Abstract
The Zebrafish Model Organism Database (ZFIN; http://zfin.org) is the central resource for genetic and genomic data from zebrafish (Danio rerio) research. ZFIN staff curate detailed information about genes, mutants, genotypes, reporter lines, sequences, constructs, antibodies, knockdown reagents, expression patterns, phenotypes, gene product function, and orthology from publications. Researchers can submit mutant, transgenic, expression, and phenotype data directly to ZFIN and use the ZFIN Community Wiki to share antibody and protocol information. Data can be accessed through topic-specific searches, a new site-wide search, and the data-mining resource ZebrafishMine (http://zebrafishmine.org). Data download and web service options are also available.
ZFIN collaborates with major bioinformatics organizations to verify and integrate genomic sequence data, provide nomenclature support, establish reciprocal links and participate in the development of standardized structured vocabularies (ontologies) used for data annotation and searching. ZFIN-curated gene, function, expression, and phenotype data are available for comparative exploration at several multi-species resources.
The use of zebrafish as a model for human disease is increasing. ZFIN is supporting this growing area with three major projects: adding easy access to computed orthology data from gene pages, curating details of the gene expression pattern changes in mutant fish, and curating zebrafish models of human diseases.
Keywords: curation, ontology, data mining, human disease models
1 Introduction
The Zebrafish Model Organism Database (ZFIN; http://zfin.org) is the central repository and hub of global genetic, genomic, and phenotypic data from zebrafish (Danio rerio) research (Bradford et al., 2011; Howe et al., 2013). ZFIN staff curate and integrate data from a large number of sources, producing a value-added and highly interconnected data resource with stringent data quality control. Data at ZFIN are free to access and are presented in a format that makes them accessible to a wide range of users. Some of the most recent additions to the ZFIN resource include a data mine that supports custom queries (http://zebrafishmine.org), downloadable search results, web services data access, and a new site search mechanism that promotes rapid searching and data browsing. In addition to these new features, the ZFIN database continues to provide detailed data including gene expression, phenotypes, mutations, reporter lines, gene ontology annotations, orthology, antibodies, and knockdown reagents.
2 Data displayed in ZFIN
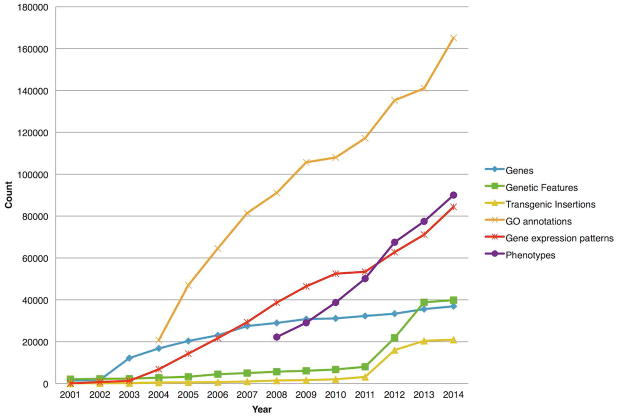
ZFIN integrates information from a wide array of sources, but the majority of the data come from publications or are directly submitted from research laboratories. The types of data displayed in ZFIN are summarized in Table 1. The growth of several major data types in ZFIN is illustrated in Figure 1, and a more comprehensive table can be accessed at ZFIN (http://zfin.org/zf_info/zfin_stats.html).
Table 1.
Data Types in ZFIN
| Data Category | Data Types | Metadata |
|---|---|---|
| Sequence Features | genes, pseudogenes, microRNAs, transcripts, BACs, PACs, fosmids, cDNA clones, ESTs, morpholinos, SSLPs, RAPDs, STSs, engineered regions and foreign genes | chromosomal location, mapping data, relationships between sequence features, sequence accession IDs |
| Alleles and Variations | insertions, point mutations, inversions, transgenic insertions, translocations, deletions, small deletions, substitutions, indels, SNPs, allele designations | genotype zygosity, chromosomal location, availability from resource centers and labs, mapping data |
| Sequence Targeting Reagents | morpholinos, CRISPRs and TALENs | sequences, phenotypes and expression in fish treated with reagent, genotypes created with reagent, transgenic constructs used to create transgenic insertions |
| Transgenic Constructs and Lines | Transgenic constructs, line designations | relationships between sequence features, genotype zygosity, availability from resource centers and labs, expression and phenotype data, transgenic constructs used to create transgenic insertions |
| Phenotypes | phenotype curated from papers and loaded through collaborations with users providing phenotype data | developmental stage, experimental conditions, genotype, anatomy, GO, MPATH, spatial and quality ontology terms |
| Genotypes | single/double/triple/etc. mutants, transgenic fish, wild-types, complex rearrangements, morpholino/TALEN/CRISPR genotypes | parental genotype, phenotypes and expression that use genotypes, genotype availability from resource centers and labs |
| Gene Expression | expression curated from papers and loaded through collaborations with users providing expression data | genotype, experimental conditions, developmental stage, anatomical structures, image sizes, dimensions, and contents, captions and labels for phenotypes and expression that use figures/images |
| Antibodies | gene expression patterns and antibody staining patterns | labeled cellular and anatomical structures, stages, targeted genes, sources and availability, host organism, antibody type and isotype, assays |
| Experimental Conditions | changes to temperature, salinity, chemical, etc. conditions | Specifics about experimental conditions such as increased or decreased temperature, which chemical was used, etc., phenotypes, expression that use the experimental condition |
| Ontologies | gene ontology (GO), quality, anatomy, MPATH neoplasms, spatial, developmental stage ontology, UBERON (Mine only), MEDIC (Mine only) | terms for phenotype, expression, and gene data, term definition, relationships between terms |
| Homologues | curated zebrafish homologues in mouse, human, and fly, predicted homologues from the PANTHER data source (Mine only) | homologue names, symbols and chromosome locations, human disease information from OMIM |
