Abstract
In Drosophila, two Piwi proteins, Aubergine (Aub) and Argonaute-3 (Ago3) localize to perinuclear ‘nuage’ granules and use guide piRNAs to target and destroy transposable element transcripts. We find that Aub and Ago3 are recruited to nuage by two different mechanisms. Aub requires a piRNA guide for nuage recruitment, indicating that its localization depends on recognition of RNA targets. Ago3 is recruited to nuage independently of a piRNA cargo and relies on interaction with Krimper, a stable component of nuage that is able to aggregate in the absence of other nuage proteins. We show that Krimper interacts directly with Aub and Ago3 to coordinate the assembly of the ping-pong piRNA processing (4P) complex. Symmetrical dimethylated arginines are required for Aub to interact with Krimper, but are dispensable for Ago3 to bind Krimper. Our study reveals a multi-step process responsible for the assembly and function of nuage complexes in piRNA-guided transposon repression.
INTRODUCTION
Small RNA pathways exist in many organisms and have multiple functions in sequence-specific regulation of gene expression. The three classes of small RNA are microRNA, short interfering RNA (siRNA) and Piwi-interacting RNA (piRNA). piRNAs associate with Piwi clade Argonaute proteins and together repress the proliferation of transposable elements (TE) in the germline of sexually reproducing animals (Aravin et al., 2001; Vagin et al., 2006). In a Piwi-piRNA complex, the piRNA is responsible for identification of target transcripts and the Piwi protein fulfills effector functions (Aravin et al., 2007; Siomi et al., 2011). D. melanogaster has three Piwi proteins, Piwi, Aubergine (Aub), and Argonaute-3 (Ago3), that share similar domain architectures, with PAZ and Mid domains responsible for the binding of guide piRNA, and a conserved DDH triad in the RNAse H-like fold of the PIWI domain required for endonuclease activity (Ma et al., 2004; Ma et al., 2005; Song et al., 2004; Wang et al., 2009). Despite their similarity, the three Piwi proteins function non-redundantly to repress transposons (Brennecke et al., 2007; Li et al., 2009; Lin and Spradling, 1997; Schmidt et al., 1999). Piwi, itself, localizes to the nucleus and is involved in transcriptional repression through installation of the repressive H3K9me3 chromatin mark on target loci (Le Thomas et al., 2013; Rozhkov et al., 2013; Shpiz et al., 2011; Sienski et al., 2012). In contrast to Piwi, both Aub and Ago3 are cytoplasmic and endonucleolytically cleave transcripts complementary to their piRNA guide (Brennecke et al., 2007; Gunawardane et al., 2007; Huang et al., 2014). Aub and Ago3 are targets for symmetric dimethylation of arginine (sDMA) modification that allow for interactions with proteins containing Tudor domains (Chen et al., 2011; Liu et al., 2010; Nishida et al., 2009).
Aub and Ago3 localize to a distinct subcellular compartment termed ‘nuage’. Nuage surrounds the nuclei of nurse cells of the Drosophila ovary, however are not separated from the cytoplasm by a membrane (Eddy, 1975; Mahowald, 1971). Germline granules similar to Drosophila nuage were first described more than 100 years ago and were identified in germ cells of almost all Metazoa (Eddy, 1975; Hegner, 1911, 1912). However, the molecular function of nuage remained rather cloudy. The recent findings that many additional proteins involved in piRNA-mediated silencing such as Vasa, Tejas, Kumo/Qin, Spindle-E, Krimper, and Tudor are also localized in nuage, suggested that this compartment is responsible for piRNA biogenesis and TE repression (Lim and Kai, 2007; Malone et al., 2009; Nishida et al., 2009; Patil and Kai, 2010; Xiol et al., 2014; Zhang et al., 2011). Nuage might be a location where mRNAs exiting the nucleus are scanned through a quality control process before gaining access to ribosomes for translation in the cytoplasm. Despite the progress in understanding the composition of nuage, the role that nuage assembly plays in piRNA repression remained unknown.
Although Aub and Ago3 have very similar domain organization and subcellular localization, the two proteins play non-redundant roles in germline transposon repression. Analysis of piRNAs associated with each protein revealed the basis of the unique functions of Aub and Ago3. While Aub-bound piRNA are predominantly antisense to TE sequences, Ago3-bound piRNAs are predominantly in sense orientation and therefore cannot be involved in repression directly (Brennecke et al., 2007; Gunawardane et al., 2007; Li et al., 2009). These and other results led to the proposal of the so-called ping-pong model, which suggests an intimate relationship between Aub and Ago3 with cleavage products generated by one protein passed to the other (Brennecke et al., 2007; Gunawardane et al., 2007). Specifically, cleavage of transposon mRNAs by Aub creates the 5′ end of new secondary piRNAs that are loaded into Ago3 (Brennecke et al., 2007; Gunawardane et al., 2007; Li et al., 2009). Similarly, Ago3 target cleavage can direct the loading of new antisense piRNAs into Aub, leading to amplification of piRNAs that target active TEs (Huang et al., 2014). The ping-pong model has very strong support from analysis of piRNA sequences in Aub and Ago3 complexes, however the molecular mechanism the protein complexes involved in the individual steps of ping-pong processing remain unknown.
Using static and dynamic imaging, we found that Aub and Ago3 have different sub-compartments within nuage. We reveal that Aub and Ago3 are recruited to nuage through two different mechanisms. Ago3 is recruited to nuage through direct interaction with Krimper (Krimp), a stable component of nuage granules that is able to form granules in the absence of other nuage components. Furthermore, Krimp coordinates assembly of the ping-pong piRNA processing (4P) complex by directly interacting with both Ago3 and methylated Aub. Together, our results lead to a model that explains nuage assembly in the context of the piRNA pathway and reveals molecular details of Aub and Ago3 interaction during ping-pong.
RESULTS
Aub and Ago3 have distinct perinuclear localizations in nuage
Aub and Ago3 are the central components of the cytoplasmic piRNA pathway, as both proteins are non-redundantly required for the repression of TEs (Brennecke et al., 2007; Li et al., 2009; Malone et al., 2009). Analysis of the piRNA sequences associated with Aub and Ago3 led to the ping-pong model, which proposes that the RNAs produced by Aub-dependent cleavage are transferred into unloaded Ago3 protein. However, the molecular mechanism of this transfer remained unknown. We wanted to understand the mechanism of interaction between Aub and Ago3 in the piRNA pathway. We started by examining the subcellular localization of Aub and Ago3 in nurse cells of Drosophila ovaries.
Previous studies that examined the subcellular localization of Aub and Ago3 proteins using immunofluorescence microscopy led to the conclusion that both proteins are co-localized to the same subcellular compartment, perinuclear nuage granules (Brennecke et al., 2007; Gunawardane et al., 2007; Harris and Macdonald, 2001; Li et al., 2009; Malone et al., 2009). To precisely characterize the subcellular localization of Aub and Ago3, we imaged both proteins fused to fluorescent protein tags. We generated transgenic flies expressing Aub and Ago3 fused to GFP, or the red fluorescent protein, mKate2 (mK2) and expressed them in germ cells of fly ovary using the Maternal α-Tubulin 67C (MαG4) driver (Bossing et al., 2002) (Figure 1A). Tagged Aub and Ago3 are associated with piRNAs (Figure 2A) and the profile of piRNA associated with tagged proteins was identical to those associated with endogenous proteins (data not shown). We also expressed GFP-tagged Aub under the control of its native regulatory region: this transgene rescues the sterility of aubHN/QC mutant females indicating that it is fully functional. Importantly, we found that the subcellular localizations and dynamics of GFP-Aub driven by the MαG4 driver or its native regulatory regions were identical and are not affected by its overexpression using the driver (Figure S1A, S2D, S2E). Therefore, GFP-tagged Aub and mK2-tagged Ago3 provide us with a tool to investigate their subcellular localization with exceptional quality.
Figure 1. Aub and Ago3 have distinct subcellular localizations.
A) The localization pattern of fluorescent protein tagged Aub and Ago3 in Drosophila ovary. Aub and Ago3 are localized in the perinuclear compartment in nurse cells called nuage. Aub appears as a smooth perinuclear layer, while Ago3 forms punctate granules around nuclei. GFP-Aub and mK2-Ago3 were co-expressed in nurse cells of ovaries using the Maternal α-Tubulin VP16-Gal4 (MαG4) driver. DNA is stained with DAPI (blue). Scale bar: 20µm.
