbc.M802934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie K, Lang C, Devgan G, Azare J, Berishaj M, Gerald W, Kim YB, Paz K, Darnell JE, Albanese C, et al. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of Transcription 3. Cancer Res. 2006;66:2544–2552. doi: 10.1158/0008-5472.CAN-05-2203. [DOI] [PubMed] [Google Scholar]
- Lu D, Wolfgang CD, Hai T. Activating Transcription Factor 3, a stress-inducible gene, suppresses ras-stimulated tumorigenesis. J. Biol. Chem. 2006;281:10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- Marampon F, Casimiro MC, Fu M, Powell MJ, Popov VM, Lindsay J, Zani BM, Ciccarelli C, Watanabe G, Lee RJ, et al. Nerve growth factor regulation of Cyclin D1 in PC12 cells through a p21RAS extracellular signal-regulated kinase pathway requires cooperative interactions between Sp1 and Nuclear Factor-{kappa}B. Mol. Biol. Cell. 2008;19:2566–2578. doi: 10.1091/mbc.E06-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos P, Jordan P. Expression of Rac1b stimulates NF-kappaB-mediated cell survival and G1/S progression. Exp. Cell Res. 2005;305:292–299. doi: 10.1016/j.yexcr.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Matsumura I, Kitamura T, Wakao H, Tanaka H, Hashimoto K, Albanese C, Downward J, Pestell RG, Kanakura Y. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999;18:1367–1377. doi: 10.1093/emboj/18.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlvang J, Kriajevska M, Vandewalle C, Chernova T, Sayan AE, Berx G, Mellon JK, Tulchinsky E. Direct repression of Cyclin D1 by SIP1 attenuates cell cycle progression in cells undergoing an epithelial mesenchymal transition. Mol. Biol. Cell. 2007;18:4615–4624. doi: 10.1091/mbc.E07-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R, Das B. Activation of STAT5-cyclin D1 pathway in chewing tobacco mediated oral squamous cell carcinoma. Mol. Biol. Rep. 2005;32:159–166. doi: 10.1007/s11033-005-0754-9. [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Ebus ME, Koster J, van Sluis P, van Noesel CJ, Versteeg R, Caron HN. Cyclin D1 and CDK4 activity contribute to the undifferentiated phenotype in neuroblastoma. Cancer Res. 2008;68:2599–2609. doi: 10.1158/0008-5472.CAN-07-5032. [DOI] [PubMed] [Google Scholar]
- Motokura T, Arnold A. PRAD1/cyclin D1 proto-oncogene: genomic organization: 5′ DNA sequence, and sequence of a tumor-specific rearrangement breakpoint. Genes Chromosomes Cancer. 1993;7:89–95. doi: 10.1002/gcc.2870070205. [DOI] [PubMed] [Google Scholar]
- Musgrove EA. Cyclins: roles in mitogenic signaling and oncogenic transformation. Growth Factors. 2006;24:13–19. doi: 10.1080/08977190500361812. [DOI] [PubMed] [Google Scholar]
- Pal R, Khanna A. Role of smad- and wnt-dependent pathways in embryonic cardiac development. Stem Cells Dev. 2006;15:29–39. doi: 10.1089/scd.2006.15.29. [DOI] [PubMed] [Google Scholar]
- Pollock R, Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990;18:6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proweller A, Tu L, Lepore JJ, Cheng L, Lu MM, Seykora J, Millar SE, Pear WS, Parmacek MS. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 2006;66:7438–7444. doi: 10.1158/0008-5472.CAN-06-0793. [DOI] [PubMed] [Google Scholar]
- Rampalli S, Pavithra L, Bhatt A, Kundu TK, Chattopadhyay S. Tumor suppressor SMAR1 mediates Cyclin D1 repression by recruitment of the SIN3/Histone Deacetylase 1 complex. Mol. Cell. Biol. 2005;25:8415–8429. doi: 10.1128/MCB.25.19.8415-8429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recio JA, Merlino G. Hepatocyte growth factor/scatter factor activates proliferation in melanoma cells through p38 MAPK, ATF-2 and cyclin D1. Oncogene. 2002;21:1000–1008. doi: 10.1038/sj.onc.1205150. [DOI] [PubMed] [Google Scholar]
