Abstract
The Arabidopsis Information Resource (TAIR) is a continuously updated, online database of genetic and molecular biology data for the model plant Arabidopsis thaliana that provides a global research community with centralized access to data for over 30,000 Arabidopsis genes. TAIR’s biocurators systematically extract, organize, and interconnect experimental data from the literature along with computational predictions, community submissions, and high throughput datasets to present a high quality and comprehensive picture of Arabidopsis gene function. TAIR provides tools for data visualization and analysis, and enables ordering of seed and DNA stocks, protein chips and other experimental resources. TAIR actively engages with its users who contribute expertise and data that augments the work of the curatorial staff. TAIR’s focus in an extensive and evolving ecosystem of online resources for plant biology is on the critically important role of extracting experimentally-based research findings from the literature and making that information computationally accessible. In response to the loss of government grant funding, the TAIR team founded a nonprofit entity, Phoenix Bioinformatics, with the aim of developing sustainable funding models for biological databases, using TAIR as a test case. Phoenix has successfully transitioned TAIR to subscription-based funding while still keeping its data relatively open and accessible.
Keywords: genome database, plant, sustainable funding
Introduction
Arabidopsis thaliana is an annual dicotyledonous plant that serves as a model plant for thousands of researchers across the globe. The Arabidopsis Information Resource (TAIR) is a curated online database of genetic and molecular biology data for Arabidopsis thaliana (Lamesch et al. 2012). Arabidopsis is an attractive model organism due to its small genome size, experimental tractability and short generation time (Somerville and Koornneef 2002; Koornneef and Meinke 2010). Arabidopsis was the first plant genome to be sequenced, and subsequently the subject of a concerted effort to understand the functions of the more than 30,000 genes identified to date (Arabidopsis Genome Initiative 2000); https://www.arabidopsis.org/portals/masc/projects.jsp). Decades of research have produced an expansive collection of data about Arabidopsis that serves as a reference for understanding plant gene functions and ultimately unraveling mechanisms of plant physiology, biochemistry and development.
TAIR continues to grow and evolve along with its research community, data and knowledge about the organism. TAIR was launched in 1999 just prior to the release of the public genome sequence and rapidly became an indispensable resource for thousands of plant biology researchers around the world (Huala et al. 2001). In 2014, TAIR registered over 2.1 million visits with an average of 178,000 visits and 61,000 users per month (Google Analytics, accessed March 1, 2015). Visits originated from around the world with Asia accounting for 42%, Europe 28%, and the Americas 27%. As of March 2015, the number of registered TAIR users was 27,500; with about 38% of these considered to be highly active. Over the years the underlying data provided by the resource has grown in both quantity and quality. The main activity of TAIR is producing a ‘gold standard’ functional annotation for this important reference plant in order to make experimental data maximally discoverable and computable by the research community. That activity was significantly curtailed in recent years due to the loss of TAIR’s main funding source from the National Science Foundation. The most significant change in TAIR has been the transition from grant funding to subscription support from institutions and individuals worldwide. This new, international, sustainable funding model provides increased stability over traditional grant funding and ensures that TAIR can continue its mission to provide highest quality Arabidopsis genome information to the research community.
Gold Standard Annotation of the Arabidopsis Genome
A rigorously annotated reference genome dataset is essential for making inferences, producing accurate models, and generating testable hypotheses about gene functions in Arabidopsis and other plant genomes. One of the most common uses of TAIR is to extrapolate the function of genes in agriculturally important species based on orthology to Arabidopsis genes. Biological roles can be inferred with greater degree of confidence if the evidence supporting the assertion for the reference gene is experimentally based rather than the result of a computational prediction. Thus, the main activity of TAIR curators is integrating experimental data from the research literature to produce high quality annotations that enable functional and comparative genomics (Lamesch et al. 2010).
TAIR curators extract and organize a variety of data from the peer-reviewed literature (Table 1). Curated data include, but are not limited to, gene function information captured in the form of Gene Ontology (GO) annotations (Berardini et al. 2004; Gene Ontology Consortium et al. 2013), gene expression information in the form of Plant Ontology (PO) annotations (Cooper et al. 2013), gene symbol and full names, alleles, phenotypes and germplasm information, and publications. These data are carefully curated according to exacting standards and are presented in a structured way that reflects actual biological relationships. When community members submit data to TAIR, a curator reviews the submission and performs standard quality control checks before incorporation of this data into the resource.
Table 1.
Literature-derived, curated data added to TAIR since August 31, 2013 (data as of June 15, 2015)
| Data type | Number added or updated |
|---|---|
| Articles | 7428 |
| Gene Symbols | 1288 |
| Genes linked to articles | 10,818 |
| Articles linked to genes | 4019 |
| Articles used for experimentally-supported GO or PO annotations | 432 (TAIR curators + community) * |
| 215 (TAIR curators only) | |
| Experimentally-supported GO and PO annotations | 2870 (TAIR curators + community) * |
| 1120 (TAIR curators only) | |
| Alleles from the literature | 150 |
| Phenotypes from the literature | 90 |
All community submissions are reviewed by a TAIR curator prior to incorporation into the database. Occasionally, additional information that supplements or clarifies the user’s submission is added during the review process.
