Abstract
Basal breast cancer, common among patients presenting with inflammatory breast cancer, has been shown to be resistant to radiation and enriched in cancer stem cells. The Notch pathway plays an important role in self-renewal of breast cancer stem cells and contributes to inflammatory signaling that promotes the breast cancer stem cell phenotype. Herein we inhibited Notch signaling using a gamma secretase inhibitor, RO4929097, in an in vitro model that enriches for cancer initiating cells (3D clonogenic assay) and conventional 2D clonogenic assay to compare the effect on radiosensitization of the SUM149 and SUM190 inflammatory breast cancer (IBC) cell lines. RO4929097 downregulated the Notch target genes Hes1, Hey1 and HeyL and showed a significant reduction in anchorage independent growth in SUM190 and SUM149. However, the putative self-renewal assay mammosphere formation efficiency was increased with the drug. To assess radiosensitization of putative cancer stem cells, cells were exposed to increasing doses of radiation with or without 1uM RO4929097 in their standard (2D) and self-renewal enriching (3D) culture conditions. In the conventional 2D clonogenic assay, RO4929097 significantly sensitized SUM190 cells to ionizing radiation and has a modest radiosensitization effect in SUM149 cells. In the 3D clonogenic assays, however, a radioprotective effect was seen in both SUM149 and SUM190 cells at higher doses. Both cell lines express IL-6 and IL-8, cytokines known to mediate the efficacy of notch inhibition and to promote self-renewal of stem cells. We further showed that RO429097 inhibits normal T-cell synthesis of some inflammatory cytokines, including TNF-α, a potential mediator of IL-6 and IL-8 production in the microenvironment. These data suggest additional targeting agents may be required to selectively target IBC stem cells through notch inhibition, and that evaluation of microenvironmental influences may shed further light on the potential effects of this inhibitor.
Keywords: Notch, Cancer stem cells, R04929097, Inflammatory breast cancer, Radiation
Introduction
In breast cancer and other solid tumors, there is increasing evidence suggesting that aggressive basal tumor types are enriched for a subpopulation of cells called cancer stem cells (CSCs) or tumor-initiating cells. These cells drive tumor formation when transplanted into immunocompromised mice, undergo self-renewal and re-populate the tumor bulk and its heterogeneity. They are believed to contribute to therapeutic resistance and tumor metastasis or local recurrence [1–9]. Human breast CSCs can be enriched by cell surface markers CD44+CD24low and by aldehyde dehydrogenase (ALDH) enzyme activity as ALDH+ cells [10, 11]. In vitro propagation as mammospheres also enriches for the CSC population [12].
Inflammatory breast cancer (IBC) is the most aggressive form of breast cancer accounting for 2.5% of all breast cancer cases. It is characterized by rapid progression, local and distant metastases, younger age of onset, and lower overall survival compared with other breast cancers [13]. The prognosis of patients with IBC has only improved slightly with standard therapies over the last 10 to 15 years. Recent studies have indicated that the metastatic, aggressive behavior of IBC may be mediated by the stem cell phenotype. By using the Mary-X model of IBC, Barsky’s group showed that tumor spheroids expressed a CSC profile characterized by CD44+CD24low, ALDH1, and CD133, consistent with Mary-X spheroids having characteristics of CSCs [14]. Most recently, the Mary-X IBC model was shown to have a significant ALDH+ subpopulation which exhibited a stem cell-like phenotype including secondary spheroid formation, high tumorigenicity and self-renewal [15]. In other studies, the IBC cell line SUM149 was documented to express the putative stem cell surface markers CD44+CD24low and ALDH1 [5, 16]. Consistent with previous studies, Charafe-Jauffret et al. showed recently that cells from SUM149 and Mary X possess a subpopulation of ALDH1+ cells that mediate invasiveness and metastasis and, that the expression of ALDH1 protein in tissue sections from patients with IBC was an independent prognostic marker to predict metastasis and poor patient outcome [17]. Altogether, the studies suggest that the angioinvasive and metastatic nature of IBC may be attributed to its stem cell phenotype. Thus, targeting the CSC subpopulation in IBC is of great interest. One approach is to target signaling pathways that are critical for the self-renewal and survival of CSCs, and several candidate pathways have been identified including Notch, Wnt, and Hedgehog (reviewed in [18].
Notch proteins are a family of transmembrane receptors that play an essential role in cell fate decisions such as proliferation, differentiation and apoptosis. Notch receptors are encoded by 4 mammalian genes. Notch receptors are synthesized as a single peptide but an S1 cleavage in the Golgi separates the extracellular and transmembrane domains prior to localization to the plasma membrane. Binding one of the cognate ligands initiates an S2 cleavage by the metalloprotease TACE (tumor necrosis factor alpha-converting enzyme) freeing the extracellular domain and providing a substrate for the γ-secretase complex consisting of presenilins, APH1 and PEN2. The γ-secretase activity creates an S3 cleavage that frees the notch intracellular domain (NICD) from the membrane conferring transcriptional competence [19]. After translocating to the nucleus, NCID binds the transcription complex RBP-Jκ (also known as CSL) via the RBP-Jκ–associated module (RAM) domain displacing repressor proteins and initiating transcription that induces target gene expression including members of the Hes and Hey families of transcription factors [19] Therefore, preventing the S3 cleavage with a gamma-secretase inhibitor (GSI) will retain NICD at the membrane and prevent signaling.
Notch signaling has been shown to play an important role in stem cell self-renewal and differentiation. Dontu et al. showed that Notch signaling was important to maintaining the stem cell phenotype of mammosphere cultures [20]. Specifically, this study demonstrated addition of a Notch-activating peptide caused a 10-fold increase in secondary and tertiary mammosphere formation while inhibiting Notch signaling with blocking antibody or GSI abolished secondary mammosphere formation. Another study demonstrated that CSCs from breast cancer cell lines and patient samples expressed higher levels of Notch4, but not Notch1, and inhibition of Notch4 was more effective in reducing tumor initiation than Notch1 [21]. More recently, Grudzien et al. showed the role of Notch signaling in the maintenance of breast cancer stem-like cells through genomic and pharmacologic inhibition of Notch that reduced the stem-like population of breast cancer cells and prevented mammosphere formation [22]. In a study relating the stem cell phenotype in the IBC model of Mary-X and Notch signaling, Xiao et al. observed that the sorted ALDH1+ subpopulation exhibited enriched Notch3 downstream signaling and co-localization of Notch3 and ALDH1 were demonstrated within the lymphovascular emboli of Mary-X [15]. Inhibiting Notch3 activation in vitro with GSIs or small interfering RNA resulted in a downregulation of Notch target genes and induction of apoptosis [15]. The Notch pathway has also been linked to radiation resistance in breast cancer stem cells. Phillips and colleagues demonstrated the radioresistance of putative breast cancer stem/progenitor cells by comparing the radiosensitivity of cells derived from the CD44+CD24− subpopulation of MCF-7 cell line grown as spheres vs. monolayers and showed the Notch signaling pathway as the mediator of radioresistance [2]. Thus, the Notch pathway is a potential target to improve radiosensitization of CSCs.
Numerous studies have suggested that cell-extrinsic factors from the tumor microenvironment may be critical in controlling tumor progression [23–26]. In particular, polarized immune responses have been shown to promote metastasis [27]. Furthermore, activated immune cells produce inflammatory factors that have been associated with stem cell induction in SUM159 and SUM149 [28]. NF-κB activation by TNF-α in particular has been shown to induce a stem-like state [29]. Notch has been shown to play a role in T-cell polarization of TH2 cells [30–32] that may promote tumor growth and, in contrast, changes the balance in favor of TH1 cells [33] that are involved in the cytolytic response. TH2 cells are capable of producing small amounts of IL-6, a factor associated with stem cell regulation [28]. More importantly, TH2 cells induce IL-6 synthesis by macrophages [34]. Thus, immune cells may play a role in the induction and maintenance of stem-like cells. It has also been suggested that delta-like ligands promote notch3-mediated transcription of T-bet and therefore TH1 polarization, whereas jagged ligands promote notch 1 and 2 activation of Gata3 and the TH2 cytokine IL-4 [35]. As such, the exact role of Notch in T-cell activation remains controversial [36] but the potential for regulating microenvirornmental inflammatory cues is intriguing.
In this study we sought to inhibit Notch signaling using a potent and selective inhibitor of gamma secretase, RO4929097 [37, 38], in an in vitro model that enriches for CSCs (3D clonogenic assay) and conventional 2D clonogenic assay and assess its effect on radiosensitization of the IBC cell lines SUM149 and SUM190. Moreover, since inflammatory signaling has been associated with the promotion and maintenance of CSCs [29], we also evaluated the effect of GSI on inflammatory cytokine production to evaluate a dual tumor inhibitory function for GSI targeting both the tumor cells and the host.
Materials and Methods
Cell culture
For sphere formation, SUM149 and SUM190 cells were placed in ultralow attachment dishes (Corning, NY) in mammosphere media as previously described by Dontu et al. [12]. Briefly, single cells were seeded in 6-well ultralow attachment plates (Corning, NY) in serum free MEM media supplemented with 20 ng/ml bFGF, 20 ng/ml EGF, and B27 (Invitrogen). Adherent cells were grown on standard tissue culture plate in growth media that contains Ham’s F12 with 10% fetal bovine serum (FBS), insulin, hydrocortisone and penicillin-streptomycin.