B) Ago3 is more concentrated in nuage, while Aub is more dispersed in the cytoplasm. The fluorescence signals of GFP-Aub and GFP-Ago3 were quantified in nuage and cytoplasm (regions with identical surface area boxed in the figure – see Supplemental Experimental Procedures for details). Shown is the mean ratio of nuage to cytoplasmic signal measured for 81 and 80 individual nurse cells, respectively. Standard deviation from the mean is shown for each protein.
C) The localization of Ago3 in nuage is more granular compared to Aub. Signal intensities of GFP-tagged Aub and Ago3 were measured in the perinuclear space of 50 nurse cells. Box plots show the coefficient of variation distribution of GFP signals for individual cells (boxes show the upper and lower quartiles; whiskers depict the highest and lowest data points within 1.5-times the interquartile ranges).
D) The profiles of Aub and Ago3 localization around a single nucleus are distinct. Signal intensities of Aub (GFP-Aub) and Ago3 (mK2-Ago3) were traced along the perimeter of the nurse cell nucleus marked with an asterisk in Figure 1A. The line intersecting the perinuclear region marks the start of measurements, continuing clockwise around the perimeter. Localization of Aub and Ago3 with reciprocally swapped fluorescent tags can be found in Figure S1B.
Figure 2. Aub requires a piRNA guide for localization to nuage.
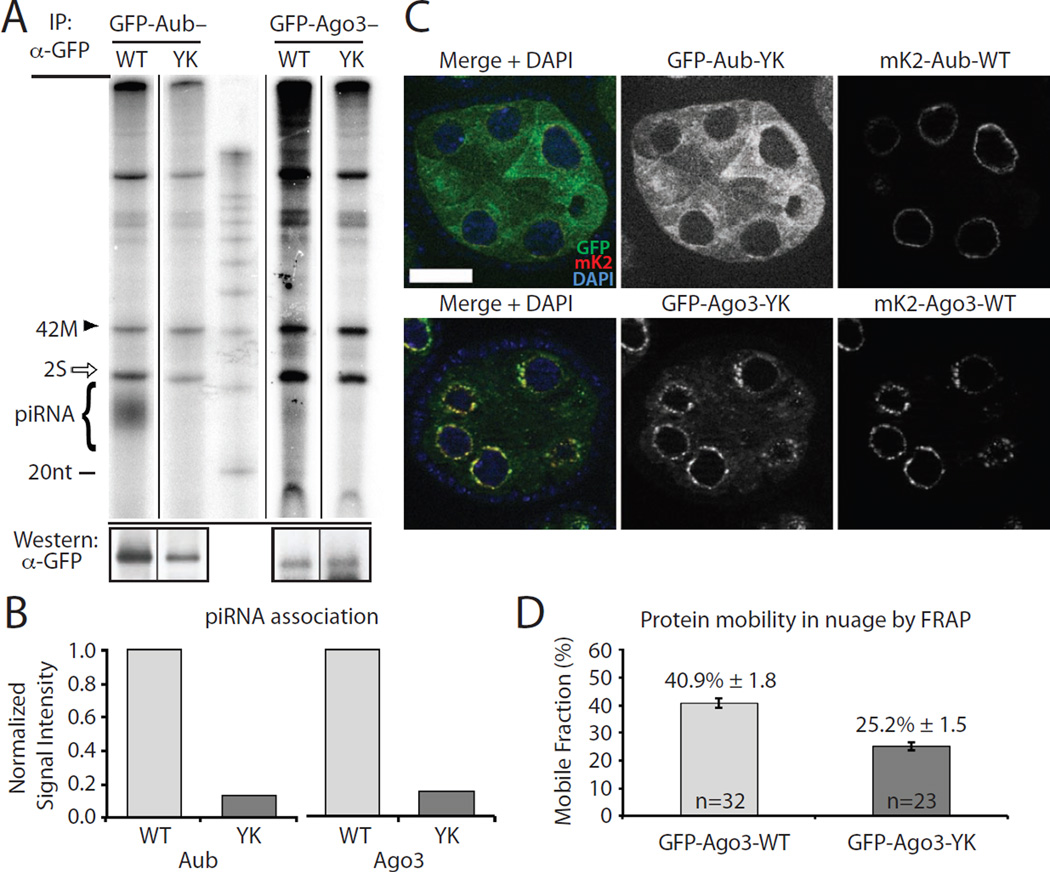
A) Aub-YK and Ago3-YK point mutants associate with less piRNAs compared to their wild type (WT) counterparts. GFP-tagged proteins and were expressed in ovaries, immunoprecipitated and their associated small RNA isolated and 5′ radiolabeled. The strong band at 30 nt is abundant 2S ribosomal RNA (rRNA) (white arrow). A 42 nt RNA (42M) was spiked into each immunopurified sample to control for labeling efficiency and RNA loss during isolation. The levels of immunopurified proteins are shown by western blot (lower panels).
B) Quantification of associated piRNA of wild type and mutant Aub and Ago3 shown on panel (A). The signals of radiolabeled piRNA were normalized to the amount of immunoprecipitated protein and to the amount of spike RNA. See Supplemental Experimental Procedures for details.
C) The piRNA-binding deficient Aub-YK does not localize to nuage, but is diffused in the cytoplasm. Wild type protein tagged with mKate2 (mK2) was co-expressed in the same cells (upper panel). In contrast to Aub, the localization of piRNA-binding deficient Ago3-YK is indistinguishable from that of wild type Ago3. DNA is stained with DAPI (blue). Scale bar: 20µm.
D) The mobility of the piRNA-binding deficient Ago3-YK mutant is decreased in nuage. The fractions of mobile wild type and Ago3-YK proteins were measured by FRAP on 32 and 23 independent cells, respectively. Bars show standard error values. See also Figure S2 for additional information on FRAP analysis.
We observed important differences in the localization of Aub and Ago3. First, unlike Ago3, Aub also exhibits a diffused cytoplasmic localization. Indeed, quantification of the signals originating from the perinuclear nuage and the cytoplasm reveal that Ago3 is more enriched in nuage compared to Aub (Figure 1B). Second, we found that the localization of the two proteins within nuage is different. Aub localizes around the nucleus as a smooth continuous layer with some regions of higher density. The localization of Ago3 is more punctate, with the protein concentrated in several distinct granules surrounding the nucleus. To quantitatively examine the granularity of Aub and Ago3 in the perinuclear region, we calculated the coefficient of variation (the ratio of standard deviation and mean fluorescence signal) around 50 nuclei for each protein. This measurement is an indicator of granularity/unevenness of the protein localization in nuage. In agreement with visual observation, the quantitative measurement showed that Ago3 localization in nuage is more granular compared to Aub (Figure 1C).
Simultaneous imaging of both proteins tagged with different fluorophores showed that Ago3 granules have high concentrations of Aub protein. Conversely, nuage regions with a significant concentration of Aub might have very low Ago3 protein levels (Figure 1D). To check if fluorophore tags affect our observations, we reversed the orientation of fluorescent tags on Aub and Ago3 and observed the same characteristic localizations of Aub and Ago3 (Figure S1B). These results indicate that Aub and Ago3 have distinct localization patterns in nuage, with Aub being distributed more uniformly, and Ago3 being concentrated to granules that also contain Aub.
Aub, but not Ago3 requires a piRNA guide for localization to nuage
The distinct localization patterns of Aub and Ago3 in nuage suggested that different mechanisms recruit each protein to this subcellular compartment. Both Aub and Ago3 are loaded with piRNAs that recognize complementary RNA molecules. We decided to test if recognition of complementary RNA targets, which was proposed to occur in nuage, is required for recruitment of Aub and Ago3 to this compartment (Klattenhoff et al., 2009; Malone et al., 2009). Argonaute proteins lacking small RNA guides are not able to recognize complementary targets. To test the role of target recognition in the localization of Aub and Ago3, we generated transgenic flies that express mutant proteins (Aub-YK: [Y568L, K572E]; and Ago3-YK: [Y578L, K582E]), which are unable to bind piRNAs due to point mutations in the conserved Y-K-K motif of the 5′ piRNA binding pocket (Djuranovic et al., 2010; Ma et al., 2005; Parker et al., 2005). Purification and labeling of RNA associated with Aub-YK and Ago3-YK showed that they indeed do not associate with piRNAs (Figure 2A, 2B). We found that piRNA-deficient Aub was completely delocalized from nuage and was instead dispersed throughout the cytoplasm (Figure 2C). Surprisingly, localization of piRNA-deficient Ago3-YK in nuage was unperturbed. These results indicate that the mechanism of Ago3 recruitment to nuage is different from that of Aub and does not require piRNA loading or interaction with target RNA molecules.