Because the Arabidopsis literature is extensive and TAIR’s curation resources are limited, we established a triage system that enables us to focus our literature curation efforts on articles with novel or high impact results (Li et al. 2012). Each month, approximately 350 new articles indexed by PubMed contain the word ‘Arabidopsis’ in the title, abstract or MeSH headings. Of those, about 60% contain information about one or more Arabidopsis genes. TAIR curators use a semiautomated entity recognition and linking method to associate genes to research articles (Yoo et al. 2006). Curators read the abstracts of all new articles, review computationally generated links, and validate, invalidate or manually add new links between genes and articles. As a result, the growing literature corpus is accurately associated to individual genes and becomes accessible from the corresponding gene detail pages. A subset of these articles, such as those that contain experimental results on gene function for previously uncharacterized genes, is read in depth and experimentally derived results are added to TAIR in a structured manner.
Gene Function
Gene function information is captured in the form of Gene Ontology (GO) annotations that describe the molecular function, biological role and subcellular localization of gene products (The Gene Ontology Consortium et al. 2000). The ontologies themselves are controlled vocabularies that are structured to accurately represent biology and are continuously reviewed and updated. TAIR curators play an important role in maintaining the ontologies and ensuring that plant biology terms and relations are incorporated correctly. TAIR is the main source of manual GO annotations for Arabidopsis. Since 2001 TAIR has created tens of thousands of annotations from the literature. In the current set of valid annotations, TAIR’s contribution includes 91,445 manual GO annotations to 18,932 distinct Arabidopsis gene products accounting for 80.6% of all publically available experimentally based (i.e. manual) GO annotations for Arabidopsis thaliana (http://www.ebi.ac.uk/GOA/arabidopsis_release, GOA Arabidopsis (version 118), released on 27 May, 2015). Annotations are curated according to a rigorously defined set of standards that have been extensively discussed and documented by the GO consortium (Hill et al. 2008). In addition to the annotation done by TAIR curators, we integrate manual Arabidopsis thaliana annotations from UniProtKB, the GO Consortium, and contributions from our research community to present a unified view of Arabidopsis gene function. TAIR also incorporates computationally-generated annotations from UniProt that rely on InterPro domain mapping (Burge et al. 2012) as well as in-house computational inferences based on sequence features. The computational annotations are especially valuable in the case where no other information is available from the research literature. Computationally predicted gene function annotations are removed from display as new experimentally validated annotations are added, or when more accurate and up-to-date computational predictions become available. Researchers can filter annotations based on evidence to select only those annotations that are supported by experiment.
Alleles and Phenotypes
The Arabidopsis community has generated an extensive toolkit of genetic resources including numerous mutant collections. Tens of thousands of insertion lines, knockdowns, point mutations, over-expressors and more have been donated to the stock centers and many of the insertion sites have been mapped to the genome (Sessions et al. 2002; Till et al. 2003; Alonso et al. 2003; Rosso et al. 2003; Woody et al. 2007). Many of these alleles have been characterized experimentally and described in the literature. TAIR adds value to these genetic resources by gathering phenotype descriptions from the literature and linking that information to the allele record in TAIR, making phenotype data more accessible to researchers. For example, if a researcher reports the phenotype of a T-DNA insertion line generated by the Salk T-DNA Express project, TAIR curators will associate information about the molecular phenotype of the insertion (e.g. verified to be a null allele) as well as the mutant plant phenotype. Phenotype information is summarized in the form of free text descriptions that are searchable in the database. Alleles and phenotypes are linked to the source publications so that researchers can immediately find the relevant references.
Gene Expression
Researchers wishing to mine Arabidopsis gene expression data in TAIR can access data from different types of experiments ranging from large-scale, genome wide expression studies of whole plants, tissues and single cells to in-situ expression of single genes using probes or reporter genes. We integrate gene expression data using Plant Ontology (PO) annotations. The PO terms describe plant structures from the whole plant down to individual cells and plant growth and developmental stages (whole plant and plant parts) (Jaiswal et al. 2005). As with the GO annotations, the PO annotations facilitate cross experiment and cross species comparisons (Cooper, 2013). As part of our literature curation workflow, we annotate gene expression patterns reported in publications. This often adds significant granularity to what might be known from studies using microarrays or RNA-seq data. For example, a gene shown to be expressed in flowers in the AtGenExpress tool (Schmid et al. 2005) might be shown to have very tissue specific expression (e.g. only ovules) when examined by in-situ hybridization. TAIR captures and displays both of these results to present a more detailed expression profile. Literature curation adds granularity and makes experimental resources more visible by including information about the type of experiment supporting the annotation. If a gene fusion is used to localize expression, that information will be part of the annotation so a researcher can quickly determine whom to contact to obtain the construct.
Genome organization and structure; then and now
The reference genome annotation for the original Arabidopsis thaliana Col-0 genome sequence has been periodically updated to incorporate new experimental data on gene expression, update gene structures, and add splice variants and newly discovered genes. From 2005 to 2010, TAIR was responsible for updating both gene structures and gene function for the standard Arabidopsis genome release, available from NCBI’s RefSeq and many other resources. Using a combination of computational methods and manual review, TAIR produced a total of five genome releases with the most recent (TAIR10) made public in November 2010 (https://www.arabidopsis.org/portals/genAnnotation/gene_structural_annotation/annotation_data.jsp#data) (Swarbreck et al. 2008; Lamesch et al. 2012). In 2013, the newly funded Araport project took on the responsibility to provide additional Arabidopsis genome releases (Araport, www.araport.org) (Krishnakumar et al. 2015). The Araport genome releases will be made available from TAIR along with data on tagged insertion sites, protein features like predicted subcellular localization and domains, and orthology data from other organisms. As we have done with previous releases generated by TAIR, we will also produce a set of custom sequence datasets based on the Araport genome release for our sequence analysis tools.