| Data Sources | publications, laboratories, companies, people | names, PubMed IDs, addresses, phone numbers, reagents, allele registrations, web sites, lab members |
| Nomenclature | genes, pseudogenes, transgenics, mutants, constructs | line designations, gene names |
| Figures, Images, Movies | expression and phenotype images from publications, user submitted images and movies, images of people, labs and companies | image sizes, dimensions, and contents, captions and labels for phenotypes and expression that use figures/images |
| Links to Other Databases | NCBI (Gene, RefSeq, GenPept, UniGene, UniSTS, dbSNP, PubMed), OMIM, Wellcome Trust Sanger Institute (Vega, Ensembl, ZMP), MGI, FlyBase, UniProtKB, InterPro, Pfam, PROSITE, GEO, EBI, NCBO-CARO, UCSC, CreZoo, miRBASE, Addgene, MODB, Zfishbook | length and types of sequences in other databases, plasmids, protein domains |
Acronyms and abbreviations used in Table 1: Addgene: non-profit plasmid repository www.addgene.org; BAC: Bacterial Artificial Chromosome; cDNA: Complementary DNA; CreZoo: database of zebrafish CreER driver lines https://crezoo.crt-dresden.de/crezoo/; CRISPR: Clustered Regularly Interspaced Short Palindromic Repeats knockdown reagent; dbSNP: NCBI Short Genetic Variations database http://www.ncbi.nlm.nih.gov/SNP; EBI: European Bioinformatics Institute http://www.ebi.ac.uk/; Ensembl; automatically annotated eukaryotic genomes http://www.ensembl.org; EST: Expressed Sequence Tag; FlyBase: Drosophila model organism database http://flybase.org/; GenPept; NCBI Protein Sequence Database; GEO: Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/; GO: Gene Ontology; InterPro: Integrated Resource of Protein Domains and Functional Sites http://www.ebi.ac.uk/interpro/; MEDIC: disease vocabulary that includes MeSH (Medical Subject Headings) and OMIM disease terms; MGI: Mouse Genome Informatics http://www.informatics.jax.org; miRBase: miRNA sequence and annotation database http://www.mirbase.org/; MODB: MOrpholino Database http://www.morpholinodatabase.org; MPATH: Mouse Pathology Ontology; NCBI: National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/; NCBO-CARO: National Center for Biomedical Ontology - Common Anatomy Reference Ontology); OMIM: Online Mendelian Inheritance in Man http://www.ncbi.nlm.nih.gov/omim; PAC: P1 bacterophage Artificial Chromosome; PANTHER: Protein Analysis Through Evolutionary Relationships database http://pantherdb.org/; Pfam: Protein Families Database http://pfam.xfam.org/; PROSITE: Database of protein families and domains http://prosite.expasy.org/; PubMed: NCBI literature database http://www.ncbi.nlm.nih.gov/pubmed; RAPD: Rapid amplified polymorphic DNA; RefSeq: NCBI Reference Sequence database http://www.ncbi.nlm.nih.gov/refseq/; SSLP:Simple Sequence Length Polymorphism; STS: Sequence Tagged Site; TALEN: TAL Effector Nuclease; UBERON: cross-species “Uber-anatomy” ontology; UCSC: University of California Santa Cruz genome bioinformatics http://genome.ucsc.edu; UniGene: Non-redundant set of genes at NCBI http://www.ncbi.nlm.nih.gov/unigene; UniProtKB: Universal Protein Resource Knowledgebase http://www.uniprot.org/help/uniprotkb; UniSTS; NCBI STS database, contents now in Probe database; Vega: Vertebrate Genome Annotation Database http://vega.sanger.ac.uk; Zfishbook: Zebrafish protein trap consortium database http://zfishbook.org/; ZMP; Zebrafish Mutation Project at the Sanger Institute http://www.sanger.ac.uk/resources/zebrafish/zmp/
Figure 1. Selected Data Types in ZFIN.
The numbers of genes, genetic features, transgenic insertions, gene ontology (GO) annotations, gene expression patterns, and phenotypes in ZFIN from 2001 to 2014.
2.1 Data curation
We obtain publication information from PubMed using an automated script that finds all publications with ‘zebrafish’ in the title, abstract, or keywords and imports the PubMed ID, author names, title, and abstract for display. Publication pages that include links to PubMed IDs, the publisher, and to authors registered with ZFIN are then available in ZFIN for these publications. Links to a publication’s images are displayed on the publication pages only if we have permission from the journals to include them.
We attempt to secure a copy of each paper, either from journal or library websites, or by contacting the authors. Each available publication is first indexed by associating the genes, zebrafish lines, knockdown reagents (Morpholinos, TALENs and CRISPRs), and antibodies reported in the publication. Authors are contacted, as needed, to verify details of the data. Publications are then prioritized for curation; publications with novel genes, new mutants, or models of disease are given the highest priority. Publications with new expression, phenotype or functional gene information are also given a higher priority. Other publications with molecular data are curated as time permits. Publications with toxicology data are indexed but not curated in detail.
ZFIN curators verify all nomenclature used by the authors and manually curate gene expression, phenotype, gene ontology (GO), sequence, and orthology information from the publications into ZFIN using a custom web-based curation interface. To facilitate consistent and unambiguous annotation, curators use several ontologies, which are structured, hierarchical vocabularies that represent a domain of knowledge. These ontologies are either imported from external sources such as the Gene Ontology, or created by ZFIN curators, such as the Zebrafish Anatomy ontology (ZFA) and the Zebrafish Stage ontology (ZFS) (Van Slyke et al., 2014). A list of ontologies used in ZFIN is shown in Table 2. In addition to increasing the consistency of annotation and structuring the data to optimize searches, the use of ontologies facilitates cross-species analyses.
Table 2.