To further understand the role of piRNA guides in Ago3 recruitment to nuage, we used Fluorescence Recovery After Photobleaching (FRAP) to study the dynamics of wild type and piRNA-deficient Ago3 in nuage. Perinuclear GFP-Ago3 granules were photo-bleached and the recovery of fluorescence signal was measured by time-lapse imaging. Analysis of fluorescence recovery allowed us to determine the mobile fraction of Ago3 in nuage, or the percentage of protein that is free to exchange. GFP-Ago3 is dynamic in nuage with the mobile fraction being 40.9 ± 1.8%. The absence of a piRNA guide significantly decreases the mobile fraction of Ago3-YK to 25.2 ± 1.5% (Figure 2D, Figure S2). This result indicates that association with piRNA enhances the mobility of Ago3 protein to exchange between nuage and cytoplasm. Therefore, the mechanisms of Aub and Ago3 recruitment to nuage seem to be opposite: Aub requires association with piRNA, while piRNA is dispensable for Ago3 localization. In contrast to Aub that loses its nuage localization when devoid of piRNAs, Ago3 becomes more restrained in nuage in the absence of a piRNA guide.
Ago3 is recruited to nuage by Krimper
The recruitment of Ago3 to nuage does not require a piRNA guide, so we reasoned that an interaction with one or several proteins known to localize in nuage might recruit Ago3. Since we found that Aub and Ago3 have distinct localization patterns in nuage, we examined the localization of other known nuage proteins using fluorescent protein tags (Figure 3A). We quantitatively measured the granularity of their distribution in nuage: Tudor (Tud), Spindle-E (SpnE) and Vasa (Vas), three well-characterized nuage components, had smooth nuage localization similar to that of Aub. In contrast, other proteins such as Krimper (Krimp), Tejas (Tej) and Qin/Kumo (Qin) had more granular pattern that resembled the Ago3 distribution (Figure 3B). We reasoned that these proteins might be involved for the recruitment of Ago3 to nuage.
Figure 3. Localization and dynamics of Krimper in nuage.
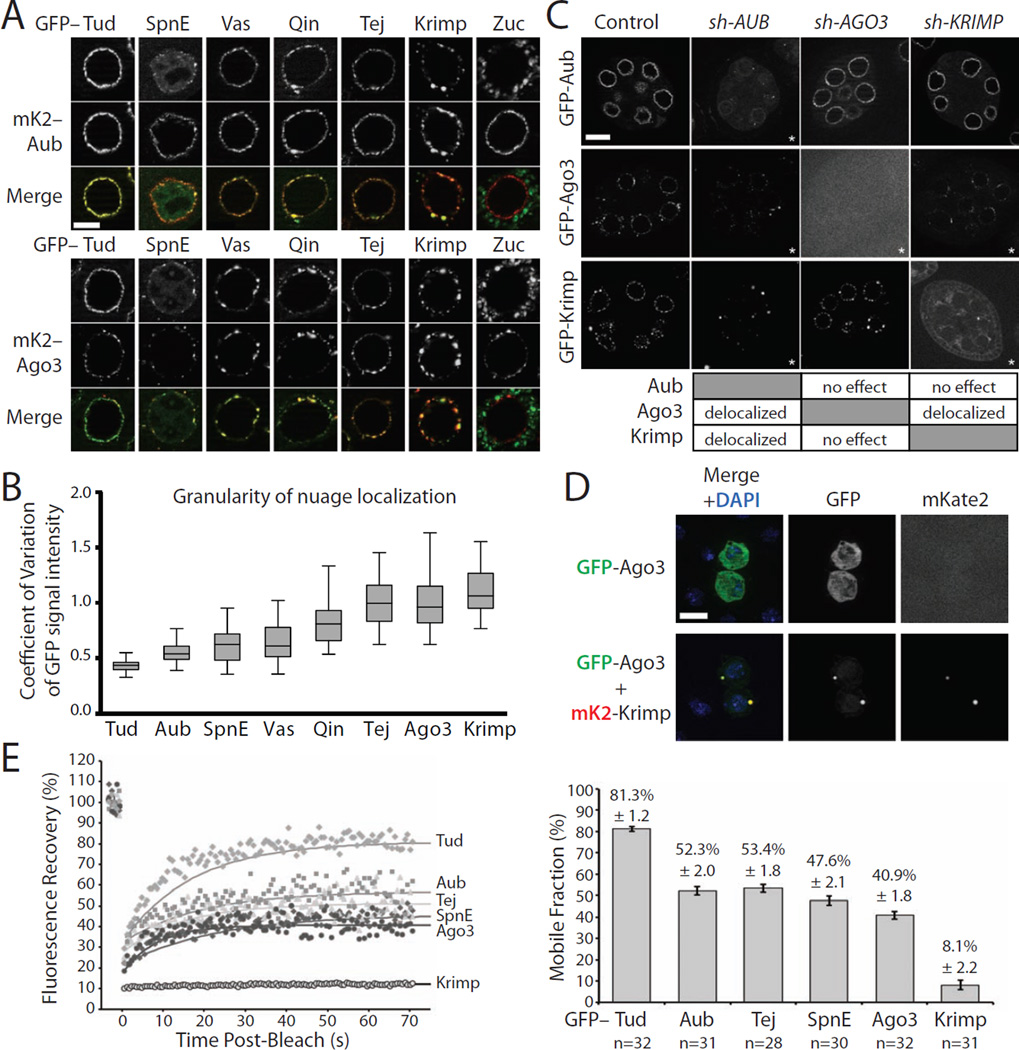
A) Co-localization of Aub and Ago3 with other nuage components. mKate2-tagged Aub (upper panel) or Ago3 (lower panel) were co-expressed in nurse cells of Drosophila ovaries with GFP-tagged piRNA pathway components (green) using the same MαG4 driver. All components with exception of Zuc co-localize with Aub and Ago3 in nuage. Scale bar: 10µm.
B) The granularity of nuage localization of different proteins measured by the coefficient of variation of GFP signal intensity. For each protein, GFP signal intensities were measured in the perinuclear space of 50 nurse cells. Box plots show the coefficient of variation distribution of GFP signals in individual cells (boxes show the upper and lower quartiles; whiskers depict the highest and lowest data points within 1.5-times the interquartile ranges). Krimp, Ago3 and Tej show uneven granular distribution in nuage, while Aub, Tud, Spn-E and Vas have a smooth distribution; Qin localization is intermediate between these two groups.
C) The genetic interaction between Aub, Ago3 and Krimp. Ovaries expressing GFP-tagged Aub, Ago3 and Krimp were reciprocally crossed to shRNA knockdown lines targeting each protein. The asterisk in each image indicates perturbed localization of the corresponding protein compared to wild type. Aub is required for Ago3 and Krimp nuage localization, while Aub localizes independently of Krimp and Ago3. Krimp is required for both localization and normal protein level of Ago3, but its localization is unchanged upon Ago3 depletion.
D) Krimp recruits Ago3 into cytoplasmic granules in S2 cells. S2 cells were transfected with plasmids encoding GFP-tagged Ago3 (green) alone or in combination with mKate2-tagged Krimp protein (red). GFP-tagged Ago3 is diffused in the cytoplasm when expressed alone. The expression of Krimp induces Ago3 recruitment into Krimp cytoplasmic granules. DNA is stained with DAPI (blue). Scale bar: 10µm.
E) Krimp is a stationary component of nuage granules. FRAP shows large differences in the mobility of different proteins in nuage. All proteins were tagged with GFP and expressed in germ cells using the MαG4 driver. Representative FRAP experiments are shown on the left. The mobile fraction of each protein was determined by modeling the recovery to an exponential recovery curve superimposed on respective datasets. The quantification of the mobile fractions for each of the proteins is shown on the right (n indicates the number of independent experiments used for FRAP measurements). More than 80% of Tud is mobile in nuage, while the mobile fractions of Aub, Tej, SpnE and Ago3 are between 40 and 50%. Krimper is the least mobile component measured, with a mobile fraction of ~8%. Bars show standard error values.