Gene Nomenclature
Each locus in Arabidopsis is assigned a unique identifier, termed the AGI locus code (AGI, Arabidopsis Genome Initiative) which consists of the prefix At, followed by the chromosome identifier (1–5 or M or C) followed by g for gene and then a unique 5 digit number (e.g. At2g46340). Typically, authors of research articles refer to a gene using a gene symbol rather than the AGI locus identifier. However, gene symbols are not unique identifiers for Arabidopsis genes; in many cases one gene symbol refers to two or more genes or the same gene has been given two or more gene symbols by different research groups. This creates challenges for both researchers and curators who want to search for or add new information to a specific gene. TAIR curators sometimes expend considerable effort to resolve these nomenclature issues and correctly link genes to publications or other information. In cases where the author does not explicitly include the unique AGI locus identifier in an article, curators must often undertake some detective work to infer which identifier should be associated to the new gene symbol and publication.
TAIR also maintains a community registry for gene symbol nomenclature (https://www.arabidopsis.org/portals/nomenclature/symbol_main.jsp) that was established to minimize duplication of gene symbols and thus make it easier to unambiguously identify genes in articles. Researchers can use this registry to ensure that the symbol they wish to use for a new gene has not already been used for a different gene, and reserve it for a future publication. Researchers who have discovered a new gene that lacks an AGI locus identifier in the most recent genome release should contact the Araport project at https://www.araport.org/contact to have one assigned. The newly assigned identifier should be included in abstracts and publications discussing the new gene.
Searching, browsing and downloading data
TAIR provides basic and advanced search tools for each of the various data types in the database (https://www.arabidopsis.org/servlets/Search?type=general&action=new_search). The header at the top of every page features a simple name search that can be used to search for specific types of data such as genes or clones. For example, a researcher looking for information about a specific locus can simply type the AGI locus identifier or symbolic name into the search box, and choose Gene from the drop down menu. For each data type there are also advanced search tools that include additional relevant search parameters such as location within the genome, associated keywords, and mutagens. Thus, a researcher interested in cell wall biosynthesis could use the advanced Gene search to identify all Arabidopsis genes annotated to this process based on experimental evidence (https://www.arabidopsis.org/servlets/Search?action=new_search&type=gene). This would be done by entering the keyword cell wall biosynthesis into the keyword field, choosing ‘contains’ search and selecting one or more types of experimental evidence (inferred from direct assay, inferred from expression pattern, inferred from genetic interaction, inferred from physical interaction, and inferred from mutant phenotype). The results can then be downloaded as a list or browsed individually.
There are several entry points for browsing different types of data. The keyword browser can be used to find annotations, loci and publications associated to GO or PO terms (https://www.arabidopsis.org/servlets/Search?action=new_search&type=keyword). Users with questions about nomenclature and gene families can browse the registry of gene class symbols (https://www.arabidopsis.org/servlets/processor?type=genesymbol&update_action=view_symbol) or community-generated gene family pages (https://www.arabidopsis.org/browse/genefamily/index.jsp).
Researchers wishing to retrieve large datasets, such as all GO annotations or upstream sequences for a set of co-expressed genes, can use the Bulk Download search and retrieval tools (https://www.arabidopsis.org/tools/bulk/index.jsp). Very large datasets, such as all GO or PO annotations, FASTA formatted BLAST datasets, and whole genome structural annotations, can be downloaded as complete files (https://www.arabidopsis.org/download/index.jsp). Custom datasets can also be created for TAIR subscribers upon request.
The Locus Detail page
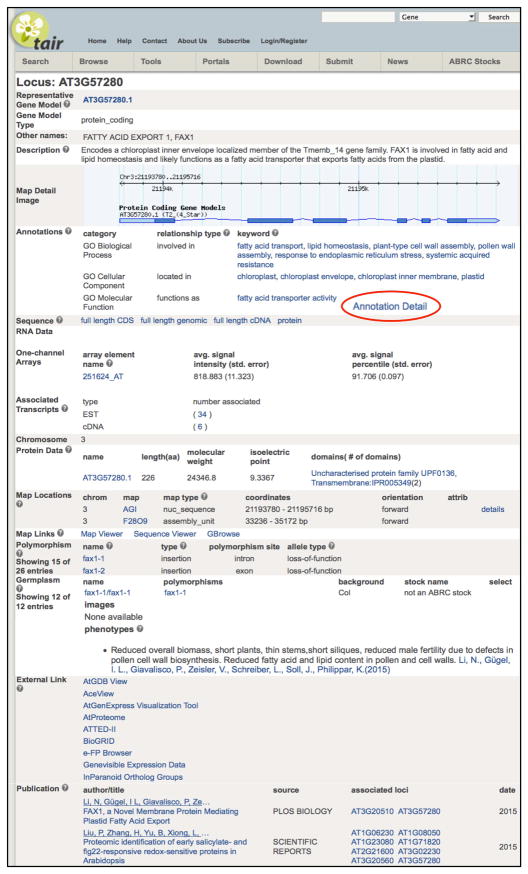
Each of the more than 30,000 genes in the Arabidopsis genome has its own locus detail page that presents an overview of the information associated with that gene. The Locus Detail Page (Figure 1) is the heart of TAIR and is the most frequently visited page in the entire resource. From this single page, users can navigate to GO and PO annotations, amino acid and DNA sequence details, RNA expression data, publication lists, and external resources that provide additional data for that particular locus. Each locus page is divided into logically grouped content areas including gene structural features such as UTRs, introns and exons; protein features such as domains and molecular weights; polymorphisms; germplasms and phenotypes; associated publications, and others. For ease of viewing, a limited amount of detail from the associated data is shown in the locus page. Clicking on a related data type opens a new detail page for that data element with additional information.