Aldefluor assay and sorting of ALDH negative/positive population
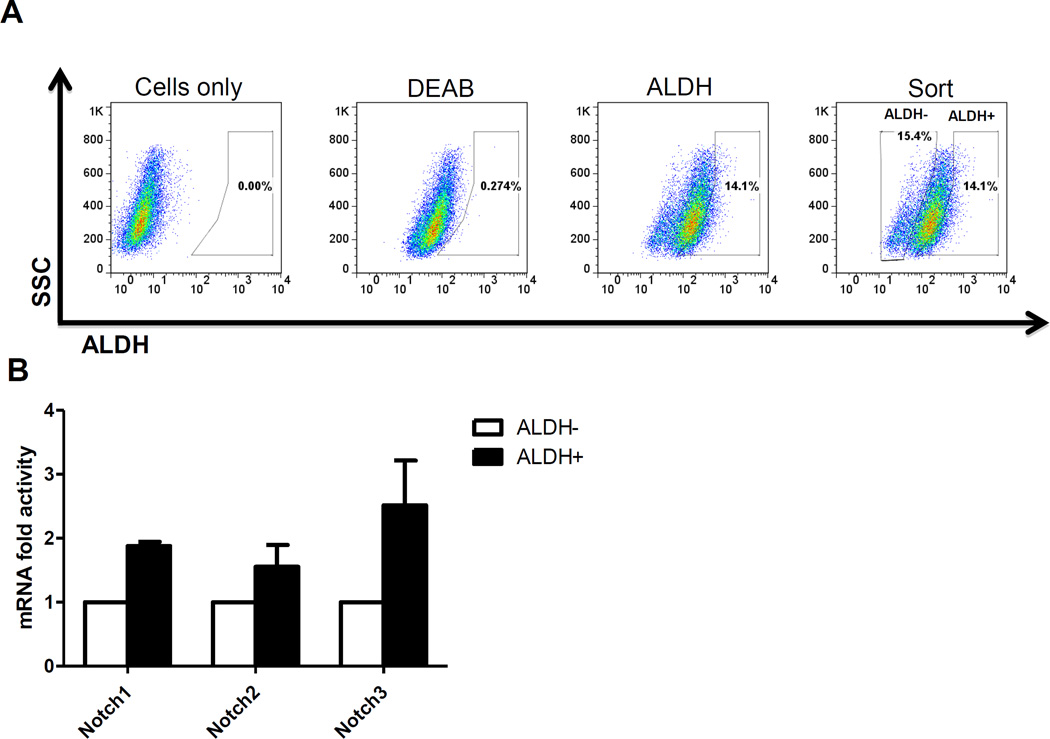
The Aldefluor assay was performed following the manufacturer’s guidelines (StemCell Technologies, Vancouver, Canada). Briefly, about 5 × 105 cells were suspended in Aldefluor assay buffer containing ALDH substrate and incubated for 30 min at 37°C. As a negative control for each sample, a sample of cells was incubated with 50 mmol/L of the specific ALDH inhibitor diethylaminobenzaldehyde (DEAB). Aldefluor fluorescence was excited at 488 nm and fluorescence emission was detected using FACSAria II flow cytometer from Becton Dickinson (BD Biocsiences, San Jose, USA. The data files were analyzed using FlowJo software (Treestar, Ashland, OR). For sorting, gates were established using ALDH-stained cells treated with DEAB as negative controls and taking the high negative and positive cells (Fig. 1A).
Figure 1. Expression of Notch receptors in ALDH+ SUM149 Inflammatory breast cancer cells.
A. Flow cytometry diagram illustrating the gating strategy used to isolate ALDH+ and ALDH− SUM149 cells based on the Aldeflour assay. Gating is set to DEAB-inhibitor control cells. SUM190 cells do not have ALDH+ cells (data not shown). B. ALDH− or ALDH+ (from A) were subjected to RT-PCR for Notch gene expression. Results indicate that ALDH+ SUM149 cells express Notch 1, 2 and 3 at slightly higher levels than ALDH− cells. Notch4 was not detected.
RNA Isolation and RT-PCR
After sorting SUM149 cells into ALDH+ or ALDH− subpopulation, total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's protocol. After treatment with DNase I (Ambion), two micrograms of the RNA samples were reverse-transcribed with random hexamers using Super Script III First-Strand Synthesis System (Invitrogen). Control reactions excluded reverse transcriptase. Quantitative PCR was performed on 7300 Real-Time PCR equipment (Applied Biosystems) with aliquots of the cDNA samples and SYBR Green/ROX qPCR Master Mix (SABiosciences) at annealing temperature of 60°C with the following primers: Notch1, forward 5’-cctgtaacgagggctccaac-3’ and reverse 5’-cagacactggcactcgaagg-3’; Notch2, forward 5’-aggaggggaggagagagtgg-3’ and reverse 5’-tcctgtgccattgtggtagg-3’; Notch3, forward 5’-agccatgctgatgtcaatgc-3’ and reverse 5’-ttggcaaagtggtccaacag-3’; GAPDH, forward 5’-cccactcctccacctttgac-3’ and reverse 5’-tggtggtccaggggtcttac-3’. To assess the effect of RO4929097 on the Notch signaling pathway,, SUM149 and SUM190 cells were seeded on a 10cm plate, left to attach overnight followed by treatment the next day. RNA was extracted using the RNeasy Mini Kit (Qiagen, City, CA) and gene expression analysis performed using SABiosciences RT2 Profiler Notch Signaling Pathway PCR Array (PAHS-059) as per the manufacturer's protocol. PCR was performed on 7300 Real-Time PCR equipment (Applied Biosystems, Foster City, CA). Data was analyzed using an Excel-based data analysis system (SABiosciences) that is based on the ΔΔCt method. Normalization of the raw data was made to one or more of the housekeeping genes in the Array.
Cell Proliferation, Anchorage Independent Growth and Invasion assay. The IBC cell lines SUM149 and SUM190 were seeded at a density of 5 × 104 cells. The next day, they were treated with vehicle or increasing doses of RO4929097, ranging from 0.1 nM to 10 uM. After 72hrs, cells were trypsinized and viable cells counted with a hemocytometer. Anchorage-independent growth was assessed by a soft agar colony assay. Briefly, this assay was performed in six-well plates with a base of 2 mL of medium containing 1% FBS and 0.5% Agarose (Fisher, BP1360-100). About 5 × 104 SUM149 and SUM190 cells were layered onto the base in 2 mL of medium containing 1% FBS and 0.35% agar. The experiments were performed in triplicate and repeated twice. An invasion assay was carried out using a modified Boyden chamber to evaluate the capability of tumor cells to migrate through Matrigel. Briefly, 2.5×104 SUM149 cells were plated onto each BD BioCoat GFR Matrigel invasion chamber insert (24-well insert; pore size, 8 mm; BD Biosciences). The medium in the upper compartment was serum-free RPMI-1640 supplemented with 0.2 µM purified Glu-plasminogen (American Diagnostica, Greenwich, CT), and 5% FBS was used as a chemoattractant in the lower compartment. After incubation for 24 hours in a humidified tissue culture incubator at 37 °C with 5% CO2, the non-invading cells were removed from the upper surface of the membrane by scrubbing. The migrated cells were fixed with 4% paraformaldehyde and stained with crystal-violet. For quantification, the average number of migrating cells was calculated by counting five random fields in the central region of the membrane using a light microscope. Data represent the mean number of migrating cells obtained from triplicate membranes.
Clonogenic/3D mammosphere formation assay
This was conducted as we previously described [39]. Briefly, monolayer cultures of both IBC cell lines were trypsinized into single cells and were seeded into individual wells of a 6-well tissue culture plate (for 2D) or ultralow attachment plates (for 3D, 20,000 cells/ml) in the presence or absence of 1 µM RO4929097. Then the 2D and 3D 6-well plates containing seeded single cells were exposed to increasing doses of irradiation (0, 2, 4, or 6 Gy) 4 hrs after plating. For Figures 4A and 4B, however, SUM149 2D monolayer cells were also pre-treated with 1 µM RO4929097 or vehicle for 24 hours to see if cell contact had an effect. 2D plates were incubated for 14 days and colonies were stained with crystal violet while 3D cells were incubated in mammosphere media for 7 days, the spheres were assessed for proliferation using the MTT assay and those with a size of 50 µM were counted using a Gelcount colony counter (Oxford Optronix, Oxford, UK). For secondary mammosphere assay, cells from primary mammospheres were dispersed with 0.05% trypsin, seeded in 6-well ultra-low attachment plates (10,000 cells/ml) in mammosphere media and counted after a week. Survival curves were generated using Sigmaplot 8.0.