To test the role of candidate proteins in Ago3 recruitment to nuage, we monitored localization of Ago3 and Aub after knocking down Vas, Tej, SpnE, Krimp and Tud in germ cells. We verified the efficiency of each knock-down using two independent methods (for details see Experimental Procedures). Among the proteins we targeted for depletion, only depletion of Krimp affected the localization of Ago3 but not that of Aub (Figure 3C). In agreement with previous studies, we found that Krimp depletion caused mislocalization of Ago3 from perinuclear nuage and a decrease in Ago3 protein level that is particularly apparent in later stages of oogenesis (Lim and Kai, 2007; Malone et al., 2009; Nagao et al., 2011). Importantly, Aub localization in nuage is not affected by Krimp depletion, supporting the conclusion that Aub and Ago3 are recruited to nuage through two different mechanisms.
Krimp was shown to be involved in piRNA-mediated repression of TEs, however, its biochemical function remained unknown (Lim and Kai, 2007; Malone et al., 2009; Nagao et al., 2011). To test if Krimp is directly responsible for recruitment of Ago3 into nuage, we used S2 cells that do not normally express Aub, Ago3, or the majority of other nuage components. First, we expressed GFP-tagged versions of each protein and monitored their subcellular localization. In contrast to germ cells, Ago3 had a uniform cytoplasmic distribution in S2 cells, indicating that it is not capable of forming granules in the absence of other nuage components or piRNA (Figure 3D). Conversely, Krimp forms prominent cytoplasmic granules when expressed in S2 cells, indicating that it is able to form granules independently of other nuage proteins and a functional piRNA pathway. It should be noted that Krimp granules observed in S2 cells are randomly distributed in the cytoplasm and are not perinuclear; therefore, they are distinct from genuine nuage granules formed in nurse cells of the ovary. Importantly, co-expression of Krimp with Ago3 in S2 cells sequesters Ago3 into Krimp cytoplasmic granules (Figure 3D). This result, together with the Krimp depletion experiments described above, demonstrate that Krimp is both necessary and sufficient for cytoplasmic granules assembly, and is able to recruit Ago3 into these compartments.
To further understand the role of Krimp in the formation of nuage, we measured the mobility of Krimp using FRAP and compared it to the mobility of other nuage components. These experiments showed that Krimp protein is extremely stable in nuage granules with only 8.1% ± 2.2% of the protein being mobile (Figure 3E). In contrast to Krimp, the mobile fractions of other tested proteins, Aub, Ago3, Tud, Tej and Spn-E, exceeded 40%. This result indicates that Krimp can be considered as a structural component of nuage granules. Together, our experiments implicate Krimp as a stable component of nuage that is required for recruitment of Ago3 into this subcellular compartment.
Tudor domains of Krimper bind the N-terminal region of Ago3
The ability of Krimp to form granules in S2 cells suggested that it is able to multimerize. Using co-immunoprecipitation of the Krimp protein tagged with two different tags we found that it is capable of forming dimeric or multimeric complexes. The N-terminal fragment of Krimp (aa 1–310), which lacks any characterized domains, co-purified and co-localized in granules with full-length Krimp, indicating that this region is required for Krimp–Krimp interactions and granule formation (Figure 4A, S3A, S3B, S4B).
Figure 4. Separate domains of Krimper are responsible for dimerization and association with Ago3.
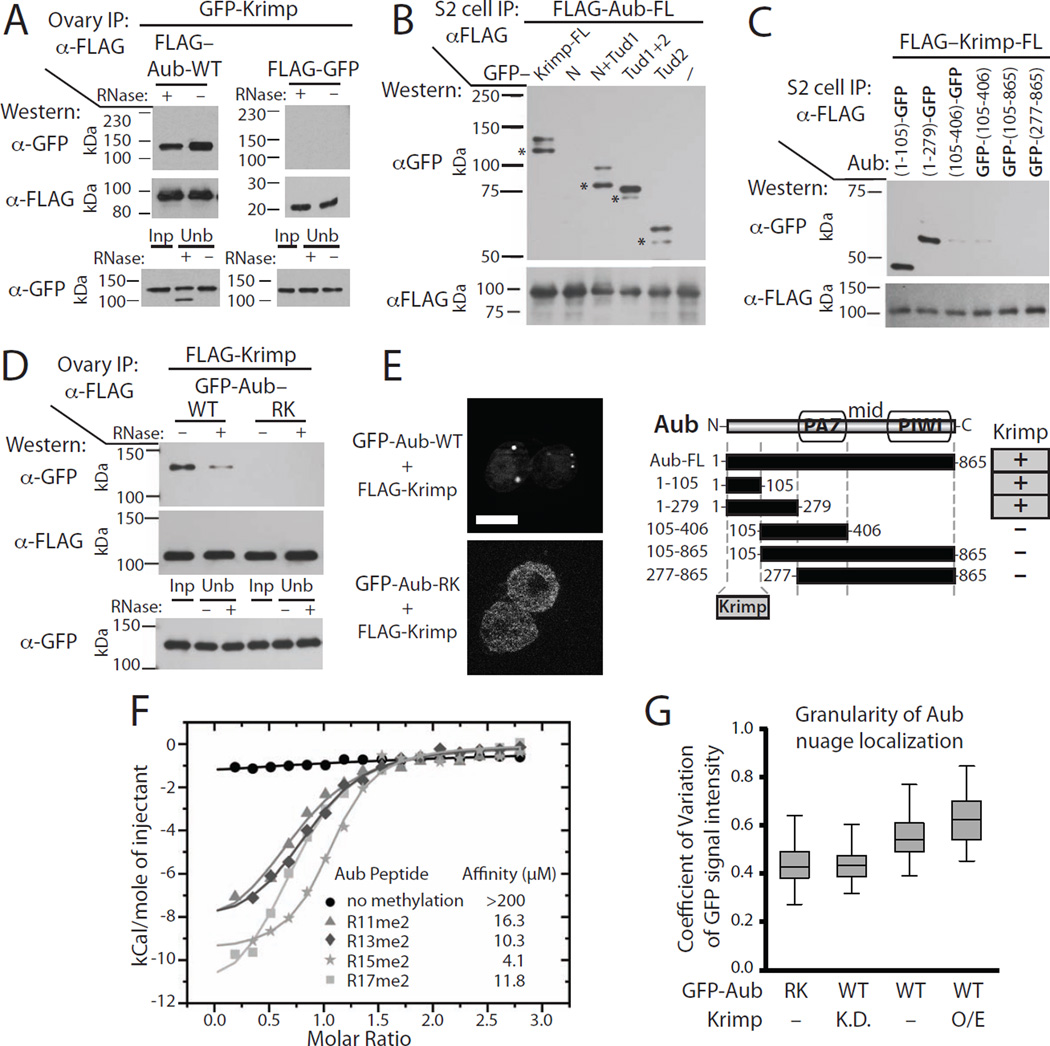
A) The N-terminal region of Krimp is required for Krimp dimerization. FLAG-tagged full-length Krimp (Krimp-FL) was co-expressed with GFP-tagged fragments of Krimp (shown in the diagram below) in S2 cells. Fragments of Krimp containing the first 310 amino acid residues co-purified with Krimp-FL, while fragments lacking N-terminal residues did not co-purify.
B) Krimp forms a complex with Ago3 in Drosophila ovary independently of arginine methylation, or RNA. FLAG-Krimp was co-expressed in fly ovaries along with wild type (GFP-Ago3-WT), arginine-methylation deficient (GFP-Ago3-RK), and piRNA-binding deficient Ago3 (GFP-Ago3-YK). Krimp was immunoprecipitated with anti-FLAG beads in the presence (+) or absence (−) of RNase A followed by Western blotting to detect GFP- and FLAG-tagged proteins. The lower panel shows Input (Inp) and the unbound fractions (Unb). Additional experimental controls are provided in Figure S3C.
C) Tudor domains of Krimp bind Ago3. FLAG-tagged full length Ago3 (FLAG-Ago3-FL) was co-expressed with GFP-tagged fragments of Krimp in S2 cells. FLAG-Ago3 was immunoprecipitated followed by Western blotting to detect GFP-Krimp fragments. Ago3 co-purifies with fragments of Krimp containing either of its two Tudor domains, but not with the N-terminal fragment responsible for dimerization.