Ontologies Used in ZFIN
| Ontology | Description | Link | Citation |
|---|---|---|---|
| Zebrafish Anatomy Ontology (ZFA) | Ontology that represents the anatomy of Danio rerio | http://www.berkeleybop.org/ontologies/zfa.obo | (Van Slyke et al., 2014) |
| Zebrafish Stage Ontology (ZFS) | Ontology that describes the developmental stages of Danio rerio | http://www.berkeleybop.org/ontologies/zfs.obo | (Van Slyke et al., 2014) |
| Biological Spatial ontology (BSPO) | Ontology that provides standardized description of spatial and topological relationships | https://biological-spatial-ontology.googlecode.com | (Dahdul et al., 2014) |
| Phenotypic Quality ontology (PATO) | Ontology of phenotypic qualities | http://wiki.obofoundry.org/wiki/index.php/PATO:Main_Page | (Mabee et al., 2007) |
| Disease Ontology (DO) | Ontology of human diseases | http://disease-ontology.org/ | (Kibbe et al. 2014) |
| Mouse Pathology ontology (MPATH) | Ontology describing the phenotype or pathology of rodent disease. ZFIN uses the Neoplasm subset. | http://www.berkeleybop.org/ontologies/mpath.owl | (Schofield et al., 2013) |
| Gene Ontology (GO) | Ontology representing gene product properties covering three domains: Cellular Component – the parts of a cell; Molecular Function-activities of gene products at the molecular level; Biological Process – sets of molecular events pertinent to the functioning of an integrated living unit that have a defined beginning and end. | http://geneontology.org/ | (Ashburner et al., 2000) |
| Sequence Ontology (SO) | Ontology representing sequence features | http://www.sequenceontology.org/ | (Eilbeck et al., 2005) |
Nomenclature
To establish proper annotation of any data from a publication, the nomenclature used throughout the manuscript must be accurate. When authors use official gene names and follow nomenclature guidelines for the genes, transgenes and reagents used in their experiments, annotation is a simple and more accurate process (https://wiki.zfin.org/display/general/ZFIN+Zebrafish+Nomenclature+Guidelines). When incorrect or outdated nomenclature is employed, the process of annotation is slowed; gene names must be confirmed by orthology (see below), knockdown reagents verified, transgene nomenclature corrected, and, in many cases, authors must be contacted for verification before data from the publication can be added to the database.
Gene expression curation
ZFIN staff began curating gene expression summaries in 2004, and detailed gene expression in 2005 (Sprague et al., 2006). Gene expression is curated by associating a gene with an anatomical term and a developmental stage. The ZFA and ZFS ontologies are used in conjunction with the Gene Ontology Cellular Component ontology (GO-CC), the Biological Spatial Ontology (BSPO) (Dahdul et al., 2014), and the Neoplasm section of the Mouse Pathology Ontology (MPATH) (Schofield et al., 2013) to annotate the time and location in which a gene product is expressed. Experimental details, including the fish genotype, knockdown reagents, experimental conditions, and expression assays, are also attached to the gene expression data.
Phenotype curation
ZFIN staff began curating detailed phenotypes in 2007 (Sprague et al., 2008). To annotate phenotype, ZFIN utilizes the Entity-Quality (EQ) and Entity-Quality-Entity (EQE) conventions (Washington et al., 2009). The “Entity” is the anatomical structure, process, or function in which the phenotype is observed. The “Quality” (Q) describes the way in which the structure, process, or function is affected. Examples of phenotype descriptions using EQ syntax are: “heart (E) increased size (Q)” and “heart development (E) disrupted (Q)”. An example of a phenotype description using EQE syntax is: “eye (E) fused with (Q) eye (E)”.
We use multiple structured, controlled vocabularies to describe Entities and Qualities. The ontologies used to describe Entities are the ZFA, the GO Biological Process (GO-BP), the GO Molecular Function (GO-MF), the GO Cellular Component (GO-CC) and the MPATH-Neoplasm ontologies. In the previous examples, “heart” is a ZFA ontology term, and “heart development” is a GO Biological Process ontology term. Qualities are chosen from the Phenotypic Quality Ontology (PATO) (Mabee et al., 2007). Experimental details, such as the fish genotype, knockdown reagents, and experimental conditions, are also attached to the phenotype data.
Gene Ontology (GO) curation
Many publications document the involvement of gene products in a particular biological process or molecular function. This information, as well as the subcellular localization of the gene product, is recorded by associating a gene with a GO term. These annotations include an “evidence code” that describes the work or analysis upon which this association is based (e.g., mutant phenotype, direct assay, etc.). These GO annotations follow the policies and guidelines established by the Gene Ontology Consortium (http://geneontology.org/)(Ashburner et al., 2000). The GO annotation policies and guidelines support consistent annotation across the multiple organizations and curators that produce GO annotations, and serve as a resource to improve understanding of the GO data. Consistent annotation is very important so that GO annotations can be used to compare and extend biological knowledge by pooling annotations from different organisms. GO annotation policies include training curators to interpret the meaning of a GO term by its definition, rather than its name. This is exemplified by the terms “integument development” (GO:0080060) and “Integument” (ZFA:0000368). The GO term “integument development” (GO:0080060) is defined as “The process whose specific outcome is the progression of the integument over time, from its formation to the mature structure. Integument is one of the layers of tissue that usually covers the ovule, enveloping the nucellus and forming the micropyle at the apex.” In contrast, “Integument” (ZFA:0000368) in the zebrafish anatomy ontology is defined as “The outer protective barrier that separates the animal from its aquatic environment.”
Consequently, the GO term “integument development” is only applicable to plants based on its definition, and it means something entirely different than the development of the integument in a zebrafish. GO policies also include extensive quality-control checks of the annotations to ensure that all GO annotations are made with the same level of quality assurance. Examples of quality checks include removing annotations that use obsolete GO terms and flagging annotations that use terms that are not applicable to the organism. These extensive GO annotation policies and quality checks have recently been described (Balakrishnan et al., 2013) and are also provided on the gene ontology web site (http://geneontology.org/page/documentation).
Orthology
Orthology data are added to ZFIN by curators conducting independent analyses while either curating published literature or integrating directly submitted data. Orthology is established based on analyses that include, but are not limited to, amino acid and nucleotide sequence identity, including reciprocal best hits, conserved map location, and topology of gene trees across a variety of species. Unpublished orthology data in ZFIN are attributed to a reference, indicating that the orthologous relationship has been established by ZFIN curators rather than reported in a scientific publication.
