Abstract
Background
Lignocellulase hypersecretion has been achieved in industrial fungal workhorses such as Trichoderma reesei, but the underlying mechanism associated with this process is not well understood. Although previous comparative genomic studies have revealed that the mutagenic T. reesei strain RUT-C30 harbors hundreds of mutations compared with its parental strain QM6a, how these mutations actually contribute to the hypersecretion phenotype remains to be elucidated.
Results
In this study, we systematically screened gene knockout (KO) mutants in the cellulolytic fungus Neurospora crassa, which contains orthologs of potentially defective T. reesei RUT-C30 mutated genes. Of the 86 deletion mutants screened in N. crassa, 12 exhibited lignocellulase production more than 25% higher than in the wild-type (WT) strain and 4 showed nearly 25% lower secretion. We observed that the deletion of Ncap3m (NCU03998), which encodes the μ subunit of the adaptor protein 3 (AP-3) complex in N. crassa, led to the most significant increase in lignocellulase secretion under both Avicel and xylan culture conditions. Moreover, strains lacking the β subunit of the AP-3 complex, encoded by Ncap3b (NCU06569), had a similar phenotype to ΔNcap3m, suggesting that the AP-3 complex is involved in lignocellulase secretion in N. crassa. We also found that the transcriptional abundance of major lignocellulase genes in ΔNcap3m was maintained at a relatively higher level during the late stage of fermentation compared with the WT, which might add to the hypersecretion phenotype. Finally, we found that importation of the T. reesei ap3m ortholog Trap3m into ΔNcap3m can genetically restore secretion of lignocellulases to normal levels, which suggests that the effect of the AP-3 complex on lignocellulase secretion is conserved in cellulolytic ascomycetes.
Conclusions
Using the model cellulolytic fungus N. crassa, we explored potential hypersecretion-related mutations in T. reesei strain RUT-C30. Through systematic genetic screening of 86 corresponding orthologous KO mutants in N. crassa, we identified several genes, particularly those encoding the AP-3 complex that contribute to lignocellulase secretion. These findings will be useful for strain improvement in future lignocellulase and biomass-based chemical production.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-015-0302-3) contains supplementary material, which is available to authorized users.
Keywords: Trichoderma reesei, RUTC30, Neurospora crassa, Adaptor protein 3 complex, Lignocellulase secretion
Background
In recent years, the use of large quantities of inexpensive plant biomass as feedstock for biofuel production has become an increasing focus of research. One of the key steps of the integrated biomass conversion process is production of lignocellulases, which are responsible for the degradation of plant cell wall material into fermentable sugars. This step is one of the major determinants of biofuel production costs [1, 2]. In nature, saprophytic fungi have evolved a highly efficient capability to secrete lignocellulases, with this property subsequently exploited in industry for lignocellulase production [3, 4].
Lignocellulases are synthesized and secreted through fungal secretory pathways [5, 6]. Generally, the nascent peptides of lignocellulases must be translocated into the endoplasmic reticulum (ER) lumen for folding and modification before delivery to the Golgi apparatus for further processing and subsequent targeting to their final proper destination (e.g., the extracellular matrix) via small secretory vesicles [7, 8]. To balance intracellular nutritional homeostasis or relieve the deleterious effects of unfolded or misfolded proteins, lignocellulases may undergo turnover via proteasome- or vacuole/lysosome-mediated degradation processes to meet the demand of nitrogen source recycling within fungal cells [9, 10]. For this reason, the dynamic competition between intracellular protein degradation and secretion may determine the final production titer of lignocellulases.