- Reddig PJ, Xu D, Juliano RL. Regulation of p21-activated kinase-independent Rac1 signal transduction by nischarin. J. Biol. Chem. 2005;280:30994–31002. doi: 10.1074/jbc.M502546200. [DOI] [PubMed] [Google Scholar]
- Renshaw MW, Price LS, Schwartz MA. Focal adhesion kinase mediates the integrin signaling requirement for growth factor activation of MAP kinase. J. Cell Biol. 1999;147:611–618. doi: 10.1083/jcb.147.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah M, Courilleau D, Mester J, Redeuilh G. Estrogen induction of the cyclin D1 promoter: Involvement of a cAMP response-like element. Proc. Natl. Acad. Sci. USA. 1999;96:11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mora N, Presmanes MC, Monroy V, Moreno N, Lara-Martinez JM, Aladro MH, Alvarez-Fernandez E. Micropapillary lung adenocarcinoma: a distinctive histologic subtype with prognostic significance. Case series. Hum. Pathol. 2008;39:324–330. doi: 10.1016/j.humpath.2007.05.029. [DOI] [PubMed] [Google Scholar]
- Schweizer L, Rizzo C, Spires T, Platero JS, Wu Q, Lin T-A, Gottardis M, Attar R. The androgen receptor can signal through Wnt/beta-Catenin in prostate cancer cells as an adaptation mechanism to castration levels of androgens. BMC Cell Biol. 2008;9:4. doi: 10.1186/1471-2121-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KD, Nath-Sain S, Agnew MD, Marignani PA. LKB1 catalytically deficient mutants enhance Cyclin D1 expression. Cancer Res. 2007;67:5622–5627. doi: 10.1158/0008-5472.CAN-07-0762. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhang Y, Uray IP, Hill JL, Kim HT, Lu C, Young MR, Gunther EJ, Hilsenbeck SG, Chodosh LA, et al. The AP-1 transcription factor regulates postnatal mammary gland development. Dev. Biol. 2006;295:589–603. doi: 10.1016/j.ydbio.2006.03.042. [DOI] [PubMed] [Google Scholar]
- Shen Q, Uray IP, Li Y, Krisko TI, Strecker TE, Kim HT, Brown PH. The AP-1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors. Oncogene. 2007;27:366–377. doi: 10.1038/sj.onc.1210643. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley MJ, Dale TC. Wnt signalling in mammalian development and cancer. Cancer Metastasis Rev. 1999;18:215–230. doi: 10.1023/a:1006369223282. [DOI] [PubMed] [Google Scholar]
- Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, Balda MS. Regulation of PCNA and Cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol. Cell. Biol. 2006;26:2387–2398. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Ge C, Shi S, Pestell RG, Stanley P. Notch1-induced transformation of RKE-1 cells requires up-regulation of Cyclin D1. Cancer Res. 2006;66:7562–7570. doi: 10.1158/0008-5472.CAN-06-0974. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Kojima M, Nakajima K, Suzuki-Migishima R, Takeuchi T. Functions of a jumonji-cyclin D1 pathway in the coordination of cell cycle exit and migration during neurogenesis in the mouse hindbrain. Dev. Biol. 2007;303:549–560. doi: 10.1016/j.ydbio.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Takasawa S, Ikeda T, Akiyama T, Nata K, Nakagawa K, Shervani NJ, Noguchi N, Murakami-Kawaguchi S, Yamauchi A, Takahashi I, et al. Cyclin D1 activation through ATF-2 in Reginduced pancreatic beta-cell regeneration. FEBS Lett. 2006;580:585–591. doi: 10.1016/j.febslet.2005.12.070. [DOI] [PubMed] [Google Scholar]