D) Krimp interacts with the N-terminal fragment of Ago3. FLAG-tagged full length Krimp was co-expressed with GFP-tagged fragments of Ago3 in S2 cells. The N-terminal Ago3 fragment containing the first 296 amino acid residues co-purifies with Krimp, while other Ago3 fragments are not able to interact with Krimp.
E) Ago3 in complex with Krimp is not bound to piRNA. Total Ago3 and Ago3-Krimp complexes were purified from Drosophila ovaries. Co-purified RNA was labeled and resolved on the gel (top panel), while the amount of Ago3 and Krimp proteins in immunoprecipitates was determined by Western blotting (middle panels). The amount of piRNA in each immunoprecipitate was normalized to the amount of Ago3 protein (lower panel). An unlabeled 42 nt RNA (42M) was spiked into each sample to control for labeling efficiency and RNA loss.
The ability of Krimp to recruit Ago3 to granules suggests that the two proteins might interact. We examined Ago3 interactions with Krimp and other components of the piRNA pathway by co-immunoprecipitations of tagged proteins from Drosophila ovaries. We found that immunoprecipitation of Krimp co-purifies Ago3, while immunoprecipitations of several other nuage components, Spn-E, Mael, Qin and Vasa, did not co-purify with Ago3 under the same conditions (Figure. 4B, S3). We also found that Krimp interacts with Ago3 when tagged proteins are transiently expressed in Drosophila S2 cells (Figure 4C, S3F, S4B). This result suggests a direct interaction between the two proteins. To further dissect the interaction between Krimp and Ago3, we mapped the interaction domains on both proteins using co-immunoprecipitation in S2 cells. The C-terminal portion of Krimp contains two putative Tudor domains. Krimp fragments that contain at least one of the two Tudor domains are able to interact with Ago3 (Figure 4C, S3F, S4B). In contrast, the N-terminal fragment of Krimp involved in its multimerization is not necessary for interaction with Ago3.
Next, we identified the region of Ago3 that interacts with Krimp. Piwi proteins contain PIWI, PAZ and Mid domains responsible for target cleavage, and binding of the 3′ and 5′ end of piRNA, respectively (Ma et al., 2004; Ma et al., 2005; Song et al., 2004). Upstream of the PAZ domain is the N-terminal region for which the structure is not known. We found that the N-terminal fragment of Ago3, consisting of the first 296 amino acids, is sufficient to interact with Krimp (Figure 4D). The N-terminal fragment of Ago3 is recruited to Krimp granules when co-expressed with full length Krimp in S2 cells supporting that this fragment mediates Krimp-Ago3 interaction (Figure S3G). Overall, we show that Krimp is able to aggregate through its N-terminal region, while its Tudor domains are responsible for binding the N-terminal region of Ago3. Other Tudor domain-containing proteins were detected to bind the symmetrically dimethylated arginine residues (sDMA) of Ago3 (Liu et al., 2011; Nishida et al., 2009). To test if arginine methylation is required for interaction between Krimp and Ago3, we mutated three arginine residues in the N-terminal region of Ago3 shown to be targets of symmetrical dimethylation (R68K, R72K, R79K). When expressed in fly ovaries, the methylation-deficient Ago3-RK mutant protein interacts with Krimp, indicating that arginine methylation of these residues is dispensable for binding Krimp (Figure 4B, S3C). Additionally, mutation of arginine methylation sites did not disrupt recruitment of Ago3 to Krimp granules in S2 cells (Figure S3H).
To determine if Ago3 associated with Krimp is loaded with piRNAs, we labeled small RNAs isolated from Krimp-Ago3 complexes purified from fly ovaries. While piRNAs can be easily detected in total Ago3 immunopurified from ovarian lysate, similar levels of Ago3 protein from Krimp immunoprecipitation were devoid of piRNA, indicating that Ago3 associates with Krimp in its unloaded state (Figure 4E). Furthermore, the Ago3 mutant deficient in piRNA binding (Ago3-YK) co-immunoprecipitates with Krimp, indicating that piRNA binding is not required for interaction between two proteins (Figure 4B, S3C). In contrast to Aub, the Ago3 mutant deficient in piRNA binding (Ago3-YK) still localizes to nuage, further supporting the idea that piRNA loading is not required for interaction between Krimp and Ago3 (Figure 2C). Together, our results suggest that the Krimp-Ago3 complex is mostly free of piRNA, and loading of piRNA into Ago3 might lead to dissociation of the two proteins.
Krimp interacts with Aub by direct binding to N-terminal methylated arginine residues
The ping-pong model proposes an interaction between Aub and Ago3 proteins, since Aub loaded with piRNA cleaves target RNA that is then loaded as piRNA substrates into Ago3 (Aravin et al., 2007; Brennecke et al., 2007; Gunawardane et al., 2007). As Krimp recruits Ago3 to nuage, we tested if it also interacts with Aub. Co-immunoprecipitations of tagged Aub and Krimp showed that they indeed interact when expressed in Drosophila ovary and S2 cells (Figure 5, S4). We further dissected the domains responsible for the interaction between each protein. Similar to the interaction between Krimp and Ago3, the N-terminal fragment of Aub interacts with the Tudor domains of Krimp and localizes to Krimp granules in S2 cells (Figure 5B, 5C, S4). However, in contrast to Ago3, the interaction between Krimp and Aub requires methylation of Aub N-terminal arginine residues, since mutating these residues to lysine (R11K, R13K, R15K, R17K, R26K) completely abolished the Aub-Krimp interaction in ovaries (Figure 5D, S4D). The interaction of Krimp with Aub is further confirmed by ability of Krimp to recruit Aub into cytoplasmic granules in S2 cells. Importantly, methylation-deficient Aub-RK mutant protein is not recruited to Krimp granules, supporting the requirement of arginine methylation for Aub to interact with Krimp (Figure 5E). To further study the interaction between Krimp and methylated Aub, we measured the affinity of symmetrically dimethylated (sDMA) and non-methylated Aub peptides with the C-terminal Tudor domain of Krimp using isothermal titration calorimetry (ITC). The direct measurement of interactions confirmed that the C-terminal Tudor domain (Tud2) of Krimp binds methylated, but not unmethylated Aub peptides, with each of the four arginine residues (R11me2; R13me2; R15me2; R17me2) being sufficient for interactions (Figure 5F, S4E).
Figure 5. Krimper binds Aub through recognition of methylated arginine residues by Krimp Tudor domain.
A) Krimp forms a complex with Aub in Drosophila ovary. FLAG-Aub and GFP-Krimp were co-expressed in fly ovaries. FLAG-Aub or control (FLAG-GFP) proteins were purified using anti-FLAG beads in the presence (+) or absence (−) of RNase A followed by Western blotting to detect GFP- and FLAG-tagged proteins. The interaction between Aub and Krimp is independent of RNA. The lower panel shows Input (Inp) and the unbound (Unb) fractions.
B) The Tudor domains of Krimp interact with Aub. FLAG-Aub was co-expressed with GFP-tagged fragments of Krimp protein in S2 cells. Krimp fragments containing either of the two Tudor domains co-purify with Aub. Asterisks indicate bands corresponding to degradation products of GFP-Krimp.
C) The N-terminal fragment of Aub binds Krimp. FLAG-Krimp was co-expressed with GFP-tagged fragments of Aub in S2 cells. The N-terminal fragment of Aub containing the first 105 amino acid residues co-purifies with Krimp, while other Aub fragments lacking this region did not co-purify.
D) Krimp interaction with Aub requires arginine methylation. FLAG-Krimp was co-expressed in fly ovaries with either wild type (GFP-Aub-WT) or arginine-methylation deficient Aub (GFP-Aub-RK). FLAG-Krimp was immunoprecipitated with anti-FLAG beads in the presence (+) or absence (−) of RNase A. The lower panel shows Input (Inp) and the unbound fractions (Unb).
E) Krimp recruitment of Aub into cytoplasmic granules in S2 cells requires arginine residues on Aub N-terminus. S2 cells were transfected with plasmids encoding GFP-tagged wild type (GFP-Aub-WT) or methylation deficient Aub (GFP-Aub-RK) in combination with FLAG-tagged Krimp protein. While GFP-tagged Aub-RK is diffused in the cytoplasm, wild type, but not arginine methylation deficient Aub-RK, is recruited to cytoplasmic Krimp granules. Scale bar: 10µm.