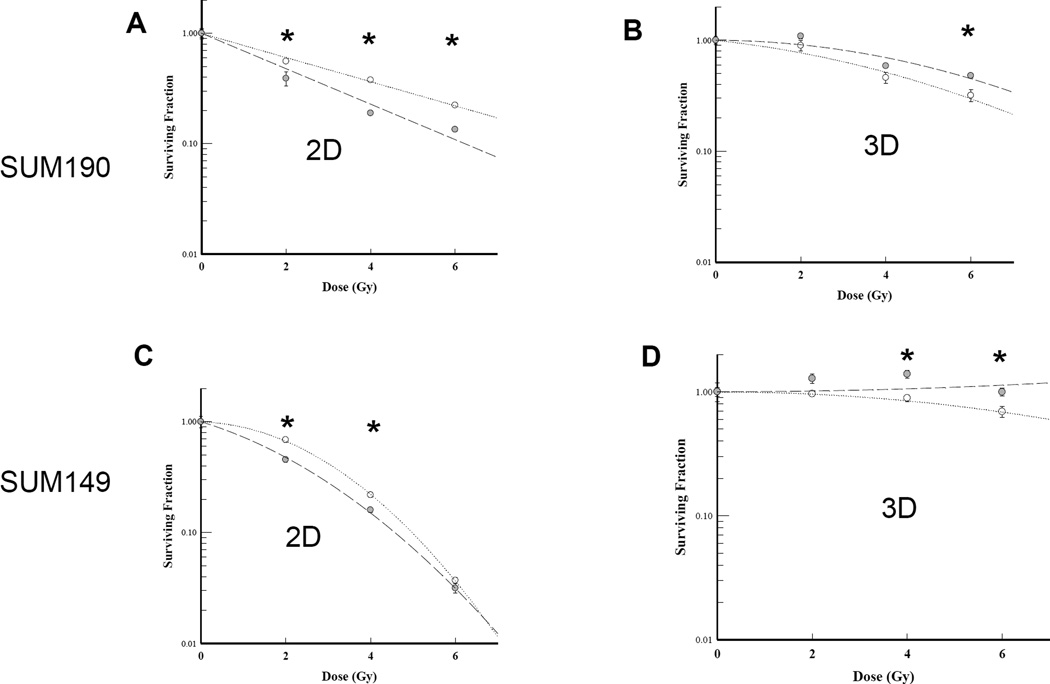
Figure 4. Impact of RO4929097 treatment on radiation response of IBC cell lines.
SUM190 and SUM149 cells were seeded as single cells in adherent (2D) or stem cell enriching culture (3D) conditions and either 1uM RO4929097 or vehicle was added to the media. Four hours after seeding, cells were irradiated with single, increasing doses (0, 2, 4, 6 Gy) of radiation. A. SUM190 monolayer (2D) cells showed radiosensitization relative to the untreated cells. B. Treated 3D SUM190 cells showed no radiosensitization effect compared to untreated cells. C. SUM149 monolayer (2D) cells showed little radiosensitization relative to the untreated cells. D. Treated 3D SUM149 cells showed radioprotection effect compared to untreated cells. Data shown are representative of at least two experiments. Dotted line: No drug control; Dashed line: 1uM RO4929097. * P<0.05.
Immunoblotting
Cell extracts were prepared from 2D monolayer and 3D sphere cultures of SUM149 cells in the presence or absence of 1uM RO4929097 and with or without exposure to 4Gy of radiation based on clonogecic data. The cells were scraped and lysed with cell lysis buffer (Cell Signaling, City, MA), supplemented with protease inhibitor (1mM phenylmethanesulfonyl fluoride, PMSF). Protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA) and 40 µg of each sample were separated using 4–20% Tris-HCl polyacrylamide gel (Invitrogen, City, CA) and transferred onto a PVDF membrane from Bio-Rad. The membranes were blocked in 5% milk powder in TBS-T for ~1 hour and were then incubated with the primary antibodies to β-catenin and survivin (Cell Signal). Membranes were washed three times and incubated with the corresponding secondary antibody conjugated with horseradish peroxidase in 5% nonfat milk at room temperature. After incubation with secondary antibody, the membranes were washed three times and immunoreactivity was detected by enhanced chemiluminescence. Anti-actin (Sigma) antibody was used as a loading control.
Cytokine measurement
The Luminex multiplex bead array was used to measure cytokines in culture supernatants of breast cancer cell lines, as we previously described [40]. Briefly, breast cancer cell lines SUM149, SUM190 and KPL4 were grown on plastic in 2D conditions. The non-IBC cell lines MCF-7, MDA-231, MDA-453, SKBR3, and BT474 were cultured with DMEM supplemented with 10% FBS and penicillin-streptomycin. Cell culture supernatants were collected when the cultures reached 80% confluence, centrifuged at 400×g to remove cellular debris, and then frozen at −80°C until analysis. Cytokine concentrations were measured using Milliplex MAP Human Cytokine/Chemokine Panel as per manufacture protocol (EMD Millipore, Billerica, MA) and analyzed with a Luminex 100 Analyzer running BioPlex 4.2 Software (Bio-Rad).
Effect of GSI RO4929097 on T-cell cytokine synthesis
Peripheral blood was collected by venipuncture from healthy volunteers. Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation over Ficoll-Paque Plus (GE Healthcare, Piscataway, NJ), washed with phosphate buffered saline (PBS) and resuspended at 1×106 per mL in RPMI + 10% FBS) with penicillin-streptomycin (RPMI and antibiotic cocktail from Cellgro by MediaTech, Inc, Manassas, VA; certified FBS from Gibco by Life Technologies, Carlsbad, CA). 1.5 × 106 PBMC were stimulated in six-well plates with 200 µg/mL immobilized anti-CD3 antibody (Beckman Coulter, Brea, CA) plus soluble anti-CD28 antibody (final concentration 1.5 µg/mL, BD Biosciences, San Jose, CA) or left un-stimulated in uncoated wells as previously published [41]. To test the effects of gamma-secretase inhibition, GSI RO4929097 or DMSO as a vehicle control was added as indicated and incubated at 37 °C in 5% CO2 for 36 hours. The Golgi transport inhibitor brefeldin A (Sigma-Aldrich, St. Louis, MO) was added at a final concentration of 10µg/mL the final 4 hours for intracellular cytokine production. Following stimulation, samples were fixed with FACS Lyse (BD Biosciences) and the cell membranes were permeablized with BD FACS Permeabilizing Solution 2. Aliqouts were stained with a cocktail of monoclonal antibodies against surface antigens (CD3, CD4, CD69) and a panel of cytokines: IFN-γ, IL-2, IL-4+IL-13 (TH2), IL-10, IL-17, or TNF-α. Unpermeablized cells were stained for the surface phenotypic markers CD3 and CD4 and the activation markers CCR7, CD25, and CD69. Four-color cytometric evaluation was acquired on a BD FACS Calibur flow cytometer and the data analyzed using FlowJo software (Treestar, Inc, Ashland, OR).
Statistical methods
Survival curves were generated using Sigmaplot 8.0 and t-test was used to compare differences at each dose following normalization to the unirradiated samples. A p-value of <0.05 in a paired two-sided t-test was considered statistically significant.
Results
Expression of Notch receptors in IBC stem cell subpopulation
The stem cell marker ALDH has been previously reported in Mary-X tumor and SUM149 cell line [14, 15, 17]. By utilizing this stem cell marker, we sorted SUM149 cells into ALDH− or ALDH+ (Fig.1A) and assessed the expression of the Notch receptors in the IBC stem cell population using RT-PCR. About 2-fold higher expression of Notch1, Notch2 and Notch3 receptors was found in the ALDH+ subpopulation vs. ALDH− subpopulation (Fig.1B). Notch4 was not detected in either of the population while SUM190 IBC cells have very low (<1%) ALDH+ subpopulation (data not shown).
Gene expression changes of Notch downstream targets by RO429097
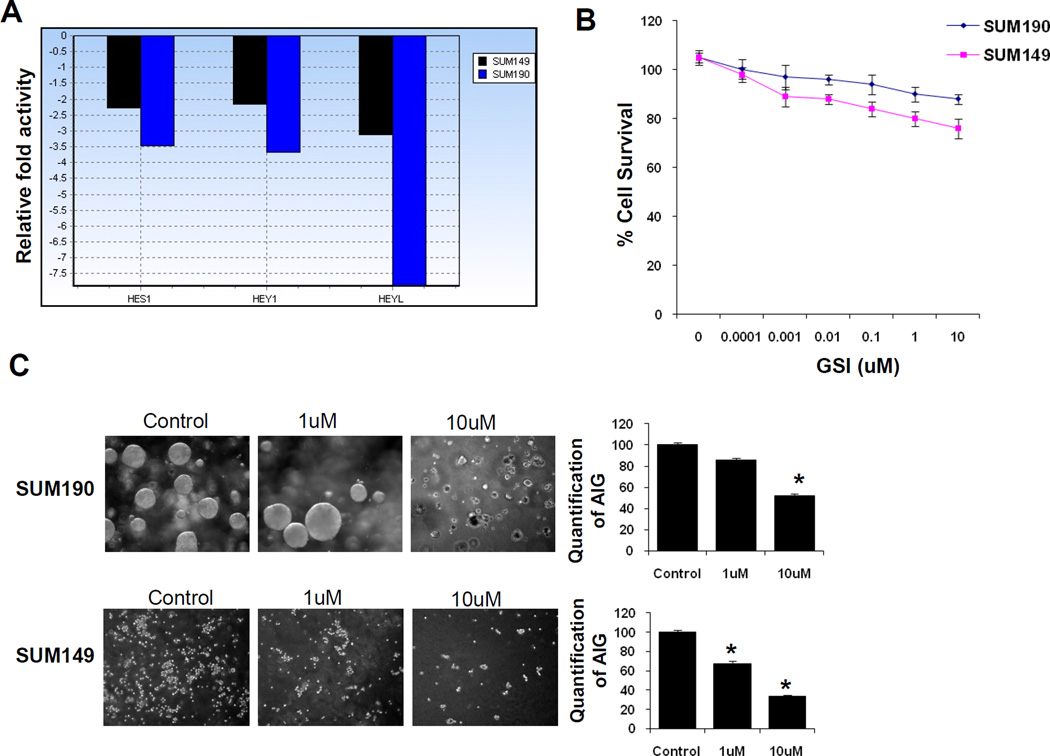
RO429097 is a gamma secretase small molecule inhibitor that prevents the proteolytic step that leads to the generation of the active intracellular domain of Notch, thus suppressing Notch signaling. In a previous study, RO4929097 inhibited Notch processing in human non–small cell lung carcinoma cells as measured by the reduction of intracellular Notch expression by Western blot and reduced expression of the Notch transcriptional target gene Hes1 [37]. To confirm whether RO429097 targets Notch downstream genes in IBC cells, gene expression analysis was performed using Notch Signaling Pathway PCR Array in both SUM149 and SUM190 cell lines in 2D cultures treated with RO4929097 and compared with vehicle-treated controls. Treatment with the inhibitor showed a 2- to 3-fold reduction in the expression of direct Notch target genes, Hes1, Hey1 and Heyl in SUM149 and a 3.5- to 8-fold decrease in expression in SUM190 cells (Fig. 2A).