2.2 Data-direct submission
In addition to curating data from the primary literature, we work with individual research labs to incorporate the gene expression and phenotype data that they have produced. Directly submitted data sets can vary greatly in size, for example from gene expression patterns for 20 genes with 175 images from the Lewis laboratory (England et al., 2014) to 383 genes with more than 5000 images from the Talbot laboratory (Rauch et al., 2003). The Christine and Bernard Thisse laboratory has submitted expression data for 8300 genes (Thisse et al., 2005, Thisse and Thisse, 2004, Thisse and Thisse, 2001). The Burgess and Lin labs have submitted information about 15,223 viral insertion mutants generated in a mutagenesis screen (Burgess and Lin 2012).
Curators collaborate with research scientists to determine which data will be incorporated and the necessary metadata that are required to store and retrieve the data. In general, metadata include information about alleles, genotypes, transgenes, anatomical structures, developmental stages in which the gene expression occurs, and the phenotypes of affected structures. Once the data set and accompanying metadata are finalized, the curator then works with ZFIN technologists to incorporate the data into the database. Direct data submissions provide the zebrafish research community access to previously unreported data, or data sets that are too large for traditional publications.
2.3 Data loads
Sequences, accession numbers, UniProt, NCBI, Ensembl
To increase data interconnectivity, accuracy, and integrity, we collaborate with other database resources to exchange data. Unambiguously matching zebrafish gene records with the corresponding gene records in NCBI, Vega, and Ensembl and the protein records in UniProt allows ZFIN to link to these other data resources as well as to extract content for display on ZFIN’s gene pages. ZFIN data content is updated to reflect changes to the external database records since the last data exchange. Data records between databases must match unambiguously. Scripts identify conflicting data, which are then reported to curators for evaluation and correction. Data in these records are not added to ZFIN until the discrepancies are examined and resolved.
Integration of genome sequence data
We actively collaborate with the Wellcome Trust Sanger Institute to integrate the gene, transcript, and clone annotations produced by the Human and Vertebrate Analysis and Annotation team (HAVANA) (Howe et al., 2013). Prior to new releases of these annotations at the Vertebrate Genome Annotation Database (Vega), the annotations are made available to ZFIN. ZFIN curators use a sequence analysis pipeline to merge, rename or add novel genes and transcripts to ZFIN. We add ZFIN and Vega database identifiers to these annotations to establish links at ZFIN, Vega, and Ensembl before public release of genome annotations at Ensembl. In addition, ZFIN curators provide nomenclature support for novel genes identified during the annotation process.
3 Accessing ZFIN data
The carefully curated and richly detailed data in ZFIN can be searched, downloaded and mined. ZFIN BLAST and the GBrowse genome browser provide access to sequence-based data. Here we detail the most recent search and download options we provide.
3.1 Single Box Search
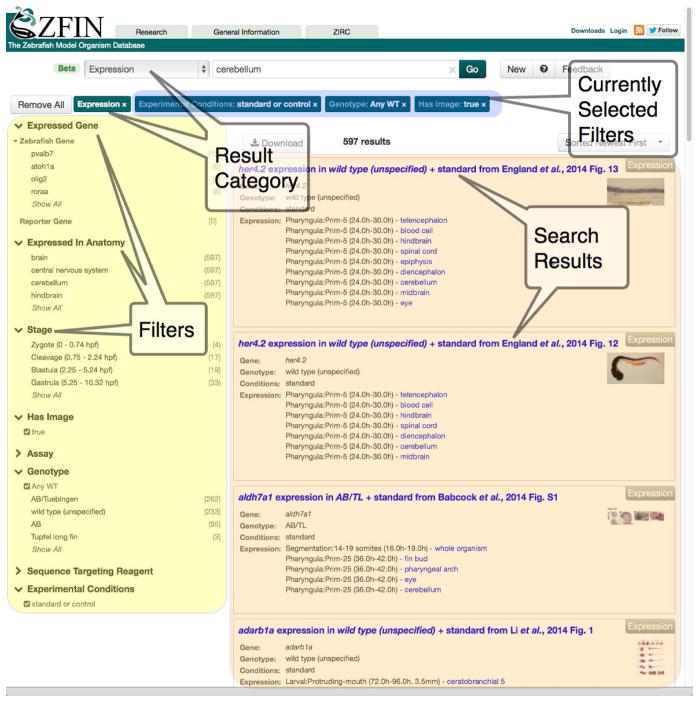
In October 2014, we released a powerful search and data-browsing interface to replace the prior ZFIN site search mechanism. This new search tool surveys nearly all of the data found in ZFIN in one search interface. The results from the search are rapidly returned and grouped into categories such as Gene / Transcript, Expression, Phenotype, Publications, etc. Filters at the left of the search results page facilitate refinement of the results (Figure 2). The interface is a standard search and filter mechanism used in many e-commerce sites as well as at Ensembl and PubMed. Simple text searches often produce the desired record at the top of the list of search results. More complex queries may require narrowing the search results with the filters located at the left of the search results screen. An alternative method of examining data in ZFIN is to enter no text, using only the filters, which provide a pure data-browsing method to find records of interest. The new search also supports advanced query methods, including Boolean logic, exclusion, and the ability to search specifically within one field of data such as anatomical structure. Tips on these advanced search methods can be found on the “ZFIN Single Box Search Help” page (https://wiki.zfin.org/display/general/ZFIN+Single+Box+Search+Help). The index upon which the search is run is currently regenerated on a daily basis.
Figure 2. ZFIN’s New Search Interface.
The new search provides rapid searching and browsing. Results can be refined using the filters on the left. In this example, the search term is “cerebellum”. The search has been narrowed by selecting the Expression category, and the results have been further refined by selecting these filters: standard/control experimental conditions, wild-type genotypes, and figures with images showing gene expression. The search results in the right-hand panel provide the requested details: figures that have images showing gene expression in the cerebellum in wild-type zebrafish in standard/control conditions.