F) Krimp Tud2 domain binds symmetrically di-methylated arginine-containing Aub N-terminal peptides. The binding affinity of unmethylated and symmetrically di-methylated Aub peptides for Krimp Tud2 domain was measured by Isothermal Titration Calorimetry (ITC). Aub peptide with symmetrically di-methylated arginine 15 (R15me2) binds with highest affinity (4.1 µM), while symmetrically di-methylated arginine 11 (R11me2) has the weakest affinity (16.3 µM) for Krimp Tud2 domain. Unmethylated peptide shows little affinity to Krimp Tud2 protein. See also Figure S4E.
G) Aub granular localization in nuage is promoted by arginine methylation and the presence of Krimp protein. The GFP signal intensities of arginine-methylation deficient (Aub-RK) or wild type (WT) Aub protein upon Krimp knock-down or overexpression was measured in the nuage of 50 nurse cells. Box plots show the coefficient of variation distribution of GFP signals in individual cells (boxes show the upper and lower quartiles; the highest and lowest data points within 1.5-times the interquartile ranges). Methylation deficient Aub-RK shows less granular distribution in nuage compared to wild type Aub. Similarly, knock-down (K.D.) of Krimp leads to less granular localization of wild type Aub, while Krimp over-expression (O/E) enhances granular localization.
Unlike Ago3, the localization of Aub in the nuage of germ cells from fly ovaries was not drastically affected by Krimp depletion (Figure 3C). To study in greater detail the role of Krimp on the localization of Aub protein in nuage, we quantitatively assessed the localization of Aub in germ cells that were depleted for, or were overexpressing Krimp protein (Figure 5G). Overexpression of Krimp caused an increase in the granular localization of Aub in nuage. Conversely, Krimp depletion led to a smoother perinuclear distribution of Aub around the nucleus. Importantly, methylation-deficient Aub-RK mutant protein also had smoother localization in nuage compared to wild type protein. This result suggests that Krimp influences Aub localization in nuage through its interactions with methylated arginine residues in the N-terminal region of Aub. Overall, our results indicate that while Krimp is not required for the initial recruitment of Aub to nuage (as it is for Ago3), Krimp does play an important role to anchor Aub in prominent granules enriched for Krimp and Ago3. Taken together, the genetic and imaging experiments are supported by biochemical data that show the interaction of Krimp with N-terminal sDMA residues of Aub is important for the formation of nuage granules.
Krimper mediates the formation of a complex between Aub and Ago3 and is essential for ping-pong piRNA biogenesis
Our data show that Krimp is able to interact with both Ago3 and Aub using its Tudor domains, while its N-terminal domain is responsible for aggregation (Figure 4A, 4C, 5B, S3, S4). Furthermore, Krimp is required for the recruitment of Ago3 to nuage and also anchors methylated Aub in prominent nuage granules (Figure 3C, 5D, 5E, S4D). Therefore, Krimp should be able to mediate formation of an Aub/Krimp/Krimp/Ago3 complex that places Aub and Ago3 in close physical proximity. To test the ability of Krimp to mediate the interaction of Aub and Ago3, we co-expressed fluorescently tagged Aub and Ago3 in S2 cells. In the absence of Krimp, both Aub and Ago3 were dispersed in the cytoplasm, however, co-expression of Krimp led to recruitment of both Aub and Ago3 into distinct cytoplasmic granules (Figure S5A). Therefore, Krimp is sufficient to mediate the co-localization and physical proximity between Aub and Ago3. To directly test if Krimp forms a complex that contains both Aub and Ago3 simultaneously, we sequentially immunopurified Aub, followed by Krimp, from ovarian lysate expressing FLAG-tagged Aub and GFP-tagged Krimp (Figure S5B). After elution of purified Aub complexes, a subsequent purification step was used to recover complexes that also contained Krimp. Western blotting revealed the presence of Ago3 protein in Aub-Krimp complexes, indicating that a triple complex containing all three proteins does indeed exist (Figure S5B).
Taken together, our data suggest that Krimp mediates interactions between Aub and Ago3 necessary for the formation of an Aub/Ago3 complex. This complex, which contains both Aub and Ago3, could be essential for a key step of the ping-pong piRNA amplification process: coupling Aub-induced RNA target cleavage with the loading of RNA into Ago3 (Figure 6A). If Krimp indeed plays this role, its deficiency should disrupt ping-pong piRNA processing. To test the effect of Krimp deficiency on the ping-pong cycle, we analyzed published piRNA profiles of Krimp, Ago3, Aub and Piwi mutants (Li et al., 2009; Malone et al., 2009). We examined the output of ping-pong processing using two different tests. The first test measures the fraction of secondary piRNAs, determined by the presence of adenine residue at position 10 and the absence of uridine residue at position one, among all piRNA (Figure 6A, 6B, S5C). The second test determines the fraction of piRNAs found in ping-pong pairs (Figure 6B, S5C) (for details see Experimental Procedures). As expected from previous studies, Piwi mutation led to a large decrease in piRNA levels, but it did not have a significant effect on ping-pong processing as measured by these two tests. In contrast to Piwi, deficiency in Aub completely disrupted ping-pong processing, while Ago3 mutation had a relatively mild effect on ping-pong: in fact, the fraction of secondary piRNA and piRNA engaged in ping-pong pairs remains mostly unperturbed in Ago3 mutants (Figure 6B, S5C). This result closely matches a previously published conclusion that heterotypic ping-pong between Aub and Ago3 observed in wild type flies is replaced by homotypic Aub-Aub ping-pong in Ago3 mutants (Li et al., 2009; Zhang et al., 2011). Similar to the three Piwi mutants, Krimp mutant showed severe defects in its piRNA profile with a large reduction in the amount of piRNA against multiple TE families. Interestingly, our analysis shows that Krimp deficiency essentially eliminates ping-pong processing as measured by a decrease in the fraction of secondary piRNA and the fraction of piRNA present in ping-pong pairs. Indeed, the effect of Krimp deficiency on ping-pong processing was similar to that observed for Aub mutant, and was significantly stronger than the effects observed for Ago3 deficiency. This result suggests that Krimp is indeed required for ping-pong amplification. Furthermore, the function of Krimp is not restricted to heterotypic Aub-Ago3 ping-pong, but is also required for homotypic Aub/Aub ping-pong. Overall, the analysis of piRNA profiles supports the central role of Krimp in ping-pong amplification. Therefore, we propose to name the complex that includes Krimp, Aub and Ago3 proteins as ping-pong piRNA processing (4P) complex (Figure 7).
Figure 6. Krimper is required for ping-pong piRNA processing.
A) Complementary piRNA pairs formed by ping-pong processing are separated by 10nt. Primary piRNA has a uridine bias at position one. Corresponding secondary piRNA has an adenine bias at position 10 and lacks nucleotide bias at the first position. The graphs below show the fraction of complementary piRNA overlap at their 5′ ends. The predominant peak at 10nt is seen in wild type piRNA populations indicating a large fraction of piRNA overlap by 10 nt at their 5′ ends. This peak is absent in piRNA from Aub and Krimp mutants indicating failure of ping-pong piRNA processing.
B) Krimp is required for ping-pong piRNA processing. The fraction of secondary piRNAs and the fraction of the ping-pong pairs that have 10nt overlap were analyzed in Krimp, Aub, Ago3 and Piwi mutants and control (heterozygous) flies. The changes in mutant compared to control for each family of transposable elements are shown on the heat map. Only TE families with high levels of piRNA expression and abundance in ping-pong pairs are shown. The same analysis for all TE families is shown in Figure S5C.