Figure 2. Effect of RO4929097 on Notch downstream targets and growth characteristics of inflammatory breast cancer cells.
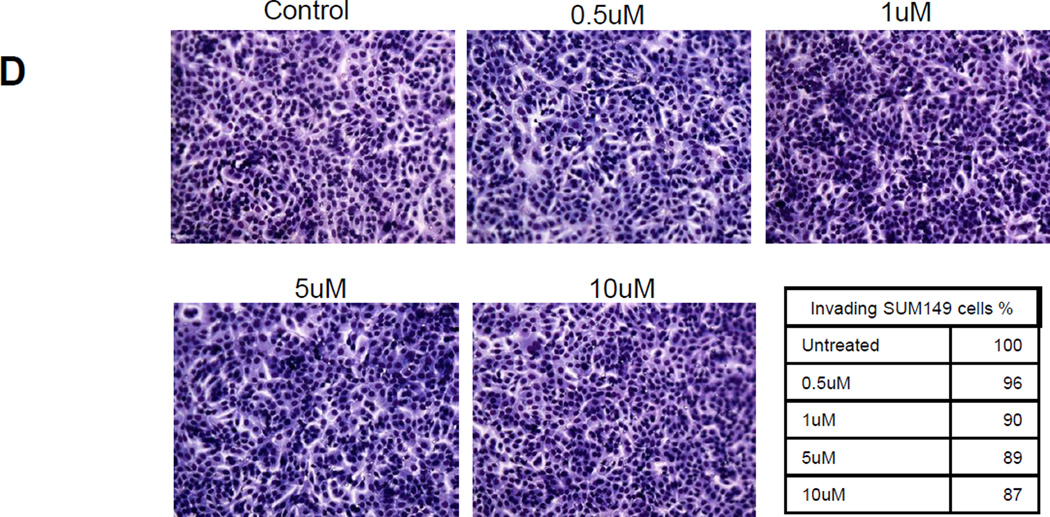
A. Real time PCR analysis of SUM149 and SUM190 cells treated with RO4929097 on the expression of HES1, HEY1 and HEYL in SUM149 and Sum190 cells. B. Proliferation assay with increasing doses of RO4929097 in SUM149 and SUM190 cells. C. Anchorage independent growth in SUM149 and SUM190 inflammatory breast cancer cell lines using RO4929097. Quantitation for each cell line was shown with the bar graphs and significant reduction was observed in both cell lines (P<0.05). D. Invasion assay in SUM149 at increasing doses of RO4929097.
RO429097 on growth characteristics of inflammatory breast cancer cell lines
We assessed the efficacy of RO4929097 on cell proliferation and anchorage independent growth in both IBC cell lines and invasion assay in SUM149 cells. For the proliferation assay, cells were treated with vehicle or increasing concentrations of the drug for 72h before the number of viable cells was counted. Each of these cell lines showed modest growth inhibition with RO4929097 in a dose dependent manner. At a concentration of 1 µM of RO4929097, growth inhibition was 20% for SUM149 and 10% for SUM190 cells, relative to vehicle-treated controls (Fig 2B). Anchorage-independent colony formation in soft agar has been used as a surrogate to assess tumorigenicity of cancer cells. As shown in Fig. 2C, a decrease in colony formation by SUM190 and SUM149 cells treated with RO4929097 was nearly 50% and 75%, respectively at 10 µM (P<0.05). The invasiveness of SUM149 cells in the presence of increasing doses of RO4929097 showed that the inhibitor does not have a marked effect in invasiveness of these cells (Fig. 2D).
RO4929097 on 2D colony and 3D mammosphere formation in IBC cell lines
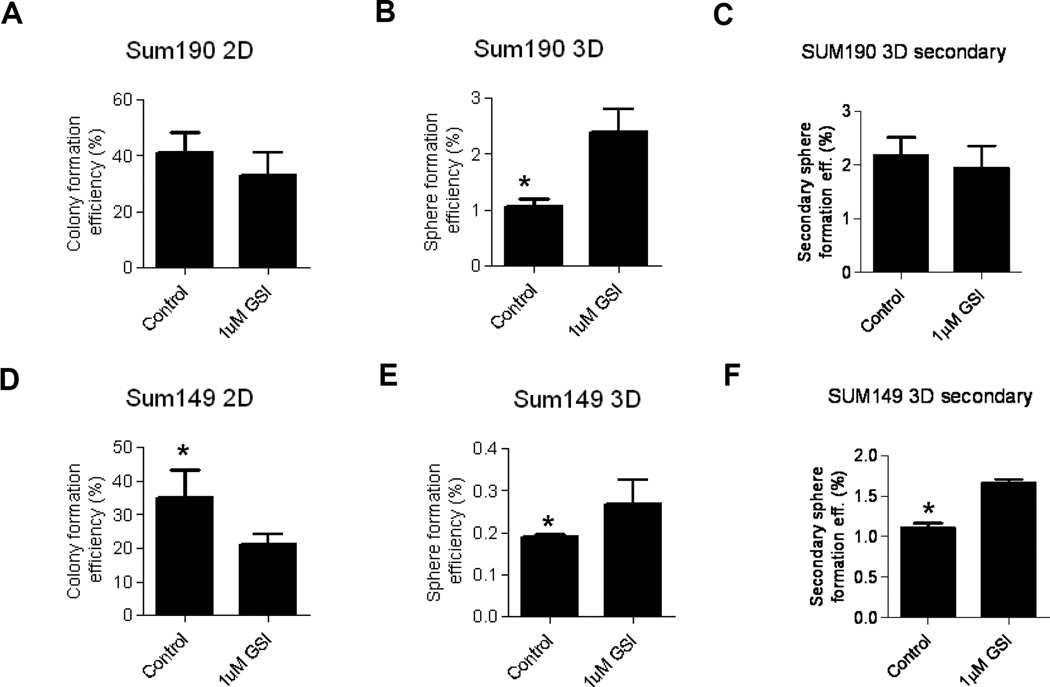
We next studied the ability of these IBC cell lines to form colonies when plated at clonal density on 6-well cell culture plates in the presence or absence of 1µM RO4929097 for 2 weeks. RO4929097 significantly reduced colony formation by both cell lines (P<0.05; Fig. 3A, 3C) with the effect being more notable in SUM149 than by SUM190 cells. CSCs are thought to possess the ability to self renew similar to normal stem cells. The generation of mammospheres from single cells cultured under non-adherent conditions in growth factor-supplemented media is a widely used approach to enrich cancer stem/progenitor cells from cell lines and primary tumor samples. To evaluate whether RO4929097 has an effect on the formation of mammospheres in vitro, we exposed single cells from SUM149 and SUM190 cell lines to a vehicle or 1 µM RO4929097 for a week and counted the resultant spheres. As shown in Fig. 3B and 3D, the inhibitor increased sphere formation significantly in both cell lines (P<0.05). We also examined secondary mammosphere formation in order to assess self-renewal ability upon passaging. A significant increase in secondary mammosphere forwation was observed in SUM149 (P<0.05) with RO4929097 treatment while no significant difference was observed in SUM190 cells.
Figure 3. Effect of RO4929097 on colony and sphere formation in IBC cell lines.
SUM190 and SUM149 cells were seeded as single cells in adherent (2D) or stem cell enriching culture (3D) conditions and either 1uM RO4929097 or vehicle was added to the media. 2D Colonies were counted 14 days later while 3D spheres were counted 7 days after seeding. A. SUM190 2D colony formation. B. SUM190 3D sphere formation assay. C. SUM190 secondary sphere formation. D. SUM149 2D colony formation. E. SUM149 3D sphere formation assay. F. SUM149 secondary sphere formation. * P<0.05.