3.2 ZebrafishMine
ZFIN offers customizable and flexible search options with the recently launched ZebrafishMine database (http://zebrafishmine.org). At ZebrafishMine, researchers can write and save their own custom searches, save sets of genes from one search and use them in another search, and download all or part of their results in a variety of formats.
ZebrafishMine is based on the InterMine biological warehousing system (http://intermine.github.io/intermine.org/) (Smith et al., 2012), and contains weekly updates of most of ZFIN’s data, as well as predicted human and mouse gene homology data from the PANTHER database (http://pantherdb.org/, Mi et al., 2013).
Data in ZebrafishMine can be accessed in multiple ways. The search box on the ZebrafishMine homepage allows simple one-word searches. The “Templates” tab houses predefined searches of varying complexity. Researchers can alter these predefined templates or write new searches using the “Query Builder”. Gene lists consisting of gene symbols, names or identifiers can be uploaded to ZebrafishMine and analyzed against other data in the mine. Gene lists can be compared, combined, subtracted, and used in searches. ZebrafishMine also offers a “My Mine” facility where users can log in and save lists and custom searches so they are easy to reuse at a later time.
ZebrafishMine is part of the InterMOD consortium (Lyne et al., 2015 this issue; Sullivan et al., 2013) that includes several other model organism Mines: FlyMine (http://www.flymine.org/, Lyne et al., 2007), YeastMine (http://yeastmine.yeastgenome.org/, Balakrishnan et al., 2012), RatMine (http://ratmine.mcw.edu/), MouseMine (http://www.mousemine.org) and WormMine (http://www.wormbase.org/tools/wormmine/). The InterMine framework allows users to navigate among the different model organism Mines and the newly developed HumanMine (http://www.humanmine.org) database via computed homology (for specific examples, see Lyne et al., 2015, this issue).
3.3 ZFIN Data Downloads
All data in ZFIN are available on the “Downloads” page (http://zfin.org/downloads). ZebrafishMine provides additional customizable options for downloading data from ZFIN. ZebrafishMine search results can be downloaded in a variety of formats, including XML, TSV, CSV, and JSON. ZebrafishMine offers data access via API web services and client libraries in Perl, Python, Ruby, and Java. The results of the single box search can be downloaded as a CSV file.
4 Services offered by ZFIN
ZFIN serves the research community by providing assistance with zebrafish gene, mutant, and transgenic line nomenclature, and with direct data submission to ZFIN (see section 2.2). ZFIN also provides a public wiki for researchers to share information.
4.1 Nomenclature
The importance of using standardized gene nomenclature cannot be overstated (Klionsky et al., 2012). When official, standardized nomenclature is used, it is immediately clear to readers which gene is being discussed. Correct and standardized nomenclature not only support accurate annotation of the data, but they also enable comparisons with gene names in other model organisms and humans. To help ensure that proper gene nomenclature is utilized, ZFIN participates in the Zebrafish Nomenclature Committee and the ZFIN Nomenclature Coordinator communicates with researchers to determine the correct nomenclature of their genes of interest. In general, the nomenclature coordinator helps researchers develop gene nomenclature that is informative and specific, and that takes into account relationships with gene families and/or the functions of the products. Inquiries regarding nomenclature should be sent to nomenclature@zfin.org.
4.2 ZFIN Community Wiki
ZFIN provides a Community Wiki for researchers to share information. The Protocols section of the Wiki includes content from The Zebrafish Book and protocols submitted by researchers. The Antibody section of the Wiki contains antibody data from ZFIN as well as antibodies submitted by the community. The wiki also supports comments from the community on protocols and antibodies.
5 Collaborations: resource centers
The global zebrafish research community now has three major resource centers: ZIRC, EZRC, and CZRC, located in the United States, Germany, and China, respectively (Table 3). These resource centers provide genetic lines and various materials and services to the zebrafish research community.
Table 3.
Zebrafish Resource Centers
| Resource Center | Location | Resources & Services Provided |
|---|---|---|
| Zebrafish International Resource Center (ZIRC) Web: http://zebrafish.org Contact: ZIRC@zebrafish.org |
Eugene, Oregon, USA | WT, mutant and transgenic fish, embryos, sperm, antibodies, probes, paramecia, Pathology and Health Services |
| European Zebrafish Resource Center (EZRC) Web: http://ezrc.kit.edu Contact: EZRC-Request@itg.kit.edu |
Karlsruhe, Germany | WT, mutant and transgenic fish, plasmids, bioinformatics support |
| China Zebrafish Resource Center (CZRC) Web: http://zfish.cn Contact: zebrafish@ihb.ac.cn |
Wuhan, China | WT, mutant and transgenic fish, knockout services, paramecia, antibodies, plasmids, cell lines, EST/cDNA |
ZIRC
Although ZFIN and ZIRC (The Zebrafish International Resource Center) are both located at the University of Oregon in Eugene, Oregon, they are distinct organizations. ZIRC, established in 1999, handles and distributes biological samples, including about 20,000 genetic strains available as sperm, embryos, or adults, as well as antibodies and probes, whereas ZFIN handles data about genes, mutants, gene expression, phenotypes, etc. ZFIN and ZIRC work closely together to exchange data, allowing ZFIN to display “Order This” links for resources that are available for distribution from ZIRC and allowing ZIRC to display data from ZFIN. Links from ZIRC to ZFIN provide detailed information about mutants, antibodies, and other resources available from ZIRC. ZIRC also provides paramecia and pathology and health services for the research community. Services include zebrafish husbandry and health consultation, histopathology for disease investigation or sentinel testing, bacteriology, and necropsy exams.
EZRC
The European Zebrafish Resource Center, established in 2012, consists of a stock center that contains several thousand mutants from Tübingen screens and the Zebrafish Mutation Project (ZMP) as well as many transgenic and wild-type lines from diverse sources. EZRC also distributes more than 2000 plasmids containing sequence from zebrafish genes. We collaborate with EZRC to provide accurate and current “Order This” links from ZFIN to mutants that are available for distribution from EZRC. EZRC links back to ZFIN for more information about genes and mutants. EZRC also offers bioinformatics support and screening services for the Sanger ZMP project mutations.