Figure 1. A TAIR Locus Detail page.
The locus page aggregates and summarizes all the information in TAIR about any given locus with the most frequently used data towards the top. Each locus is referred to by its unique AGI identifier along with other names from the literature. The graphical representation of the locus is linked to GBrowse for viewing within the chromosomal context. GO and PO annotations are presented in summary form; the detailed view is accessible from the link to annotation detail page (red circle). Similarly, links within each data subtype (e.g. Protein Data, Polymorphism) can be followed to access more detailed information from TAIR. Hyperlinks from the external links section go directly to resources outside of TAIR. Manually validated associated publications are shown at the bottom.
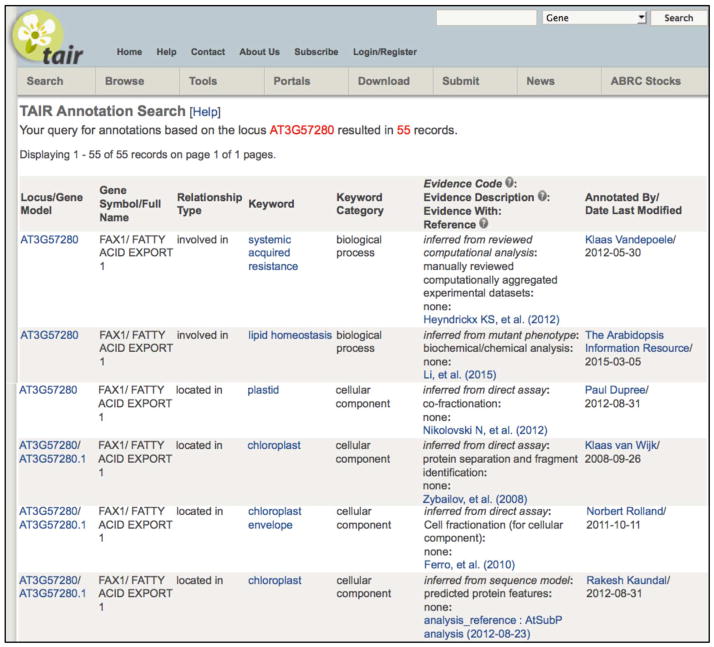
An example of a linked page providing an additional level of detail is the Annotation Detail page, which can be opened by clicking on the ‘Annotation Detail’ link on the locus page. This secondary page provides the detailed evidence and other metadata associated to individual GO and PO annotations, including the publication on which the annotation is based, the evidence code or basis for linking the gene product to the controlled vocabulary term, and the person or organization that contributed the annotation. Figure 2 illustrates annotations attributed to a variety of sources.
Figure 2. A TAIR Annotation Detail page.
This secondary page linked from the annotation summary on the locus page presents additional details and links about the origins of each annotation including publication (linked to publication detail page), keyword (linked to keyword detail page) annotation contributor (linked to the community detail page), evidence code, and evidence description.
Data visualization and analysis tools
TAIR’s main tools to visually interrogate the Arabidopsis genome are the Seqviewer (http://tairvm09.tacc.utexas.edu/servlets/sv) (Huala et al. 2001) and GBrowse, (http://tairvm17.tacc.utexas.edu/cgi-bin/gb2/gbrowse/arabidopsis/) (Stein et al. 2002). Both can be used for exploration of the A. thaliana reference sequence and mapped sequenced objects such as cDNAs, ESTs, polymorphisms, gene-disrupting T-DNA insertion sites, and markers. SeqViewer was built in house and is searchable by feature name and short (15–150 nt) sequences. It allows users to scan entire chromosomes or drill down to the nucleotide level, viewing 10 kb of nucleotide sequence at a time with gene structures and mapped objects clearly marked at the individual nucleotide level, making visualization of the locations of mapped objects very clear (Fig. 3A). GBrowse, a component of the GMOD project (Stein et al. 2002) provides views of additional data types not available in SeqViewer including methylation and phosphorylation patterns, repeat regions, and orthologous genes in other species, as well as additional expression data types including peptides and RNA-seq reads (Fig. 3B). GBrowse can also be customized to include user defined annotation tracks. Currently, the most recent genome annotation release, TAIR10 (November 2010), provides the underlying data for both browsers. The reference genome will be updated to the Araport genome annotation when this is released.
Figure 3. Genome Browsers at TAIR.
a. SeqViewer Close-Up View. Each of the five chromosomes is represented by a green line on top. A subset of any given chromosome is shown in the blue-boxed area along with the specific tracks of data selected using the check boxes in the upper left. b. SeqViewer Nucleotide View. Drilled down nucleotide level view of locus AT3G57280 showing the exact positions of exons (orange), introns (light blue), UTR (red), TDNA/transposon insertions (purple track), translation start/stop (boxed blue area) c. GBrowse showing similar data as 3a but with a different presentation. d. Additional data tracks in GBrowse. Some examples of data types are shown that are visible in GBrowse but not in SeqViewer such as phosphorylation sites and transcriptional regulatory sequences.