RO4929097 treatment on radiation response of IBC cell lines
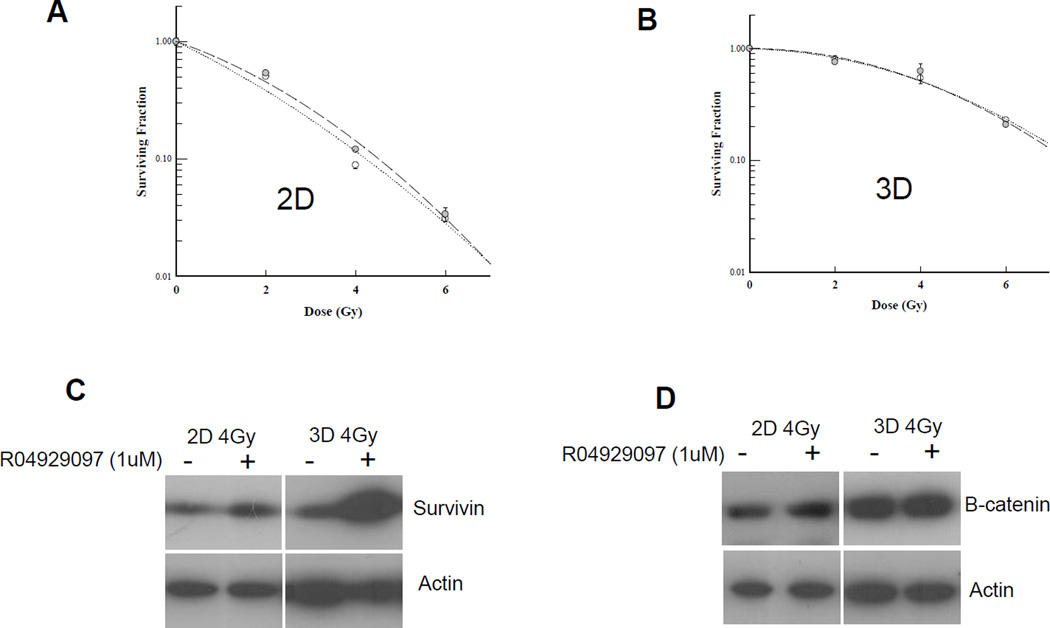
We then examined the effect of RO4929097 on sensitivity to radiation of SUM149 and SUM190 cells grown to form 2D colonies or 3D mammospheres. After treating with increasing doses of ionizing radiation in the presence or absence of the drug, 2D colonies were allowed to grow for 10–14 days while the mammospheres were permitted to grow for 1 week. At 1 µM, RO4929097 was able to sensitize adherent cells to radiation with a more significant effect seen in SUM190 than in SUM149 cells (Fig. 4A, 4C). However, the same dose of inhibitor radioprotected cells grown under conditions that favor the enrichment of the cancer stem cells at higher doses of ionizing radiation (Fig. 4B, 4D). This discrepancy between 2D and 3D cultures suggested that cell contact may be needed for a notch inhibitor to have a significant effect. To test this, we treated confluent SUM149 cells in monolayer with 1 µM RO4929097 or vehicle for 24 hrs. After 24hrs, cells were seeded as single cells in a 2D or stem cell enriching 3D conditions containing 1 µM RO4929097 or vehicle. Four hours after seeding, cells were irradiated with single, increasing doses of radiation. We did not detect radiosensitization in either culture condition (Fig. 5A, 5B). Similarly, using MCF7 spheres adding the notch inhibitor to the spheres and treating them with increasing doses of radiation was tested but did not increase radiosensitivity (data not shown). In an attempt to understand whether redundant stem cell survival pathways and downstream effectors may play a role in the radioprotection effect seen in 3D stem cell enriching cultures, protein lysates were prepared from 2D monolayer and 3D sphere cultures of SUM149 cells with and without 1 µM RO4929097 and 4Gy ionizing radiation. We found that β-catenin and survivin were upregulated in 3D culture vs. monolayer cells (Fig. 5C, 5D). Moreover, treatment with 1 µM RO4929097 significantly upregulated survivin in 3D irradiated cells compared to untreated irradiated controls (Fig. 5C).
Figure 5. RO4929097pre-treatment of monolayer Sum149 IBC cell line.
SUM149 cells were either pre-treated with 1uM RO4929097 or with vehicle for 24hrs. After 24hrs, cells were trypsinized, seeded as single cells in adherent (2D) or stem cell enriching culture (3D) conditions and either 1uM RO4929097 or vehicle was added to the media. Four hours after seeding, cells were irradiated with single, increasing doses (0, 2, 4, 6 Gy) of radiation. A. 2D clonogenic assay showed no radiosensitization effect relative to the untreated cells. B. Treated 3D SUM149 cells showed radioprotection effect compared to untreated cells. Dotted line: vehicle control; Dashed line: 1uM RO4929097. C, D. Western blot analysis of SUM149 cells irradiated with 4Gy radiation and treated with 1uM RO4929097 or vehicle. Anti-survivin, beta-catenin and actin antibodies were used.
High expression of IL-6 and IL-8 in IBC Cell Lines
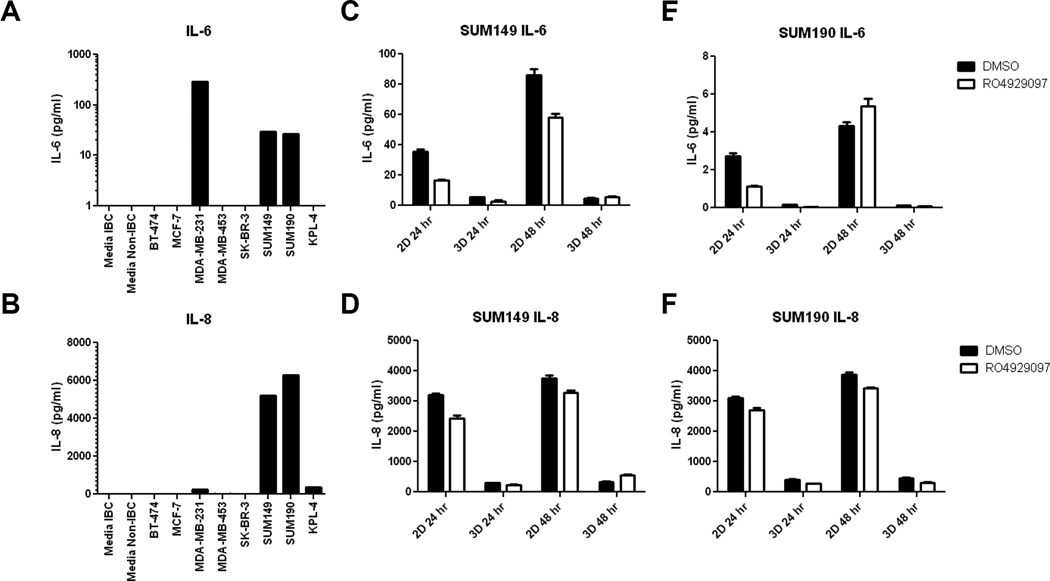
As the inflammatory cytokines IL-6 and IL-8 were recently suggested to reduce the effectiveness of RO4929097 in vivo [38], we tested the secretion of these factors by the IBC cell lines SUM149 and SUM190 vis-à-vis other non-inflammatory breast cancer lines. SUM149 and SUM190 cells were the only tested breast cancer cell lines that secreted detectable levels of both of these cytokines (IL-8 >> IL-6, Fig. 6A, 6B). We next tested the effect of RO4929097 on cytokine secretion by SUM149 and SUM190 cells in 2D and 3D. Both SUM149 and SUM190 secreted IL-6 and IL-8 in 2D culture, which persisted after treatment with RO4929097. Supernatants from cells grown in 3D had lower, but still detectable, concentrations of IL-6 and IL-8 compared to 2D cultures and were minimally decreased with RO4929097 treatment.
Figure 6. IBC Cell lines secrete high levels of IL-6 and IL-8.
A – B Cell line supernatants were collected from 80% confluent untreated cultures and assayed for soluble IL-6 and IL-8 using Luminex bead assays. A. IL-6 was detected in untreated cultures from MDA-MB-231, SUM-149 and SUM-190. B IL-8 was detected in cultures from MDA-231, SUM149, SUM190, and KPL4 cells. C–F SUM 149 and SUM-190 cells were cultured with DMSO vehicle or RO4929097 for 24 and 48 hours. Treated and untreated cells secreted similar levels of IL-6 in SUM-149 cultures (C) and SUM190 cultures (E), however 2D cultures produced more IL-6 that 3D cultures. D SUM149 cells produced similar IL-8 with RO4929097 treatment in both 2D and 3D culture. F. RO4929097 treatment had minimal effect on IL-8 production by SUM159 cells.
Effect of RO4929097 on cytokine synthesis and T-cell activation
As inflammatory cytokine signaling through IL-6 and IL-8 can be mediated in vivo by adaptive immune responses, we tested the effect of RO4929097 on cytokine production by TCR-activated T cells. To test the anti-inflammatory effects of RO4929097 on T cells, we measured intracellular cytokine production by normal PBMC following stimulation through the TCR in the presence of RO4929097. Fresh PBMC from healthy volunteers were stimulated with plate-bound anti-CD3 and soluble anti-CD28 antibodies for 36 hours in the presence of 0.1nM – 10µM RO4929097. Flow cytometry was used to quantify the percentage of T cells producing cytokines. T-cells are not a major source of IL-6 [42] or IL-8 [43], but can influence the production of these cytokines by other cells. In particular, monocytes have a strong IL-6 response to TNF-α and epithelial, endothelial and fibroblast cells often secrete IL-6 in response to IL-17. Therefore, we focused on the canonical T-cell cytokines. IL-2, IFN-γ, and TNF-α were measured to characterize a TH1 polarized response; IL-4, IL-13 and IL-10 for TH2 and IL-17 for TH17. In a serial dilution of GSI around the expected therapeutic range from 0.1 nM to 70 nm, there were no significant changes in the percentage of CD4 or CD8 T-cells producing IFN-γ, IL-2, TNF-α, IL-4/IL-13, IL-10, or IL-17. However, there was a trend for a decreasing percentage of CD8+ T cells producing IFN-γ, IL-2 and TNF-α, CD4 cells producing IFN-γ and IL-2, and a weak trend in decreasing production of the TH2 cytokines IL-4 and IL-13 but neither IL-10 nor IL-17 (data not shown).