- Tan Y, Yoshida Y, Hughes DE, Costa RH. Increased expression of hepatocyte Nuclear Factor 6 stimulates hepatocyte proliferation during mouse liver regeneration. Gastroenterology. 2006;130:1283–1300. doi: 10.1053/j.gastro.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Song X, Deng X, Xie D, Lee LM, Liu Y, Li W, Li L, Deng L, Wu Q, et al. Nuclear accumulation of epidermal growth factor receptor and acceleration of G1/S stage by Epstein-Barr-encoded oncoprotein latent membrane protein 1. Exp. Cell Res. 2005;303:240–251. doi: 10.1016/j.yexcr.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Taurin S, Sandbo N, Yau DM, Sethakorn N, Dulin NO. Phosphorylation of {beta}-catenin by PKA promotes ATP-induced proliferation of vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2008;294:C1169–C1174. doi: 10.1152/ajpcell.00096.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Toualbi K, Guller MC, Mauriz JL, Labalette C, Buendia MA, Mauviel A, Bernuau D. Physical and functional cooperation between AP-1 and [beta]-catenin for the regulation of TCF-dependent genes. Oncogene. 2006;26:3492–3502. doi: 10.1038/sj.onc.1210133. [DOI] [PubMed] [Google Scholar]
- Toualbi-Abed K, Daniel F, Guller MC, Legrand A, Mauriz J-L, Mauviel A, Bernuau D. Jun D cooperates with p65 to activate the proximal {kappa}B site of the cyclin D1 promoter: role of PI3K/PDK-1. Carcinogenesis. 2008;29:536–543. doi: 10.1093/carcin/bgm293. [DOI] [PubMed] [Google Scholar]
- Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev. Cell. 2003;5:85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- Villanueva J, Yung Y, Walker JL, Assoian RK. ERK activity and G1 phase progression: identifying dispensable versus essential activities and primary versus secondary targets. Mol. Biol. Cell. 2007;18:1457–1463. doi: 10.1091/mbc.E06-10-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Z, Fu M, Bouras T, Pestell RG. Signal transduction mediated by cyclin D1: from mitogens to cell proliferation: a molecular target with therapeutic potential. Cancer Treat. Res. 2004;119:217–237. doi: 10.1007/1-4020-7847-1_11. [DOI] [PubMed] [Google Scholar]
- Wang D, Liu Z, Li Q, Karpurapu M, Kundumani-Sridharan V, Cao H, Dronadula N, Rizvi F, Bajpai AK, Zhang C, et al. An essential role for gp130 in neointima formation following arterial injury. Circ. Res. 2007;100:807–816. doi: 10.1161/01.RES.0000261350.61711.9e. [DOI] [PubMed] [Google Scholar]
- Watanabe G, Albanese C, Lee RJ, Reutens A, Vairo G, Henglein B, Pestell RG. Inhibition of Cyclin D1 Kinase activity is associated with E2F-mediated inhibition of Cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E. Recognition of regulatory regions in genomic sequences. J. Biotechnol. 1994;35:273–280. doi: 10.1016/0168-1656(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Xiao D, Chinnappan D, Pestell R, Albanese C, Weber HC. Bombesin regulates Cyclin D1 expression through the early growth response protein Egr-1 in prostate cancer cells. Cancer Res. 2005;65:9934–9942. doi: 10.1158/0008-5472.CAN-05-1830. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Tamakawa S, Yoshie M, Yaginuma Y, Ogawa K. Neoplastic hepatocyte growth associated with cyclin D1 redistribution from the cytoplasm to the nucleus in mouse hepatocarcinogenesis. Mol. Carcinog. 2006;45:901–913. doi: 10.1002/mc.20204. [DOI] [PubMed] [Google Scholar]
- Yang C, Klein EA, Assoian RK, Kazanietz MG. Heregulin beta1 promotes breast cancer cell proliferation through rac/erk-dependent induction of Cyclin D1 and p21 Cip1. Biochem. J. 2008;410:167–175. doi: 10.1042/BJ20070781. [DOI] [PubMed] [Google Scholar]
- Zhao J, Pestell R, Guan J-L. Transcriptional activation of Cyclin D1 promoter by FAK contributes to cell cycle progression. Mol. Biol. Cell. 2001;12:4066–4077. doi: 10.1091/mbc.12.12.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Bian ZC, Yee K, Chen BPC, Chien S, Guan J-L. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of Cyclin D1 and cell cycle progression. Mol. Cell. 2003;11:1503–1515. doi: 10.1016/s1097-2765(03)00179-5. [DOI] [PubMed] [Google Scholar]