Currently, the most successful industrial hosts for lignocellulase production are primarily those originating from Trichoderma species, for example, T. reesei RUT-C30 (ATCC 56765). This hypersecretion mutant was obtained in the late 1970s via a three-step procedure [11–14]: (1) UV mutagenesis of wild-type (WT) Qm6a to generate isolate M7; (2) creation of NG14 by further mutagenesis of M7 using N-nitroguanidine; and (3) selection of RUT-C30 following another UV round of mutagenesis on the NG14 strain. Although the final secretion titer has been increased 20-fold in RUT-C30 [11, 15], little is known about the mechanism underlying the physiological hypersecretion process, especially that responsible for remodeling of the secretory pathway and associated regulation. With advances in omics technologies, recent comparative genomic studies have revealed that RUT-C30 contains a large chromosomal fragment deletion and hundreds of small mutations compared with its paternal strain QM6a. Several mutations have affected genes involved in lignocellulase regulation, such as two previously well-characterized targets: the carbon catabolite repression (CCR) regulator cre1 (tre120117), which was found to be truncated in RUT-C30 [16], and the gls2α gene encoding the glucosidase II alpha subunit and engaged in protein glycosylation, which had a frame-shift mutation in RUT-C30 [17]. In addition, several mutations potentially affecting extracellular enzyme trafficking and secretion have also been identified; examples include genes encoding a plasma membrane-related protein (tre81136), a cell wall protein (tre124295), an ARP2/3 complex protein (tre2439), and actin-interacting protein 3 (tre35386) [18, 19]. Although recently reported follow-up work has attempted to explain how these mutations affect phenotype (as defined by the transcriptome and cultivation behavior) [20], direct experimental evidence for the actual ability of each of these targets to contribute to the final protein secretion is still lacking at the cellular level. Although the generation of knockout (KO) mutants for these genes might be a direct way to check whether gene functions contribute to hypersecretion, the construction of hundreds of KO mutants in T. reesei would be time consuming and difficult to complete. Given that Neurospora crassa has a close phylogenetic relationship with T. reesei and possesses a nearly complete set of genome-wide gene deletion mutants, thereby making it a powerful tool for use in genetic studies [21], we reasoned that comparative genomic screening of N. crassa mutants could be applied as an alternative approach to study functions of mutated genes in T. reesei RUTC-30.
In the present work, systematic screening of 86 N. crassa KO mutants for mutated RUT-C30 orthologs was used to identify at least 12 genes with negative effects on lignocellulase secretion and 4 genes with positive effects. We further examined two genes that encode subunits of the adaptor protein 3 (AP-3) complex mediating hypersecretion in N. crassa and explored the possible conservation of the underlying mechanism in other ascomycetes including T. reesei. On the basis of our findings, we proposed a novel strategy to achieve the hypersecretion of lignocellulase in filamentous fungal systems by disrupting the natural balance between protein secretion and nutritionally required protein degradation.
Results
Screening of mutants
To address whether genes that bear mutations in T. reesei RUT-C30 are genuinely involved in protein secretion, we tested the secretion capacity of orthologous gene KO mutants in N. crassa. Le Crom et al. [19] previously identified 223 single nucleotide variants, 15 small insertions/deletions, and 18 larger deletions in RUT-C30, with an additional 17 mutations reported by Vitikainen et al. [18]. After excluding mutated genes shown to lack protein secretory functions in the two published studies, 164 mutated T. reesei genes were selected for ortholog calling in N. crassa using the local BLASTp program. We found 140 orthologs in N. crassa, among which 86 had homokaryotic gene KO mutants, including Δcre1 (NCU08807) [22]. We further screened these 86 mutants by determining their lignocellulase secretion capacity through batch culturing with microcellulose (2% [w/v] Avicel) as the carbon source and yeast extract (0.75% w/v) as the nitrogen source. Similar to T. reesei, lignocellulases accounted for most of the secretome (91% by weight) in N. crassa, with the four major components (CBH-1, CBH-2, EG-1, and BG-2) representing about 65% of the total cellulase cocktail proteins [23, 24]. Measured concentrations of extracellular proteins, used to reflect lignocellulase secretion capacity, are shown in Fig. 1 and Table 1. We found lignocellulase production to be reduced by more than 25% in 4 mutants compared with the wild-type strain (WT), with secretions elevated by more than 25% in 12 strains. Similar to Δcre1, 5 of these 12 mutants had markedly increased secretion of lignocellulases, including NCU07880 (annotated as a protein kinase), whose deletion increased the amount of secreted protein by approximately 35% compared with WT N. crassa and NCU01242 (encoding a protein predicted as a G2/mitotic-specific cyclin), whose deletion increased protein secretion by about 32%. Both these genes are involved in cell cycle-related functions. In addition, deletion of NCU01161 (encoding a protein functionally similar to actin polymerization protein Bzz1 and associated with endo- or exocytic pathways) increased protein secretion by approximately 34%. Loss of NCU07492, encoding a hypothetical protein, enhanced protein secretion by more than 30%. Finally, loss in N. crassa of the NCU03998 gene, whose counterpart in T. reesei RUT-C30, tre53811, has a mutation in its exon that changes serine73 to leucine [18], increased secreted protein levels up to 42% compared with the WT; this mutant exhibited the highest protein secretion among tested strains. NCU03998 was predicted to encode the μ subunit of the AP-3 complex. Because the way in which the AP-3 complex affects lignocellulase secretion has not been previously reported, we designated the gene at locus NCU03998 as Ncap3m in this study and focused on its functional characterization.