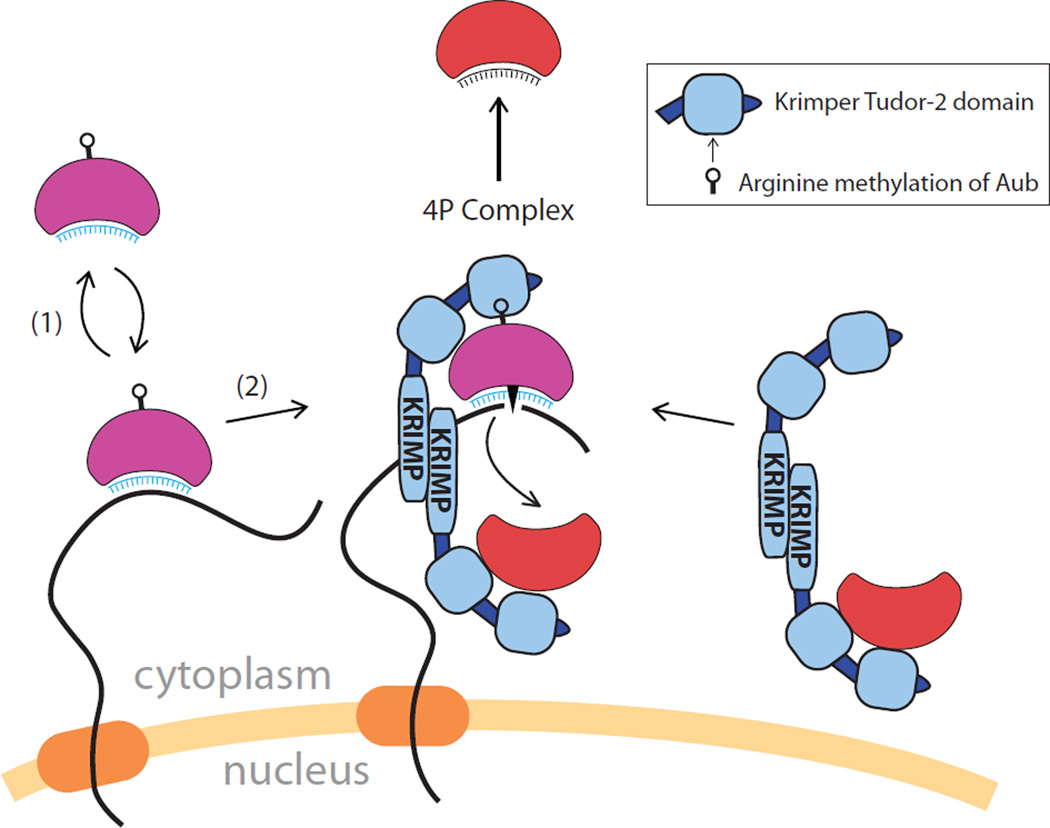
Figure 7. A model for recruitment of Aub and Ago3 to nuage and formation of the ping-pong piRNA processing (4P) complex.
Aub-piRNA complexes shuttle between the cytoplasm and nuage interrogating RNA transcripts as they exit the nucleus. The recognition of complementary RNA targets by the piRNA guide recruits Aub to nuage. Ago3 that is devoid of piRNA is recruited to nuage through interaction with Krimp, a stable component of nuage granules. Upon recognition of a complementary target by the Aub-piRNA complex, the target is cleaved, followed by release of Aub-piRNA from nuage (1). Alternatively, after Aub engaged to an RNA target, it interacts with the Ago3-Krimp complex through sDMA residues on the Aub binding of Krimp Tudor domain to form the 4P complex (2). In the 4P complex, the Krimp dimer places unloaded Ago3 protein in proximity to methylated Aub associated with target RNA. Endonucleolytic cleavage of target RNA by Aub generates the RNA substrate that is loaded into Ago3, resulting in the 10nt overlap between Aub primary piRNA and Ago3 secondary piRNA. Once bound to RNA, Ago3 may be released from the 4P complex.
DISCUSSION
Krimper assembles Ago3/Aub complexes for ping-pong processing
The interaction between Aub and Ago3 lies at the heart of the ping-pong piRNA processing model, however, the molecular mechanism of this process remained poorly understood (Brennecke et al., 2007; Gunawardane et al., 2007). No direct association between Aub and Ago3 was ever reported, which suggested that if Aub and Ago3 form a complex, this interaction is likely transient and mediated by other proteins (Zhang et al., 2011). Genetic studies identified several genes implicated in the ping-pong process (Ipsaro et al., 2012; Lim and Kai, 2007; Malone et al., 2009; Nagao et al., 2011; Nishida et al., 2009; Patil and Kai, 2010; Zhang et al., 2011). The molecular function of these genes remained largely unknown: some of the encoded proteins might promote physical interactions between Aub and Ago3, while others might be working in other steps of ping-pong processing. The protein that promotes assembly of the ping-pong complex is expected to fulfill several criteria. First, this protein must directly interact with both Aub and Ago3. Second, it must be able to form a complex that contains Aub and Ago3 simultaneously, where Aub and Ago3 must be in a state that is compatible with progression of the ping-pong cycle (i.e. if Aub is loaded with piRNA then Ago3 should be unloaded). Third, mutation of the corresponding gene for this protein should lead to disruption of ping-pong piRNA processing. On a cellular level the protein is expected to co-localize with both Aub and Ago3. Finally, it might be expected to be a stable component of nuage that is capable of recruiting one or both Piwi proteins to this compartment.
According to our results, Krimp fulfills all the criteria as a factor that promotes the assembly of the ping-pong piRNA processing (4P) complex. First, Krimp directly interacts with two ping-pong partners, Aub and Ago3. Interestingly, Krimp uses different mechanisms to bind each Piwi proteins: binding to Aub depends on methylation of N-terminal arginine residues (Figure 5, S4), while binding to Ago3 is independent of sDMA (Figure 4B, S3C). Although we were not able to directly compare the strength of binding between Krimp and the two Piwi proteins, the pattern of subcellular localization of the three proteins in germ cells suggest that Krimp predominantly binds Ago3 in a stable complex, while interactions with Aub are transient. Due to the ability of Krimp to aggregate, Krimp dimers can simultaneously interact with Aub and Ago3, allowing for the formation of transient Aub/Krimp/Krimp/Ago3 complex. Importantly, Ago3 associates with Krimp in its unloaded state indicating that it is ready to receive a substrate from Aub cleavage (Figure 4E). Krimp mutation disrupts heterotypic (Aub-Ago3) and homotypic (Aub-Aub) ping-pong (Figure 6). Krimp has low mobility in nuage and is able to form granules by itself in the absence of other nuage proteins. Krimp is indispensable for recruitment of Ago3 to nuage (Figure 3C) and, although it is not required for recruitment of Aub per se, it promotes concentration of Aub in prominent nuage granules together with Ago3 (Figure 5E, 5G). Overall our data indicate that Krimp is a stable component of nuage that recruits unloaded Ago3 into this cellular compartment. The Krimp-Aub interaction assembles the 4P complex that coordinates the passage of cleaved RNA fragments from Aub to unloaded Ago3, defining the essential step in the ping-pong cycle (Figure 7). The 4P complex seems to be transient and disassembles rapidly upon loading of Ago3 with secondary piRNA. While the ping-pong mechanism is conserved in mouse, there is no direct ortholog of Krimp in mammals. However, several proteins with one or more Tudor domains are implicated in the piRNA pathway and TE repression in mouse (Chen et al., 2011; van der Heijden and Bortvin, 2009). It is possible that one or more Tudor-domain proteins that are able to aggregate and interact with Piwi proteins could play a role that is analogous to Krimp in flies.
Previously, Qin/Kumo was reported to mediate the interaction between Aub and Ago3 and it fulfills many criteria expected of a component responsible for the assembly of the ping-pong complex (Zhang et al., 2011). RNA helicase Vasa was also proposed to coordinate the assembly of the ping-pong (Amplifier) complex in silkmoth cells (Xiol et al., 2014). Vasa might be involved in promoting transition of intermediate ping-pong complex to the next step by unwinding piRNA from target RNA after cleavage by Aub (Nishida et al., 2015). In the future, it will be important to determine how Krimp, Qin/Kumo and Vasa cooperate in the ping-pong pathway.
A model for nuage assembly and function in the piRNA pathway
Previous studies showed that Aub, Ago3, and several other proteins that are required for piRNA repression, localize to the distinct subcellular compartment: perinuclear nuage granules. Furthermore, several mutations that disrupt nuage also cause failure in piRNA biogenesis and result in derepression of TEs. These observations led to the hypothesis that nuage is the subcellular compartment where piRNA processing, target recognition and repression take place. The mechanism by which nuage components assemble and recruit Piwi proteins remained unknown. Surprisingly, we found that Aub and Ago3 are recruited to nuage through different molecular mechanisms. The main factor determining Aub localization to nuage is its ability to bind a piRNA guide (Figure 2C). The requirement of piRNA for Aub localization in nuage helps to explain the results of previous studies, which found that deficiencies in nuclear proteins Rhino, UAP56, and Cutoff, that vital for early steps of piRNA biogenesis, cause delocalization of Aub from nuage (Klattenhoff et al., 2009; Pane et al., 2011; Zhang et al., 2012). The dependence on a piRNA guide for Aub localization suggests that recognition of target RNA molecules might tether Aub in nuage. Alternatively, binding of piRNA might lead to conformational changes in Aub that promote its interaction with other proteins in nuage.