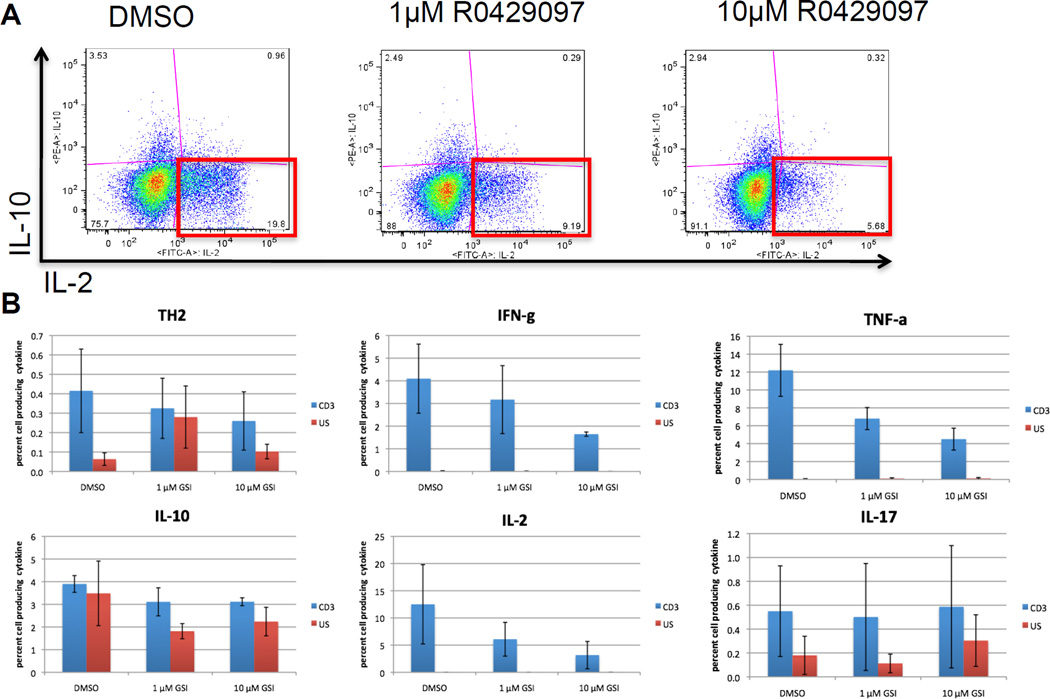
To test the effect of high-dose RO4929097 on T-cell cytokine production, activated PBMC from two healthy donors were treated 1 µM RO4929097, 10 µM RO4929097 or DMSO vehicle control. There was a reproducable trend in towards a decrease in the percentage of CD3 T cells that synthesized IFN-γ, IL-2 and most notebaly TNF-α. After treating with DMSO vehicle, an average of 12.2% ± 2.91% (mean ± s.e.m.) of TCR-activated T cells produced TNF-α compared with 6.8% ± 1.23% treated with 1µM GSI and 4.5% ± 1.2% treated with 10µM RO4929097. Although the percentages of cells synthezing TH2 cytokines were low, a decreasing trend was observed in the percentage of CD3 cells that synthesized the TH2 cytokines IL-4 + IL-13 (Fig. 7). The percentage of cells that syntheized IL-10 or IL-17 did not seem to be effected by RO4929097 treatement. These data suggest that treatment with RO4929097 can reduce the production of inflammatory cytokines by T-cells. Furthermore, with RO4929097 treatment, there was a shift in favor of TH2 over TH1 cytokines. As IL-6 is promoted by TH2 conditions, this would suggest that T-cell activtion induced IL-6 produciton would be increased with RO4929097.
Figure 7. Effect of RO4929097 on cytokine synthesis by TCR stimulated T cells.
Fresh healthy donor PBMC were left unstimulated or activated overnight through the TCR with plate-bound anti-CD3 plus soluble anti-CD28 in the in the presence of 1µm or 10µm R0429097 or DMSO vehicle control. BFA was added the final three hours. Intracellular cytokine synthesis was measured by flow cytometry analysis. Replicates are from different donors. A. A representative histogram showing the percentage of CD3 gated cells that produced IL-2, IL-10 or both. B. Percentage of anti-CD3 stimulated cells (blue) and unstimulated cells (red) that produced TH2 cytokines (IL-4+IL-13), IL-10, IFN-γ, IL-2, TNF-α, or IL-17. There was a significant decrease in the percentage of CD3 cells producing TNF-α.
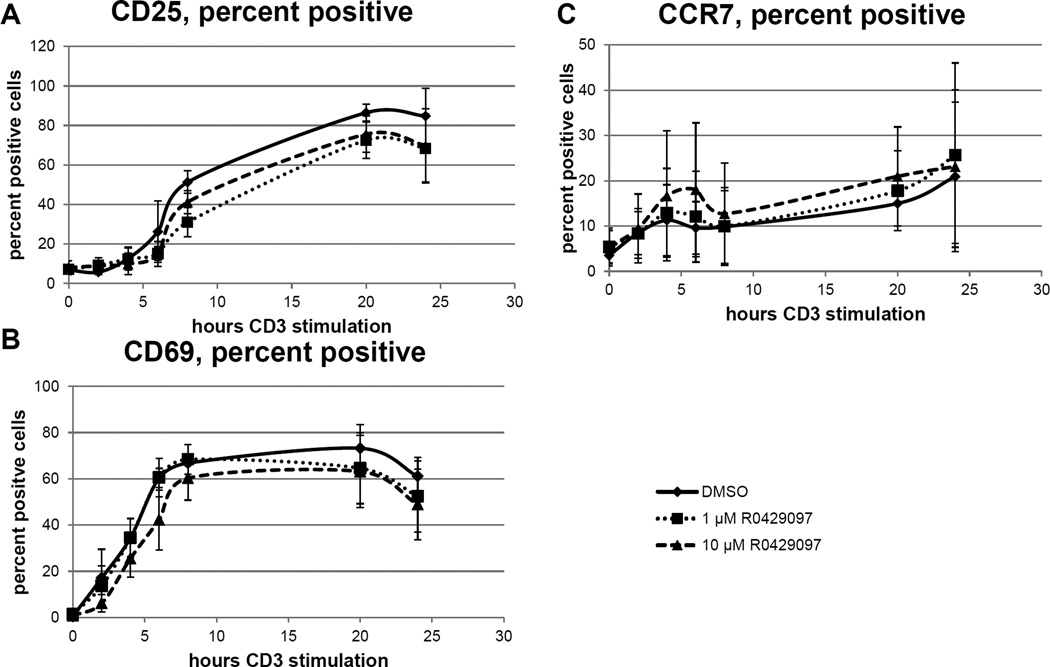
Next, to test if the reduction in inflammatory cytokine production was related to the activation of T-cells, we quanitfied the expression of the surface activation markers CD25 and CD69 on T cells stimulated through the TCR in the presence of RO4929097. Additionaly, we quantified CCR7 expression, which has been shown to be directly regulated by Notch signaling [44]. Following 24 hours of stimulation, there were no significant differences in the percentage of TCR-activated T cells expressing CD25, CD69 or CCR7. However, there was a reproducible decrease in the percent of TCR-activated T cells that expressed CD25 and CD69 after treatment with either 1 µM or 10 µM RO4929097 suggesting that RO4929097 infereres with T-cell activation. Interestingly, the percentage of TCR-activated T cells expressing the chemokine receptor CCR7 was slightly increased following exposure to RO4929097 (Fig. 8). These data suggest that RO4929097 inhibits T-cell activation, but paradoxically increases expression of CCR7 even though both T-cell activation and expression of CCR7 are under direct transctiptional control by Notch and normally up-regulated in activated T-cells.
Figure 8. Effect of RO4929097 on T cell activation.
Fresh healthy donor PBMC were activated through the TCR with plate-bound anti-CD3 plus soluble anti-CD28 in the in the presence of 1µm or 10µm R0429097 or DMSO vehicle control. The percent of CD3 gated T cells expression the surface marker expression of CD25, CD69 and CCR7 were measured by flow cytometry analysis following 0, 2, 4, 6, 8, 20, or 24 hours of activation. All time points were analyzed concurrently. Replicates are from 3 different donors, error bars are S.E.M. A. CD69 and B. CD25 expression is slightly, but significantly inhibited by GSI, while CCR7 expression was slightly enhanced (C).
Discussion
Numerous lines of evidence suggest targeting notch signaling may specifically target breast cancer stem cells. RO4929097 is a potent and selective gamma secretase small-molecule inhibitor that targets cellular Notch processing and was reported to decrease NICD production and mRNA expression of Hes1 as well as to have anti-tumor activity in human lung cancer cells in a preclinical evaluation of the drug [37]. In the present study we similarly found a reduction in the expression levels of Notch downstream target genes Hes1, Hey1 and HeyL in IBC cell lines. However, selective killing of putative cancer stem cell populations with drug alone or in combination with radiation was not observed. We demonstrate that this could be due to a redundancy and cross-talk in alternative pathways known to be involved in stem-cell self renewal. We also demonstrated that IL-6 and IL-8 may further limit the efficacy of RO4929097 as a single agent in this IBC model system. Further, we demonstrated that RO4929097 inhibits production of the IL-8 mediator TNF-α from T cells, suggesting that testing with models that permit rigorous interrogation of microenvironmental influences on tumor efficacy may prove critical to fully evaluating this targeting strategy.