CZRC
The China Zebrafish Resource Center, established in 2012, is focused on collecting existing zebrafish mutants and transgenic lines, developing new lines, and providing technical and informatics support for the Chinese and global zebrafish research communities. CZRC collaborates with ZFIN to provide links from CZRC to ZFIN for more detailed information about genes, transgenic constructs, and phenotypes. Additionally, “Order This” links are updated daily at ZFIN to connect users to resources currently available for distribution by CZRC.
6 Interconnections
ZFIN collects, curates, and integrates a large amount of data about zebrafish genetics and genomics, and provides these data to the biological research community. These data are also acquired by and integrated into other databases, further expanding the availability and usefulness of the data.
ZFIN provides NCBI (http://www.ncbi.nlm.nih.gov/) with zebrafish gene nomenclature and ZFIN-curated GO data that are displayed on NCBI zebrafish Gene pages (Maglott et al., 2011). The Ensembl Genome Browser integrates ZFIN-curated phenotype data as well as gene nomenclature, with Biomart providing zebrafish phenotype data and GO annotations. The Neuroscience Information Framework (NIF, http://www.neuinfo.org/), a comprehensive collection of neuroscience resources (Gupta et al., 2008), displays ZFIN-curated phenotype, gene expression, and antibody data. The PhenomicDB (http://phenomicdb.info/) a multi-species genotype and phenotype database (Groth et al. 2013) includes genotype and phenotype data from ZFIN. The Bgee database (http://bgee.unil.ch/bgee/bgee) allows the comparison of gene expression patterns among animal species (Bastian et al., 2008). ZFIN provides Bgee with zebrafish expression and developmental stage data that are analyzed to determine comparable anatomical structures (Niknejad et al., 2012).
The Gene Ontology (GO) Consortium (http://geneontology.org/) integrates subcellular localizations and functions of gene products from many databases. ZFIN is the official distributor of the GO annotations associated with genes from zebrafish. Many of these annotations are generated via literature curation at ZFIN. Other manually curated or electronically generated annotations are imported into ZFIN from external sources such as the Phylogenetic Annotation and Inference Tool (PAINT), and the Gene Ontology Annotation Database at UniProt (GOA; http://www.ebi.ac.uk/GOA), and missing Biological Process and Cellular Component annotations are inferred by a script based on existing Molecular Function annotations. The script and inferred annotations are provided by the GO Consortium (Gaudet et al., 2011; Huntley et al., 2014). Manually curated annotations imported into ZFIN from GOA originate from a number of third party sources including curators at AgBase, the British Heart Foundation, Ensembl, the Human Gene Nomenclature Committee, IntAct, and the Mouse Genome Informatics database. ZFIN curators contribute to the development of GO by generating detailed term requests when a new term is suggested by the literature. ZFIN also participates in Gene Ontology focus groups targeting specific aspects of biology relevant to zebrafish.
Several biological resources utilize ZFIN-curated phenotype data to focus on phenotypic comparisons among species that foster understanding the evolution of development or the identification of human disease candidate genes. These resources include Phenoscape (Dahdul et al., 2010), PhenomeNET (Hoehndorf et al., 2015), PhenoDigm (Smedley et al., 2013), and the Monarch Initiative (http://monarchinitiative.org/). The Phenoscape group (http://phenoscape.org/) annotates character states from the systematics literature using ontologies and integrates these data with model organism mutant phenotypes, with the goal of making it possible to navigate phenotypic data across species computationally. PhenomeNet (http://phenomebrowser.net/) serves as a queryable cross-species phenotype network that allows users to query using disease, gene, genotype, and allele names to explore phenotype. PhenoDigm (https://www.sanger.ac.uk/resources/databases/phenodigm/) is a database that utilizes semantic mappings between human clinical phenotype observations and mouse and zebrafish phenotypes to compare human diseases and gene models of disease. The Monarch Initiative (http://monarchinitiative.org/) focuses on tools that allow phenotypes to be aggregated and compared across species. Monarch has developed a semantic framework and interface for comparing biological data from diverse species. Users can query for genes, phenotypes, and human diseases, and explore animal models of human disease. Monarch utilizes gene, genotype, phenotype, orthology, and publication data produced by ZFIN.
7 Future directions
The ZFIN team continuously updates and improves ZFIN, prioritizing work based on the needs of the research community as assessed by direct user input and surveys of database users. In response to the increasing use of zebrafish as a model for human disease, new features being developed at ZFIN include projects that support this rapidly growing area of research. Three major projects are poised to make a significant impact on our support of zebrafish models of human disease: 1) increased support for curated and computed orthology data, 2) curation of gene expression pattern changes in mutants, and 3) curation of zebrafish models of human diseases to enhance the discovery and use of these disease models.
7.1 Computed orthology
We are in the process of increasing the robustness of ZFIN orthology data by supplementing the existing curated orthology data with computationally derived orthology from the Ensembl Compara and PANTHER databases. In cases where no curated human or mouse orthology is associated with a zebrafish gene, the computed orthology data may provide a starting point for the identity or function of the gene and its relationship to other genes or gene families. If curated orthology is available for a zebrafish gene, the addition of computed orthology may either provide supplemental support for that relationship or alert the user that the relationship to human or mouse may warrant further attention due to a conflict between the curated and computed orthologs. Further, the breadth of species contained in computed orthology database resources provides the opportunity to incorporate orthology data for a number of other vertebrates, including other fishes and basal chordates, which are impractical to curate manually.
7.2 Expression patterns as phenotypes
Gene expression patterns are often the sole or key features of reported phenotypes. Changes in gene expression in genetically modified animals can provide significant insight into disease etiology and developmental mechanisms. Our aim is to capture basic gene expression pattern phenotypes including changes in expression level or tissue-specific localization of transcripts or proteins. These data will be available together with the other expression and phenotype data already provided by ZFIN.