TAIR features other specialized tools that are developed in-house or draw upon data only available in TAIR. The chromosome map tool is used for drawing positions of loci on the five Arabidopsis chromosomes (https://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp). Users upload a list of loci that are then marked on the map and the image can be saved and used for publications. TAIR also offers a variety of sequence analysis programs including NCBI BLAST (https://www.arabidopsis.org/Blast/index.jsp) (Altschul et al. 1990) and WU-BLAST (https://www.arabidopsis.org/wublast/index2.jsp) for identification of sequence similarity, PatMatch for short sequence pattern matching (https://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl) (Yan et al. 2005), and statistical software for finding common motifs in upstream sequences (https://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp). These sequence analysis programs are paired with datasets that are generated in-house (https://www.arabidopsis.org/help/helppages/BLAST_help.jsp#datasets), some of which are exclusively available at TAIR.
Stock ordering
The Arabidopsis research community has generated an extensive collection of biological resources that have been made available via the major stock centers, the Arabidopsis Biological Resource Center (ABRC; https://abrc.osu.edu/) and the Nottingham Arabidopsis Stock Center (NASC; http://arabidopsis.info/). Stocks include all types of germplasms ranging from natural variants to single and multiply mutated lines, insertion mutants and other transgenics, as well as DNA stocks (clones) and protein chips. TAIR and ABRC have a long-standing collaboration in which TAIR provides the database infrastructure and software tools to facilitate ordering of seed and DNA stocks, protein chips and other experimental resources for the research community from the ABRC. The high level of integration with ABRC means that stocks are linked to relevant data to facilitate discovery and are not just independently searchable and browseable from within TAIR. If researcher wants to identify knockout lines to study the phenotype of a gene of interest, there are several ways to find and order the relevant stock. Starting from a locus, all associated alleles and germplasms can be viewed on the locus detail page (Fig 1). Alternatively the Polymorphism search (https://www.arabidopsis.org/servlets/Search?action=new_search&type=polyallele) can be used to identify specific types of alleles for a given locus, or alleles can be identified based on physical location using SeqViewer or GBrowse. If TAIR has curated phenotype descriptions for a stock, that information will be displayed in the Germplasm record. Users must have a basic (free) TAIR account and be affiliated with an active laboratory to order stocks from ABRC; a subscription to TAIR is not required for searching for, browsing through or ordering stocks.
Community curation in TAIR
As a community resource, TAIR is proactive in encouraging users to contribute expertise and data. Because our community members are the experts in their fields, we actively solicit their contributions of GO and PO annotations by directly contacting authors of articles published in Plant Physiology and The Plant Journal (Berardini et al. 2012). TAIR has well-established collaborations with these journals which provide us with data identifying authors of recently accepted articles, so that we may contact them and remind them to submit data to TAIR. An additional eight journals (Plant Cell, Journal of Integrative Plant Biology, Environmental and Experimental Botany, Molecular Plant, Plant, Cell and Environment, Journal of Experimental Botany, Plant Science, and Plant Physiology and Biochemistry) include instructions to submit data to TAIR in their instructions to authors or as a part of the manuscript submission process. To make this process as simple as possible, we developed an online data submission tool, TOAST (TAIR Online Annotation Submission Tool), that guides researchers through the use of controlled vocabularies (https://www.arabidopsis.org/doc/submit/functional_annotation/123) (Berardini, 2012). We also incorporate submissions from researchers in spreadsheet format. Community contributed annotations are attributed to the submitter and are linked to the publication record in TAIR as well as the locus. As of June 2015, a total of 24,379 GO and 57,788 PO have been submitted to TAIR by 730 authors derived from experimental evidence described in 1124 articles and published in 80 journals. Another way that registered community members can contribute data is by adding informal comments about any data object from the object detail (e.g. locus or germplasm detail) page. These comments are visible to everyone and are an important way of sharing information. For example, if a researcher orders a T-DNA insertion line from ABRC and finds that the insertion is not actually present, that information can be added to the comments so that the community is aware of the finding which may otherwise not ever be published.
Community Engagement
TAIR staff members provide many other important services to the research community in addition to biocuration. TAIR is a member of the Gene Ontology Consortium and actively participates both in the development of the ontologies to ensure plant biology is accurately represented and in the specification of annotation standards. Curators attend professional meetings such as the International Society of Biocurators annual conference, to learn about and follow best practices and ensure that TAIR adheres to the highest quality standards for data curation. Curators also staff the TAIR helpdesk which receives and replies to an average of 10 queries a week, ranging from requests for custom data sets to help using tools, nomenclature questions, subscription enquiries, and requests for collaboration. We respond to questions not only about TAIR-specific data and tools, but also general questions from researchers and students on a wide variety of topics. Researchers also contact us to point out errors or omissions and make requests to add certain links or datasets. This feedback from our community is one of the very important ways in which we maintain the accuracy and integrity of information in the database and informs our decision making regarding priority areas for data curation.
As a central organizing resource for the community, we strive to help foster connections among researchers and disseminate information of interest to them. Researchers can promote conferences and list job opportunities through TAIR; we typically post several on the TAIR site every week and also funnel them into our Twitter feed (@tair_news). Job seekers can subscribe to the job posting RSS feed to be notified when new opportunities are posted.