Our results show that nuage is a heterogenous compartment that can be further subdivided into two structurally and functionally different sub-compartments: one predominantly containing Aub alone, and another where Aub and Ago3 co-localize. Aub covers the entire nuclear periphery, the compartment we call Aub-nuage, while Ago3 localizes to a few distinct granules embedded in the smooth Aub-nuage (Figure 1D, S1B). To effectively cleave transposon transcripts, Aub must scan all RNA transcripts exiting the nucleus. Therefore, we propose that smooth Aub-nuage is a compartment where cleavage of TE transcripts takes place. It is important to note that this step alone should be sufficient to repress TEs without the need of the ping-pong mechanism. Accordingly, the majority of Aub molecules might not be involved in ping-pong interactions with Ago3, but instead dissociate from nuage after successful cleavage of TE targets transcripts. Yet, some Aub complexes proceed to assemble transient complexes with Ago3 and Krimp to form Aub-Ago3 nuage granules that occupy a territory distinct from Aub-nuage. Assembly of the transient Aub/Krimp/Ago3 4P complex provides an opportunity for the ping-pong cycle to take effect. Thus, in contrast to smooth Aub-nuage, the function of the distinct Ago3/Aub granules is to provide a subcellular compartment for ping-pong piRNA processing by accommodation of all players in one place. Together, our data can be integrated into a dynamic model that explains the existence of two distinct nuage compartments, the forces that drive recruitment of Piwi proteins to nuage and the function of Aub and Ago3 at the different steps of the piRNA pathway (Figure 7).
EXPERIMENTAL PROCEDURES
Fly Stocks & Generation of Transgenic Fly Lines
Short hairpin RNA lines were obtained from the Bloomington Drosophila Stock Center. cDNA were obtained by RT-PCR from ovarian RNA of Oregon R strain. Point mutations were engineered by overlap PCR. cDNAs were cloned into the pUASP-Gateway-phiC31 vector with GFP, mKate2 or Strep-FLAG tags using the Gateway cloning system. See the Supplemental Experimental Procedures for a detailed description of fly stocks.
Fluorescence Recovery After Photobleaching (FRAP)
FRAP experiments using live ovaries expressing a single GFP-tagged transgene were acquired on a Zeiss LSM710 confocal microscope. A bleach region equal to 0.49 µm × 0.49 µm was bleached by a single iteration of 100% laser power. Five initial pre-bleach images and 115 subsequent post-bleach images were acquired and normalized fluorescence recovery was analyzed using FIJI software (http://fiji.sc/). A detailed protocol is provided in the Supplemental Experimental Procedures.
Co-immunoprecipitation and Western Blots
Dissected ovaries were lysed in NT2 buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 1mM MgCl2, 0.05% Igepal, EDTA-free Complete Protease Inhibitor Cocktail). Lysate was incubated in the presence or absence of 100µg/mL RNAse A, cleared by centrifugation and incubated with anti-FLAG M2 beads (Sigma Aldrich) at 4°C for 90 min., follow ed by washing and elution in reducing SDS buffer. Antibodies used for Western Blots were rabbit anti-GFP (Covance), mouse anti-GFP B-2 (Santa Cruz Biotechnology) or anti-FLAG M2 (Sigma Aldrich) at 1:3,000 concentration. Anti-Krimper rabbit polyclonal antibody generously provided by Dr. T. Kai was used at a concentration of 1:20,000. A variant protocol for S2 cell co-immunoprecipitation and other details are provided in the Supplemental Experimental Procedures.
piRNA isolation from immunopurified protein complexes
Immunopurified protein-RNA complexes were spiked with 5 pmol of 42-nt RNA oligonucleotide, followed by proteinase K digestion and and phenol extraction. Isolated RNA was CIP-treated, radiolabeled using PNK, and labeled with gamma-P32 ATP, and run on a 15% urea-PAGE gel. See the Semi-Quantitative piRNA Binding Analysis provided in Supplemental Experimental Procedures for details.
Protein cloning, expression, and Isothermal Titration Calorimetry
The Krimper Tudor2 domain (562–746) fused with an N-terminal hexa-His tag and a Maltose-Binding Protein tag was transformed into E. coli BL21(DE3)-RIL strain (Stratagene) and expression induced by IPTG. The His-MBP tagged protein was purified using a HisTrap FF column (GE Healthcare) and the tag was cleaved by TEV protease followed by further column purifications. ITC was conducted at a Microcal calorimeter ITC 200 instrument at 20°C u sing unmethylated and symmetrically di-methylated Aubergine peptides that correspond to residues 6 to 18. Binding curves were analyzed by non-linear least-squares fitting of the data using Origin 7.0 software. Additional details are provided in the Supplemental Experimental procedures
Sequence analysis of total piRNA libraries
Sequenced total small RNA libraries from Aub, Krimp and Piwi mutant and heterozygous Drosophila ovaries published in Malone et al. and from Ago3 mutant and heterozygous ovaries published in Li et al. were obtained from the NCBI GEO database (Li et al., 2009; Malone et al., 2009). The analysis is described in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Katalin Fejes Toth and members of the Aravin and Fejes Toth labs for discussion and critical review of this manuscript. The authors would like to thank Andres Collazo and the Beckman Imaging Facility at the California Institute of Technology and Peter Rapp for assistance in developing our FRAP protocol; Georgi Marinov for assistance with piRNA analysis; and Igor Antoshechkin of the Millard and Muriel Jacobs Genetics and Genomics Laboratory. We are grateful to Julius Brennecke, Phillip Zamore, Zhao Zhang, and Toshie Kai, for fly stocks and other reagents. This work was supported by grants from the National Institutes of Health (R01 GM097363 and DP2 OD007371A) and by the Searle Scholar and the Packard Fellowship Awards to A.A.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Bossing T, Barros CS, Brand AH. Rapid tissue-specific expression assay in living embryos. Genesis. 2002;34:123–126. doi: 10.1002/gene.10145. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nature reviews. Molecular cell biology. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Zinchenko MK, Hur JK, Nahvi A, Brunelle JL, Rogers EJ, Green R. Allosteric regulation of Argonaute proteins by miRNAs. Nat Struct Mol Biol. 2010;17:144–150. doi: 10.1038/nsmb.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hegner RW. The Germ Cell Determinants in the Eggs of Chrysomelid Beetles. Science. 1911;33:71–72. doi: 10.1126/science.33.837.71. [DOI] [PubMed] [Google Scholar]
- Hegner RW. The History of the Germ Cells in the Paedogenetic Larva of Miastor. Science. 1912;36:124–126. doi: 10.1126/science.36.917.124. [DOI] [PubMed] [Google Scholar]
- Huang H, Li Y, Szulwach KE, Zhang G, Jin P, Chen D. AGO3 Slicer activity regulates mitochondria-nuage localization of Armitage and piRNA amplification. J Cell Biol. 2014;206:217–230. doi: 10.1083/jcb.201401002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010;24:1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 2011;138:1863–1873. doi: 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP. Polar granules of drosophila. IV. Cytochemical studies showing loss of RNA from polar granules during early stages of embryogenesis. The Journal of experimental zoology. 1971;176:345–352. doi: 10.1002/jez.1401760309. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A, Sato K, Nishida KM, Siomi H, Siomi MC. Gender-Specific Hierarchy in Nuage Localization of PIWI-Interacting RNA Factors in Drosophila. Frontiers in genetics. 2011;2:55. doi: 10.3389/fgene.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Iwasaki YW, Murota Y, Nagao A, Mannen T, Kato Y, Siomi H, Siomi MC. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep. 2015;10:193–203. doi: 10.1016/j.celrep.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011;30:4601–4615. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Palumbo G, Bozzetti MP, Tritto P, Pimpinelli S, Schafer U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics. 1999;151:749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39:8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature reviews. Molecular cell biology. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Bortvin A. Defending the genome in tudor style. Dev Cell. 2009;17:745–746. doi: 10.1016/j.devcel.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Coute Y, Conn S, Kadlec J, Sachidanandam R, et al. RNA clamping by vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157:1698–1711. doi: 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol Cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.