We and others have previously reported the relative radioresistance of SUM149 cells [45, 46]. Under adherent culture conditions, these cells were radiosensitied by a combination treatment of radiation and ErbB inhibitor [45]. In this study, we explored the effect of RO4929097 on radiation sensitivity of SUM149 and SUM190 grown under culture conditions that promote 3D colony stem cell growth or standard 2D non-stem cell monolayer growth. We found that RO4929097 sensitized SUM190 cells to ionizing radiation in 2D culture while it radioprotected both SUM149 and SUM190 cells in 3D culture. Moreover, while the inhibitor reduced 2D colony formation, it increased mammosphere formation in both cell lines. The increased stem cell activity observed in notch inhibitor-treated cells may be due to redundant self-renewal pathways and we have shown that β-catenin and its downstream effector survivin are upregulated in Notch inhibitor-treated stem cells after radiation (Fig. 4C/D). In support of our data, Bouras et al. [47] have shown that inhibition of the Notch pathway in mammary stem cells enhances their stem cell potential. Two other studies have also demonstrated that Notch negatively regulates β-catenin protein levels in stem/progenitor cells [48, 49]. In these studies, an increase in TCF/β-catenin-dependent luciferase activity was observed in Notch siRNA-treated neural stem cells as well as in mouse cardiac progenitor cells lacking Notch1 in vivo and in vitro, indicating that Notch1 may function to negatively regulate β-catenin levels in stem-cell populations [48, 49]. Thus, combination strategies with additional targeting agents should be explored.
Another explanation for the increase in mammosphere formation and radioprotection effect seen with RO4929097 treatment in our study could be due to the high expression and/or induction of the IL-6 and IL-8 cytokines. In a previous study with RO4929097, the IL-6 receptor was upregulated in a microarray expression data in preclinical studies of human non–small cell lung carcinoma cells [37] and recently the same group reported that high tumor levels of IL-6 and IL-8 abrogated the preclinical efficacy of RO4929097 [38]. Consistent with this observation, we found high levels of IL-6 and IL-8 produced in culture supernatants of both IBC cell lines used in this study compared to those of non-IBC cells (Fig. 6). Sansone et al showed that treatment with IL-6 triggered Notch3-dependent upregulation of the Notch ligand JAG1 and promoted primary human mammospheres and MCF-7-derived mammosphere growth [50]. Thus, IL-6 supports cancer stem cell renewal in breast cancer mammospheres via Notch3 [50] and the increase in the IL-6 receptor following RO4929097 treatment may represent a regulatory compensation for the loss of Notch signaling. Barsky’s group has also recently reported the enrichment of Notch3 in the CSC-like subpopulation and its co-localization with ALDH1 within the lymphovascular emboli of Mary-X [15]. IL-6 also enhanced the proliferation of colon CSCs and lung CSCs [51, 52]. Moreover, there is evidence that increased IL-6/IL-8 signaling is associated with radioresistance in Stat1-overexpressing tumor cells and treatment with neutralizing antibodies to both cytokines is associated with activation of apoptotic cell death and sensitization to radiation [53]. Furthermore, another interesting recent finding demonstrated that IL-6 can reprogram differentiated cells to the cancer stem cell-like phenotype in breast and prostate cell lines and in cells derived from human breast tumors [54]. Thus, the failure of the Notch inhibitor RO4929097 to radiosensitize and inhibit mammosphere formation in IBC cell lines may be attributed to increased IL-6/IL-8 signaling in these cell lines which promoted CSC features. Future experiments should test the effect of tumor derived IL-6/IL-8 on mammosphere formation.
Charafe-Jauffret et al showed that IL-8 could increase tumorsphere formation and Aldefluor positivity in the metaplastic SUM159 cell line [5, 16]. However, a concentration of 100 ng/ml was required for a significant increase in stem cell activity. In contrast, although IL-8 was detected in the supernatants of SUM159 cultures/3D mammospheres in the current study, the levels were about 20 times lower than the exogenous concentrations required for increased sphere formation. We also observed lower concentrations of both IL-6 and IL-8 in 3D cell culture, but these cultures contained fewer viable cells. However, the biologic activity of endogenous paracrines may be observed at lower concentrations than that of exogenous factors since it is released in close proximity to the receptor. Furthermore, the high level of cell aggregates in 3D culture might produce locally high concentrations of IL-6 and IL-8. Therefore, the lower levels of IL-6 and IL-8 observed in 3D culture supernatants may not reflect the optimal biologic activity of these factors.
Cytokine profiling from the serum of IBC patients has shown increased levels of IL-6 and IL-8 [55]. As signaling through IL-6 and IL-8 can be mediated in vivo by adaptive immune responses, we tested the effect of RO4929097 on cytokine production by TCR-activated T cells. This would suggest greater efficacy in vivo could be expected if the effect of RO4929097 on T cells might reduce the antagonistic effects of IL-6 and IL-8. We found that RO4929097 can reduce the production of inflammatory cytokines by activated T cells, particularly TNF-α. Although T cells contribute only a minor portion of the TNF-α within a tissue or in circulation, in an inflammatory environment, innate immune cells can potentially produce the majority of inflammatory cytokines. In IBC, Van Laere showed that overexpression of NF-κB is a hallmark of the disease [56, 57]. One of the most potent activators of NF-κB is TNF-α. In particular, TNF-α (and IL-1β) can elicit expression of IL-8 [58, 59].
T cells are responsible for coordinating the antigen-specific inflammatory response by producing pro-inflammatory TH1 cytokines such as IFN-γ, IL-2 and TNF-α or anti-inflammatory TH2 cytokines such as IL-4, IL-5, IL-10, and IL-13. Both CD8 T-cytotoxic cells and CD4 T-helper cells can produce TNF-α following activation through the TCR [60]. Notch has long been known as essential in T-cell development; however, more recent work has shown that Notch signaling may play a critical role in both T-cell activation and TH differentiation [61]. There are putative Notch/ RBP-J binding sites in the promoter of the TH2 transcription factor Gata3 [62] and the TH1 transcription factor T-bet [63]. Therefore, we hypothesized that in vivo, inhibition of Notch signaling with the gamma-secretase inhibitor RO4929097 could decrease the production of inflammatory cytokines by T cells that promote breast cancer stem cell maintenace. Indeed, our in vitro results confirm inhibition of T-cell activiation by RO4929097, specifically of TNF-α which may increase the enthusiasm for further in vivo testing in immunocompetant animal models and further underscores the need for an an immune competant animal model for IBC.
It has yet to be determined whether RO4929097 inhibits all four Notch family members or not. Differences in the expression levels of these Notch members in breast CSCs have been observed in different studies. For example, Harrison et al [21] reported that sphere-derived stem cells and CD44+CD24− cells from breast cancer cell lines and patient samples possessed elevated expression of Notch4, but not Notch-1. They found Notch-4 inhibition was more effective in reducing tumor initiation than targeting Notch-1 suggesting breast CSCs rely preferentially on Notch4 signaling [21]. In another study, Xiao et al reported that ALDH1+ subpopulation exhibited enriched Notch3 downstream signaling and co-localization of Notch3 and ALDH1 were demonstrated within the lymphovascular emboli of Mary-X [15]. Detailed understanding of the role of Notch signaling in breast CSC survival and self-renewal depends on studying each receptor and ligand and their specific interactions and the relative roles of specific Notch members in individual cancers.
In summary, the Notch inhibitor RO4929097 did not inhibit putative cancer stem cells alone or with ionizing radiation in the inflammatory breast cancer cell lines tested. This may be due to redundant self-renewal pathways or secretion of self-renewal promoting cytokines either intrinsically or extrinsically. However, the in vivo effects may mediate be sensitive to microenvironment alternations such as the decrease in inflammatory factors by immune cells shown here. Careful assessment of multiple target strategies and rigorous modeling of the microenvironmental influences may shed further light on the potential of targeting notch using RO4929097.