Recently we have begun using social media more extensively and have reached over 2,400 Facebook followers (https://www.facebook.com/tairnews) and over 850 Twitter followers (https://twitter.com/tair_news). Our Facebook community increased by 150% after implementing a global ad campaign in October of 2014 that was used to reach out to more members of the research community. We regularly post articles of interest to both Facebook and Twitter with positive engagement from our followers.
We also engage directly with community members at conferences such as the Plant and Animal Genome meeting, the annual meeting of the American Society for Plant Biology, and the International Conference for Arabidopsis Research. Through a combination of workshops, exhibit hall booths, posters and both spontaneous and planned one-on-one encounters, we gather information on the needs of our scientific community as well as the new trends in plant biology both in research areas and technology. We also conduct occasional online surveys to solicit community input on various aspects of TAIR usage and future directions.
TAIR’s place in the plant biology research landscape
TAIR is part of an extensive and evolving ecosystem of databases and tools that researchers can use to access Arabidopsis data. The main function of a model organism database is to serve as a trusted, integrated source of information for the organism and TAIR has served that role for many years. In the time since TAIR was launched, the amount of information available has grown tremendously along with the number of places where that information resides. Gene and protein information about Arabidopsis, including data from TAIR, can be found in the widely-used NCBI and UniProt databases. Examples of other popular resources that include Arabidopsis data are: Genevisible (http://genevisible.com/) (Zimmermann et al. 2004), the electronic Fluorescent Pictograph (eFP) Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (Winter et al. 2007), and MapMan (http://mapman.gabipd.org/web/guest/mapman) (Thimm et al. 2004) for gene expression data; AraCyc for metabolic pathways (Rhee et al. 2006); the Bio-Analytic Resource (BAR; http://bar.utoronto.ca/welcome.htm) and Membrane Based Interactome Database (MIND; https://associomics.dpb.carnegiescience.edu/Associomics/Home.html) for interaction data; 1001 genomes for natural variations (http://1001genomes.org/) and Aramemnon (http://aramemnon.uni-koeln.de/) for analyzing proteins based on subcellular localization. TAIR’s role within this rapidly expanding data ecosystem is to provide researchers with a dataset of experimentally verified results along with their associated sources and experimental methods that accurately and consistently represents the current state of knowledge about Arabidopsis gene function. To accomplish this, TAIR focuses primarily on extraction of experimental results and associated information from the Arabidopsis research literature and representation of these results in both computable (as Gene Ontology annotations) and human-readable (as a series of conveniently organized web pages) formats. No other resource in the current data ecosystem has taken on this charge, and TAIR has been and still remains the ultimate source of a large majority (~80%) of this type of data at all the other resources within our data landscape, with the remainder coming from multispecies curation efforts at UniProt/SwissProt.
The loss of NSF funding for TAIR and the perceived consequences of closure were deeply felt by the research community which recognized a continuing need for an Arabidopsis resource and made recommendations that ultimately led to the formation of AraPort (International Arabidopsis Informatics Consortium 2010; Krishnakumar et al. 2015). TAIR’s decision, in 2013, to pursue alternative funding models rather than shut down was a response both to the Arabidopsis community, which voiced strong support for TAIR (https://www.arabidopsis.org/doc/about/tair_survey/411), and the need to find scalable solutions to broadly address the question of database sustainability. Ultimately this decision benefits the community because our emphasis on functional annotation and continuous curation of the literature complements Araport’s focus on genome annotation and analytics. These two Arabidopsis resources reinforce each other and help ensure that the plant research community continues to have access to the highest quality data and most completely and accurately annotated plant genome possible.
The sustainability crisis and TAIR’s transition to subscription support
Historically, biological databases have been funded by grants from national funding agencies, either as stand-alone projects or as outputs of broader research efforts. Yet, despite the important role of these databases in supporting research, there are currently no established mechanisms to supply long-term funding. With the exception of a very few repositories with funding directly from line-item federal budget allocations, ongoing support generally depends on repeatedly applying for, and successfully acquiring, competitive grant funds (Merali and Giles 2005; Abbott 2009). While the number of repositories has increased, there has been no commensurate increase in funding, as agency budgets have remained relatively flat over the last decade (AAAS Society 2014).
In response to this sustainability crisis, and feedback from our community TAIR established a new user-based funding mechanism. From 1999, when the resource was established, to August 2013, TAIR was supported primarily by grants from the National Science Foundation. In September 2013, the remaining TAIR team founded a 501(c)(3) nonprofit entity, Phoenix Bioinformatics, with the explicit aim of developing sustainable funding models for data repositories, starting with TAIR as a test case. As a popular and highly used database, TAIR was in a good position to pioneer a new approach to funding scientific resources based on contributions from users. TAIR began requiring subscriptions for companies in October 2013 and for academic/nonprofit researchers in April 2014. As of June 2015, TAIR is supported by a large base of subscribers including two country-wide subscriptions (China and Switzerland), 140 individual academic institutions, four academic consortia and approximately 200 individual researchers. To enforce access restrictions, Phoenix developed a subscription management system customized for TAIR’s needs. The user-based subscription funding model provides greater stability and longevity for fundamentally important datasets, better alignment of project goals with the needs of the research community, and the ability to scale up funding support in direct proportion to the demand for a dataset or tool. A subscription model also provides a way to fund valuable activities that are difficult to support via traditional grant funding, including manual curation of research literature.