7.3 Human disease
Increasingly, researchers are developing and reporting zebrafish models of human disease (Phillips and Westerfield, 2014; Ablain and Zon, 2013; Goldsmith and Jobin, 2012; Santoriello and Zon, 2012; Ingham 2009; Lieschke and Currie, 2007). For example, Danilova et al. (2014) report using zebrafish deficient in rps19 and rpl11 as models of Diamond Blackfan anemia. Lyon et al. (2013) report a zebrafish model of spinal muscular atrophy, and Novorol et al. (2013) report several zebrafish models of microcephaly. To leverage these data effectively, we are developing better support for curation and searching of this information. To facilitate curation of zebrafish models of human disease, ZFIN will use the Disease Ontology (DO) (Kibbe et al. 2014) to annotate reported zebrafish models of human diseases. The DO is an ontology that provides definitions of diseases and references to other resources such as the Medical Subject Headings (MeSH), The Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT), the Unified Medical Language System (UMLS), the International Classification of Diseases (ICD), the National Cancer Institute Thesaurus (NCI Thesaurus), and the Online Mendelian Inheritance in Man (OMIM). In addition to annotating zebrafish models of human disease, ZFIN will display and report this information on disease term pages that will provide information about the human disease, including a definition of the disease, human genes associated with the disease, the orthologous zebrafish genes, reported zebrafish models, and citations. In addition, we are working with the Monarch Initiative (http://monarchinitiative.org/) to utilize the Monarch Phenotype Grid Widget, which was developed to identify and visualize mutated genes that produce phenotypes in model organisms similar to human disease symptoms. ZFIN curators currently link zebrafish publications that model a disease to the Disease Ontology. Full support for zebrafish disease model curation is expected to be completed in the Fall of 2015.
8 Conclusions
ZFIN is the preeminent resource for gold standard genetic, genomic, and phenotypic data from zebrafish research, and an essential hub in the landscape of highly interconnected and interdependent biological databases. ZFIN achieves this by continuously curating detailed data from publications, incorporating directly submitted data from laboratories, establishing connections to major public databanks and resource centers around the world, collaborating with organizations to develop standardized and universal vocabularies and new data access options, and providing data to other databases. To serve the zebrafish and wider biological research communities even better, we have recently expanded the options for searching, browsing, and downloading ZFIN data, and aim to facilitate the use of zebrafish as a model for human diseases.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by the National Human Genome Research Institute (HG002659, and HG004834) of the National Institutes of Health.
LITERATURE CITED
- Ablain J, Zon LI. Of fish and men: Using zebrafish to fight human diseases. Trends in Cell Biology. 2013;23(12):584–586. doi: 10.1016/j.tcb.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan R, Harris MA, Huntley R, Van Auken K, Cherry JM. A Guide to best practices for Gene Ontology (GO) manual annotation. Database (Oxford) 2013;2013:bat054. doi: 10.1093/database/bat054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan R, Park J, Karra K, Hitz BC, Binkley G, Hong EL, Sullivan J, Micklem G, Cherry JM. YeastMine-An integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012;2012:bar062. doi: 10.1093/database/bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian F, Parmentier G, Roux J, Moretti S, Laudet V, Robinson-Rechavi M. Bgee: Integrating and comparing heterogeneous transcriptome data among species. Lecture Notes in Computer Science (including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2008:124–131. [Google Scholar]
- Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SAT, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper HB, Schaper K, Shao X, Singer A, Sprague J, Sprunger B, Van Slyke C, Westerfield M. ZFIN: Enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 2011:39. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Lin S. Viral Insertion Mutants Overwrite Data. ZFIN Direct Data Submission. 2012 ( http:zfin.org)
- Dahdul WM, Balhoff JP, Engeman J, Grande T, Hilton EJ, Kothari C, Lapp H, Lundberg JG, Midford PE, Vision TJ, Westerfield M, Mabee PM. Evolutionary characters, phenotypes and ontologies: curating data from the systematic biology literature. PLoS One. 2010;5:e10708. doi: 10.1371/journal.pone.0010708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdul WM, Cui H, Mabee PM, Mungall CJ, Osumi-Sutherland D, Walls RL, Haendel MA. Nose to tail, roots to shoots: spatial descriptors for phenotypic diversity in the Biological Spatial Ontology. J Biomed Semantics. 2014;5:34. doi: 10.1186/2041-1480-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Bibikova E, Covey TM, Nathanson D, Dimitrova E, Konto Y, Lindgren A, Glader B, Radu CG, Sakamoto KM, Lin S. The role of DNA damage response in zebrafish and cellular models of Diamond Blackfan Anemia. Dis Model Mech. 2014;7:895–905. doi: 10.1242/dmm.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England S, Hilinski W, de Jager S, Andrzejczuk L, Campbell P, Chowdhury T, Demby C, Fancher W, Gong Y, Lin C, Machikas A, Rodriguez-Larrain G, Roman Rivera V, Lewis KE. Identifying Transcription Factors Expressed by Ventral Spinal Cord Interneurons. ZFIN Direct Data Submission. 2014 ( http://zfin.org)
- Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith JR, Jobin C. Think small: zebrafish as a model system of human pathology. J Biomed Biotechnol. 2012;2012:817341. doi: 10.1155/2012/817341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Bug W, Marenco L, Qian X, Condit C, Rangarajan A, Müller HM, Miller PL, Sanders B, Grethe JS, Astakhov V, Shepherd G, Sternberg PW, Martone ME. Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF) Neuroinformatics. 2008;6:205–17. doi: 10.1007/s12021-008-9033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehndorf R, Schofield PN, Gkoutos GV. PhenomeNET: A whole-phenome approach to disease gene discovery. Nucleic Acids Research. 2011;39(18) doi: 10.1093/nar/gkr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SAT, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper K, Shao X, Singer A, Sprunger B, Van Slyke CE, Westerfield M. ZFIN, the Zebrafish Model Organism Database: Increased support for mutants and transgenics. Nucleic Acids Res. 2013:41. doi: 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch GJ, White S, Chow W, Kilian B, Quintais LT, Guerra-Assunção Ja, Zhou Y, Gu Y, Yen J, Vogel JH, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Elliot D, Eliott D, Threadgold G, Harden G, Ware D, Begum S, Mortimore B, Mortimer B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Lloyd C, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire S, Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper JD, Cooper J, Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Ürün Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberländer M, Rudolph-Geiger S, Teucke M, Lanz C, Raddatz G, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Schuster SC, Carter NP, Harrow J, Ning Z, Herrero J, Searle SMJ, Enright A, Geisler R, Plasterk RHa, Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nüsslein-Volhard C, Hubbard TJP, Roest Crollius H, Rogers J, Stemple DL. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, O’Donovan C. The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res. 2014;43:D1057–63. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW. The power of the zebrafish for disease analysis. Human Molecular Genetics. 2009;18(R1) doi: 10.1093/hmg/ddp091. [DOI] [PubMed] [Google Scholar]
- Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D, Parkinson H, Schriml LM. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Research. 2014;2014:gku1011. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Bruford EA, Cherry JM, Hodgkin J, Laulederkind SJF, Singer AG. In the beginning there was babble. Autophagy. 2012;8:1165–1167. doi: 10.4161/auto.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8(5):353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Lyne R, Sullivan J, Butano D, Contrino S, Heimbach J, Hu F, Kalderimis A, Lyne M, Smith RN, Štěpán R, Balakrishnan R, Binkley G, Harris T, Karra K, Moxon SAT, Motenko H, Neuhauser S, Ruzicka L, Cherry M, Richardson J, Stein L, Westerfield M, Worthey E, Micklem G. Cross Organism Analysis Using InterMine. Genesis. 2015 doi: 10.1002/dvg.22869. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, Mclaren P, North P, Rana D, Riley T, Sullivan J, Watkins X, Woodbridge M, Lilley K, Russell S, Ashburner M, Mizuguchi K, Micklem G. FlyMine; an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8(7):R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon AN, Pineda RH, Hao LT, Kudryashova E, Kudryashov DS, Beattie CE. Calcium binding is essential for plastin 3 function in Smn-deficient motoneurons. Hum Mol Genet. 2013;23(8):1990–2004. doi: 10.1093/hmg/ddt595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabee PM, Ashburner M, Cronk Q, Gkoutos GV, Haendel M, Segerdell E, Mungall C, Westerfield M. Phenotype ontologies: the bridge between genomics and evolution. Trends Ecol Evol. 2007;22:345–50. doi: 10.1016/j.tree.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: Gene-centered information at NCBI. Nucleic Acids Res. 2011:39. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niknejad A, Comte A, Parmentier G, Roux J, Bastian FB, Robinson-Rechavi M. vHOG, a multispecies vertebrate ontology of homologous organs groups. Bioinformatics. 2012;28:1017–20. doi: 10.1093/bioinformatics/bts048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novorol C, Burkhardt J, Wood KJ, Iqbal A, Roque C, Coutts N, Almeida AD, He J, Wilkinson CJ, Harris WA. Microcephaly models in the developing zebrafish retinal neuroepithelium point to an underlying defect in metaphase progression. Open Biol. 2013;3(10):130065. doi: 10.1098/rsob.130065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JB, Westerfield M. Zebrafish models in translational research: tipping the scales toward advancements in human health. Dis Model Mech. 2014;7:739–743. doi: 10.1242/dmm.015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch GJ, Lyons DA, Middendorf I, Friedlander B, Arana N, Reyes T, Talbot WS. Submission and Curation of Gene Expression Data. ZFIN Direct Data Submission. 2003 ( http://zfin.org)
- Santoriello C, Zon LI. Hooked! modeling human disease in zebrafish. Journal of Clinical Investigation. 2012;122(7):2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PN, Sundberg JP, Sundberg BA, McKerlie C, Gkoutos GV. The mouse pathology ontology, MPATH; structure and applications. J Biomed Semantics. 2013;4:18. doi: 10.1186/2041-1480-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Oellrich A, Köhler S, Ruef B, Westerfield M, Robinson P, Lewis S, Mungall C. PhenoDigm: Analyzing curated annotations to associate animal models with human diseases. Database. 2013:bat025. doi: 10.1093/database/bat025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RN, Aleksic J, Butano D, Carr A, Contrino S, Hu F, Lyne M, Lyne R, Kalderimis A, Rutherford K, Stepan R, Sullivan J, Wakeling M, Watkins X, Micklem G. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012;28:3163–5. doi: 10.1093/bioinformatics/bts577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, Mani P, Moxon SAT, Pich C, Ramachandran S, Schaper K, Segerdell E, Shao X, Singer A, Song P, Sprunger B, Van Slyke CE, Westerfield M. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, Schaper K, Segerdell E, Song P, Sprunger B, Taylor S, Van Slyke CE, Westerfield M. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–5. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J, Karra K, Moxon SAT, Vallejos A, Motenko H, Wong JD, Aleksic J, Balakrishnan R, Binkley G, Harris T, Hitz B, Jayaraman P, Lyne R, Neuhauser S, Pich C, Smith RN, Trinh Q, Cherry JM, Richardson J, Stein L, Twigger S, Westerfield M, Worthey E, Micklem G. InterMOD: integrated data and tools for the unification of model organism research. Sci Rep. 2013;3:1802. doi: 10.1038/srep01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402) ZFIN Direct Data Submission. 2001 ( http://zfin.org)
- Thisse B, Thisse C. Fast Release Clones: High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 ( http://zfin.org)
- Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. 2004 ( http://zfin.org)
- Van Slyke CE, Bradford YM, Westerfield M, Haendel MA. The zebrafish anatomy and stage ontologies: representing the anatomy and development of Danio rerio. J Biomed Semantics. 2014;5:12. doi: 10.1186/2041-1480-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.