Acknowledgements
The small molecule inhibitor RO4929097 was kindly provided by Dr. John Boylan of Roche Pharmaceuticals. This work was supported by grants from the National Institute of Health R01CA138239-01; The State of Texas Grant for Rare and Aggressive Cancers; The University of Texas MD Anderson Cancer Center Institutional Research Grant; The University of Texas Health Sciences Center KL2 RR024149 and Susan G. Komen Breast Cancer Foundation grant KG081287; Assessment of Circulating Breast Cancer Stem Cells To Predict Recurrent Disease, W81XWH-09-1-0031 01, DOD.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Li F, Tiede B, Massague J, Kang Y. Beyond tumorigenesis: cancer stem cells in metastasis. Cell Res. 2007;17:3–14. doi: 10.1038/sj.cr.7310118. [DOI] [PubMed] [Google Scholar]
- 2.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 3.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895-1886. [DOI] [PubMed] [Google Scholar]
- 4.Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 7.Balic M, Lin H, Young L, Hawes D, Giuliano A, McNamara G, Datar RH, Cote RJ. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12:5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 8.Reuben JM, Lee BN, Gao H, Cohen EN, Mego M, Giordano A, Wang X, Lodhi A, Krishnamurthy S, Hortobagyi GN, et al. Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44(+)CD24(lo) cancer stem cell phenotype. Eur J Cancer. 2011 doi: 10.1016/j.ejca.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodward WA, Gao H, Cohen EN, Li L, Xu W, Debeb BG, Jimenez CA, Krishnamurthy S, Tucker SL, Hortobagyi G, Cristofanilli M, Buchholz T, Reuben J. Percentage of CD45−CD326+ CD44+CD24−/lo cells in pleural effusion fluid of patients with metastatic breast cancer predicts for overall survival. Interventional Oncology Society Journal. 2011;1 [Google Scholar]
- 10.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, Kamrudin S, Krishnamurthy S, Le-Petross H, Bidaut L, Player AN, et al. Inflammatory breast cancer: the disease, the biology, the treatment. CA Cancer J Clin. 2010;60:351–375. doi: 10.3322/caac.20082. [DOI] [PubMed] [Google Scholar]
- 14.Xiao Y, Ye Y, Yearsley K, Jones S, Barsky SH. The lymphovascular embolus of inflammatory breast cancer expresses a stem cell-like phenotype. Am J Pathol. 2008;173:561–574. doi: 10.2353/ajpath.2008.071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y, Ye Y, Zou X, Jones S, Yearsley K, Shetuni B, Tellez J, Barsky SH. The lymphovascular embolus of inflammatory breast cancer exhibits a Notch 3 addiction. Oncogene. 2011;30:287–300. doi: 10.1038/onc.2010.405. [DOI] [PubMed] [Google Scholar]
- 16.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakarala M, Wicha MS. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol. 2008;26:2813–2820. doi: 10.1200/JCO.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radtke F, Schweisguth F, Pear W. The Notch 'gospel'. EMBO Rep. 2005;6:1120–1125. doi: 10.1038/sj.embor.7400585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grudzien P, Lo S, Albain KS, Robinson P, Rajan P, Strack PR, Golde TE, Miele L, Foreman KE. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res. 2010;30:3853–3867. [PubMed] [Google Scholar]
- 23.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. The Journal of clinical investigation. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel SA, Dave MA, Murthy RG, Helmy KY, Rameshwar P. Metastatic breast cancer cells in the bone marrow microenvironment: novel insights into oncoprotection. Oncology reviews. 2011;5:93–102. doi: 10.1007/s12156-010-0071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiao SL, Ganesan AP, Rugo HS, Coussens LM. Immune microenvironments in solid tumors: new targets for therapy. Genes & development. 2011;25:2559–2572. doi: 10.1101/gad.169029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guise T. Examining the metastatic niche: targeting the microenvironment. Seminars in oncology. 2010;37(Suppl 2):S2–S14. doi: 10.1053/j.seminoncol.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Johansson M, Denardo DG, Coussens LM. Polarized immune responses differentially regulate cancer development. Immunological reviews. 2008;222:145–154. doi: 10.1111/j.1600-065X.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, Korkaya H, Heath A, Dutcher J, Kleer CG, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer research. 2011;71:614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storci G, Sansone P, Mari S, D'Uva G, Tavolari S, Guarnieri T, Taffurelli M, Ceccarelli C, Santini D, Chieco P, et al. TNFalpha up-regulates SLUG via the NF-kappaB/HIF1alpha axis, which imparts breast cancer cells with a stem cell-like phenotype. J Cell Physiol. 2010;225:682–691. doi: 10.1002/jcp.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J, Krawczyk CJ, Pearce EJ. Suppression of Th2 cell development by Notch ligands Delta1 and Delta4. Journal of immunology. 2008;180:1655–1661. doi: 10.4049/jimmunol.180.3.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubo M. Notch: filling a hole in T helper 2 cell differentiation. Immunity. 2007;27:3–5. doi: 10.1016/j.immuni.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 33.Pei J, Tang Z, Zang G, Yu Y. Blockage of Notch1 signaling modulates the T-helper (Th)1/Th2 cell balance in chronic hepatitis B patients. Hepatology research : the official journal of the Japan Society of Hepatology. 2010;40:799–805. doi: 10.1111/j.1872-034X.2010.00680.x. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa H, Mukai K, Kawano Y, Minegishi Y, Karasuyama H. Th2-inducing cytokines IL-4 and IL-33 synergistically elicit the expression of transmembrane TNF-alpha on macrophages through the autocrine action of IL-6. Biochemical and biophysical research communications. 2012;420:114–118. doi: 10.1016/j.bbrc.2012.02.124. [DOI] [PubMed] [Google Scholar]
- 35.Amsen D, Spilianakis CG, Flavell RA. How are T(H)1 and T(H)2 effector cells made? Current opinion in immunology. 2009;21:153–160. doi: 10.1016/j.coi.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annual review of immunology. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 37.Luistro L, He W, Smith M, Packman K, Vilenchik M, Carvajal D, Roberts J, Cai J, Berkofsky-Fessler W, Hilton H, et al. Preclinical profile of a potent gamma-secretase inhibitor targeting notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 2009;69:7672–7680. doi: 10.1158/0008-5472.CAN-09-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He W, Luistro L, Carvajal D, Smith M, Nevins T, Yin X, Cai J, Higgins B, Kolinsky K, Rizzo C, et al. High tumor levels of IL6 and IL8 abrogate preclinical efficacy of the gamma-secretase inhibitor, RO4929097. Mol Oncol. 2011 doi: 10.1016/j.molonc.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debeb BG, Xu W, Mok H, Li L, Robertson F, Ueno NT, Reuben J, Lucci A, Cristofanilli M, Woodward WA. Differential radiosensitizing effect of valproic acid in differentiation versus self-renewal promoting culture conditions. Int J Radiat Oncol Biol Phys. 2010;76:889–895. doi: 10.1016/j.ijrobp.2009.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuben JM, Lee BN, Li C, Gomez-Navarro J, Bozon VA, Parker CA, Hernandez IM, Gutierrez C, Lopez-Berestein G, Camacho LH. Biologic and immunomodulatory events after CTLA-4 blockade with ticilimumab in patients with advanced malignant melanoma. Cancer. 2006;106:2437–2444. doi: 10.1002/cncr.21854. [DOI] [PubMed] [Google Scholar]
- 41.Gao H, Lee BN, Talpaz M, Donato NJ, Cortes JE, Kantarjian HM, Reuben JM. Imatinib mesylate suppresses cytokine synthesis by activated CD4 T cells of patients with chronic myelogenous leukemia. Leukemia. 2005;19:1905–1911. doi: 10.1038/sj.leu.2403933. [DOI] [PubMed] [Google Scholar]
- 42.Villiger PM, Cronin MT, Amenomori T, Wachsman W, Lotz M. IL-6 production by human T lymphocytes. Expression in HTLV-1-infected but not in normal T cells. J Immunol. 1991;146:550–559. [PubMed] [Google Scholar]
- 43.Chen JQ, Russo J. ERalpha-negative and triple negative breast cancer: molecular features and potential therapeutic approaches. Biochim Biophys Acta. 2009;1796:162–175. doi: 10.1016/j.bbcan.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buonamici S, Trimarchi T, Ruocco MG, Reavie L, Cathelin S, Mar BG, Klinakis A, Lukyanov Y, Tseng JC, Sen F, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459:1000–1004. doi: 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao GS, Murray S, Ethier SP. Radiosensitization of human breast cancer cells by a novel ErbB family receptor tyrosine kinase inhibitor. Int J Radiat Oncol Biol Phys. 2000;48:1519–1528. doi: 10.1016/s0360-3016(00)01358-4. [DOI] [PubMed] [Google Scholar]
- 46.Woodward WA, Debeb BG, Xu W, Buchholz TA. Overcoming radiation resistance in inflammatory breast cancer. Cancer. 2010;116:2840–2845. doi: 10.1002/cncr.25173. [DOI] [PubMed] [Google Scholar]
- 47.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011 doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11:951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3:e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Efimova EV, Liang H, Pitroda SP, Labay E, Darga TE, Levina V, Lokshin A, Roizman B, Weichselbaum RR, Khodarev NN. Radioresistance of Stat1 over-expressing tumour cells is associated with suppressed apoptotic response to cytotoxic agents and increased IL6–IL8 signalling. Int J Radiat Biol. 2009;85:421–431. doi: 10.1080/09553000902838566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A. 2011;108:1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen EN, Lee BN, Gao H, Andreopoulou E, Jackson SA, Parker CA, Tin S, Li Y-D, Galland MM, Cristofanilli M, Reuben JM. Soluble Factors and Circulating Tumor Cells in Inflammatory Breast Cancer. Cancer Res. 2009;69(24 Suppl) Abstract nr 2135 2009. [Google Scholar]
- 56.Van Laere S, Van der Auwera I, Van den Eynden GG, Fox SB, Bianchi F, Harris AL, van Dam P, Van Marck EA, Vermeulen PB, Dirix LY. Distinct molecular signature of inflammatory breast cancer by cDNA microarray analysis. Breast Cancer Res Treat. 2005;93:237–246. doi: 10.1007/s10549-005-5157-z. [DOI] [PubMed] [Google Scholar]
- 57.Van Laere SJ, Van der Auwera I, Van den Eynden GG, Elst HJ, Weyler J, Harris AL, van Dam P, Van Marck EA, Vermeulen PB, Dirix LY. Nuclear factor-kappaB signature of inflammatory breast cancer by cDNA microarray validated by quantitative real-time reverse transcription-PCR, immunohistochemistry, and nuclear factor-kappaB DNA-binding. Clin Cancer Res. 2006;12:3249–3256. doi: 10.1158/1078-0432.CCR-05-2800. [DOI] [PubMed] [Google Scholar]
- 58.Murugan V, Peck MJ. Signal transduction pathways linking the activation of alveolar macrophages with the recruitment of neutrophils to lungs in chronic obstructive pulmonary disease. Exp Lung Res. 2009;35:439–485. doi: 10.1080/01902140902759290. [DOI] [PubMed] [Google Scholar]
- 59.Strieter RM, Kunkel SL, Bone RC. Role of tumor necrosis factor-alpha in disease states and inflammation. Crit Care Med. 1993;21:S447–S463. doi: 10.1097/00003246-199310001-00006. [DOI] [PubMed] [Google Scholar]
- 60.Naing A, Reuben JM, Camacho LH, Gao H, Lee BN, Cohen EN, Verschraegen C, Stephen S, Aaron J, Hong D, et al. Phase I Dose Escalation Study of Sodium Stibogluconate (SSG), a Protein Tyrosine Phosphatase Inhibitor, Combined with Interferon Alpha for Patients with Solid Tumors. J Cancer. 2011;2:81–89. doi: 10.7150/jca.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu Rev Immunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 62.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, et al. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6:680–688. [PubMed] [Google Scholar]