Assuring broad access to TAIR data
To ensure that Arabidopsis data remain open and accessible while still retaining an incentive to subscribe, we adopted the following set of guiding principles to maximize data availability: 1) TAIR data are made freely available to all after one year to facilitate reuse by other repositories (ftp://ftp.arabidopsis.org/home/tair/TAIR_Public_Releases/); 2) subscriptions for more recent data must be affordable and offered at a range of levels including country, consortium, institution and individual to maximize subscription options and coverage of researchers; 3) researchers can access a few pages per month of recent data without subscribing, to facilitate access by occasional users (e.g. animal researchers); 4) students taking a course in which TAIR is a course material are provided with free access; 5) free access is provided to the lowest income countries https://www.arabidopsis.org/doc/about/tair_subscriptions/413). Datasets resulting from our grant-funded Gene Ontology data curation and collection work are released on a monthly basis to the Gene Ontology Consortium without delay and are available at http://geneontology.org/.
Current and future directions for TAIR and Phoenix
In the first few months following the end of TAIR’s federal funding in late 2013, the remaining TAIR staff laid the groundwork for future efforts by establishing Phoenix Bioinformatics as a separate 501(c)(3) nonprofit organization located in Redwood City, California. As subscription revenue increased over the course of Phoenix’s first year of operations in 2014, we were able to add additional staff and shift our focus from maintaining a basic level of curation and software support to enhancing TAIR with an increase in both the number of articles curated and the amount and variety of data extracted. Specifically, we have doubled the number of papers curated per month and resumed curating new alleles and phenotypes which had been dramatically reduced during the past two years. Going forward, TAIR will expand efforts to capture and integrate experimental data from the literature for better coverage of new research results especially as new technologies are adopted by the community. We have also begun making long overdue improvements to the TAIR software, including streamlining our registration process, restructuring parts of the locus detail pages and other parts of the TAIR site for improved usability, and enhancing our internal curation software to improve the efficiency of our curation efforts.
As we move into our second year of user-supported funding, we will continue to provide new and updated functional information for Arabidopsis genes and add to the corpus of allele, phenotype and publication information in TAIR. New reference datasets will be integrated, including the new genome release expected from Araport. With a recent grant from the Alfred P. Sloan Foundation, we have begun developing a new cloud-based subscription management system capable of providing subscription services to additional biological databases and other research resources in need of stable financial support. In keeping with the nonprofit mission and vision of Phoenix Bioinformatics, our new home, we will continue to explore sustainable funding models, apply our lessons learned to other databases in need of help, and support the community of biological databases and the researchers that rely on them for their work.
Acknowledgments
TAIR is supported by national, academic institutional, corporate, and individual subscriptions (see, https://goo.gl/NYY6cf for a list of institutional subscribers). Gene Ontology annotations reported in this publication were generated with support from the National Human Genome Research Institute (NHGRI) of the National Institutes of Health under award number U41HG002273. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. TAIR is administered by the 501(c)(3) non-profit Phoenix Bioinformatics (www.phoenixbioinformatics.org).
References
- AAAS Society. Historical Trends in Federal R&D. 2014 http://www.aaas.org/page/historical-trends-federal-rd.
- Abbott A. Plant genetics database at risk as funds run dry. Nature News. 2009;462:258–259. doi: 10.1038/462258b. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Li D, Muller R, Chetty R, Ploetz L, Singh S, Wensel A, Huala E. Assessment of community-submitted ontology annotations from a novel database-journal partnership. Database: The Journal of Biological Databases and Curation 2012. 2012:bas030. doi: 10.1093/database/bas030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, Moseyko N, Yoo D, Xu I, Zoeckler B, Montoya M, Miller N, Weems D, Rhee SY. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiology. 2004;135:745–755. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S, Kelly E, Lonsdale D, Mutowo-Muellenet P, McAnulla C, Mitchell A, Sangrador-Vegas A, Yong S-Y, Mulder N, Hunter S. Manual GO annotation of predictive protein signatures: the InterPro approach to GO curation. Database: the journal of biological databases and curation 2012. 2012:bar068. doi: 10.1093/database/bar068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper L, Walls RL, Elser J, Gandolfo MA, Stevenson DW, Smith B, Preece J, Athreya B, Mungall CJ, Rensing S, Hiss M, Lang D, Reski R, Berardini TZ, Li D, Huala E, Schaeffer M, Menda N, Arnaud E, Shrestha R, Yamazaki Y, Jaiswal P. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant & Cell Physiology. 2013;54:e1. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Blake JA, Dolan M, Drabkin H, Hill DP, Li N, Sitnikov D, Bridges S, Burgess S, Buza T, McCarthy F, Peddinti D, Pillai L, Carbon S, Dietze H, Ireland A, Lewis SE, Mungall CJ, Gaudet P, Chrisholm RL, Fey P, Kibbe WA, Basu S, Siegele DA, McIntosh BK, Renfro DP, Zweifel AE, Hu JC, Brown NH, Tweedie S, Alam-Faruque Y, Apweiler R, Auchinchloss A, Axelsen K, Bely B, Blatter M-C, Bonilla C, Bouguerleret L, Boutet E, Breuza L, Bridge A, Chan WM, Chavali G, Coudert E, Dimmer E, Estreicher A, Famiglietti L, Feuermann M, Gos A, Gruaz-Gumowski N, Hieta R, Hinz C, Hulo C, Huntley R, James J, Jungo F, Keller G, Laiho K, Legge D, Lemercier P, Lieberherr D, Magrane M, Martin MJ, Masson P, Mutowo-Muellenet P, O’Donovan C, Pedruzzi I, Pichler K, Poggioli D, Porras Millán P, Poux S, Rivoire C, Roechert B, Sawford T, Schneider M, Stutz A, Sundaram S, Tognolli M, Xenarios I, Foulgar R, Lomax J, Roncaglia P, Khodiyar VK, Lovering RC, Talmud PJ, Chibucos M, Giglio MG, Chang H-Y, Hunter S, McAnulla C, Mitchell A, Sangrador A, Stephan R, Harris MA, Oliver SG, Rutherford K, Wood V, Bahler J, Lock A, Kersey PJ, McDowall DM, Staines DM, Dwinell M, Shimoyama M, Laulederkind S, Hayman T, Wang S-J, Petri V, Lowry T, D’Eustachio P, Matthews L, Balakrishnan R, Binkley G, Cherry JM, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hitz BC, Hong EL, Karra K, Miyasato SR, Nash RS, Park J, Skrzypek MS, Weng S, Wong ED, Berardini TZ, Huala E, Mi H, Thomas PD, Chan J, Kishore R, Sternberg P, Van Auken K, Howe D, Westerfield M. Gene Ontology annotations and resources. Nucleic Acids Research. 2013;41:D530–535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DP, Smith B, McAndrews-Hill MS, Blake JA. Gene Ontology annotations: what they mean and where they come from. BMC Bioinformatics. 2008;9(Suppl 5):S2. doi: 10.1186/1471-2105-9-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, Mueller LA, Bhattacharyya D, Bhaya D, Sobral BW, Beavis W, Meinke DW, Town CD, Somerville C, Rhee SY. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Research. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Arabidopsis Informatics Consortium. An International Bioinformatics Infrastructure to Underpin the Arabidopsis Community. The Plant Cell. 2010;22:2530–2536. doi: 10.1105/tpc.110.078519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal P, Avraham S, Ilic K, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M, Stein L, Stevens P, Vincent L, Ware D, Zapata F. Plant Ontology (PO): a Controlled Vocabulary of Plant Structures and Growth Stages. Comparative and Functional Genomics. 2005;6:388–397. doi: 10.1002/cfg.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Meinke D. The development of Arabidopsis as a model plant. The Plant Journal: For Cell and Molecular Biology. 2010;61:909–921. doi: 10.1111/j.1365-313X.2009.04086.x. [DOI] [PubMed] [Google Scholar]
- Krishnakumar V, Hanlon MR, Contrino S, Ferlanti ES, Karamycheva S, Kim M, Rosen BD, Cheng C-Y, Moreira W, Mock SA, Stubbs J, Sullivan JM, Krampis K, Miller JR, Micklem G, Vaughn M, Town CD. Araport: the Arabidopsis Information Portal. Nucleic Acids Research. 2015;43:D1003–D1009. doi: 10.1093/nar/gku1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic acids research. 2012;40:D1202–10. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Dreher K, Swarbreck D, Sasidharan R, Reiser L, Huala E. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Current Protocols in Bioinformatics. 2010;30(1.11):1.11.1–1.11.51. doi: 10.1002/0471250953.bi0111s30. [DOI] [PubMed] [Google Scholar]
- Li D, Berardini TZ, Muller RJ, Huala E. Building an efficient curation workflow for the Arabidopsis literature corpus. Database: The Journal of Biological Databases and Curation 2012. 2012:bas047. doi: 10.1093/database/bas047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merali Z, Giles J. Databases in peril. Nature. 2005;435:1010–1011. doi: 10.1038/4351010a. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Zhang P, Foerster H, Tissier C. AraCyc: Overview of an Arabidopsis Metabolism Database and its Applications for Plant Research. In: Saito PDK, Dixon PDRA, Willmitzer PDL, editors. Plant Metabolomics. Springer; Berlin Heidelberg: 2006. pp. 141–154. [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, Clarke JD, Cotton D, Bullis D, Snell J, Miguel T, Hutchison D, Kimmerly B, Mitzel T, Katagiri F, Glazebrook J, Law M, Goff SA. A High-Throughput Arabidopsis Reverse Genetics System. The Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Koornneef M. A fortunate choice: the history of Arabidopsis as a model plant. Nature Reviews Genetics. 2002;3:883–889. doi: 10.1038/nrg927. [DOI] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. The Generic Genome Browser: A Building Block for a Model Organism System Database. Genome Research. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, Radenbaugh A, Singh S, Swing V, Tissier C, Zhang P, Huala E. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Research. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal: For Cell and Molecular Biology. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, Henikoff JG, Comai L, Henikoff S. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Research. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. In: Baxter I, editor. PLoS ONE. Vol. 2. 2007. p. e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. Journal of Plant Research. 2007;120:157–165. doi: 10.1007/s10265-006-0048-x. [DOI] [PubMed] [Google Scholar]
- Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY. PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Research. 2005;33:W262–W266. doi: 10.1093/nar/gki368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D, Xu I, Berardini TZ, Rhee SY, Narayanasamy V, Twigger S. PubSearch and PubFetch: a simple management system for semiautomated retrieval and annotation of biological information from the literature. Current Protocols in Bioinformatics. 2006;13(9.7):9.7.1–9.7.27. doi: 10.1002/0471250953.bi0907s13. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]