Fig. 1.
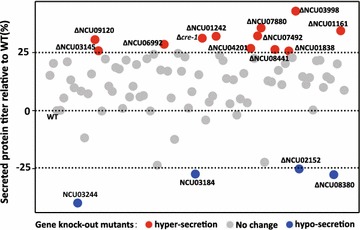
Percentages of secreted protein in 86 Neurospora crassa knockout (KO) mutants relative to the wild type (WT). After inoculating conidia from each strain into Avicel medium and culturing for 7 days, the secreted protein titers were measured and are displayed on the scatter plot. Mutants with secretion capacities altered by more than 25% compared with the WT are indicated as follows: red dots hypersecretion; blue dots hyposecretion.
Table 1.
List of extracellular proteins produced by Neurospora crassa mutants for genes orthologous to mutation-hit targets in Trichoderma reesei RUTC30
| No. | Mutant (FGSC) | Orthologs in N. crassa a | RUT C30 targetsb | Gene product names | Protein conc. (mg/L) | Increased vs WT (%)c |
|---|---|---|---|---|---|---|
| 1 | 11701 | NCU03998 | jgi|Trire2|53811| | Adaptor protein complex 3 Mu3A | 821.38 ± 21.83 | 42.9 |
| 2 | 17965 | NCU07880 | jgi|Trire2|124172| | Protein kinase | 780.01 ± 32.87 | 35.64 |
| 3 | 19384 | NCU01161 | jgi|Trire2|68926| | Actin polymerization protein Bzz1 | 773.06 ± 31.51 | 34.44 |
| 4 | 19393 | NCU07492 | jgi|Trire2|122689| | Hypothetical protein | 758.58 ± 21.01 | 32.21 |
| 5 | 15743 | NCU01242 | jgi|Trire2|69437| | G2/mitotic-specific cyclin | 759.76 ± 41.93 | 32.06 |
| 6 | 10372 | NCU08807 | jgi|Trire2|120117| | cre-1 | 755.26 ± 51.79 | 31.22 |
| 7 | 11964 | NCU09120 | jgi|Trire2|76515| | Lysine-specific histone demethylase Aof2 | 750.7 ± 20.09 | 30.6 |
| 8 | 15360 | NCU06992 | jgi|Trire2|31118| | DNA repair protein Nse1 | 737.31 ± 37.42 | 28.59 |
| 9 | 16681 | NCU04201 | jgi|Trire2|106234| | Signal transduction protein Syg1 | 729.66 ± 38.29 | 26.84 |
| 10 | 12039 | NCU08441 | jgi|Trire2|123786| | Non-ribosomal peptide synthetase | 729.27 ± 112.17 | 26.36 |
| 11 | 12276 | NCU03145 | jgi|Trire2|78301| | Vacuolar membrane zinc transporter | 723.79 ± 40.24 | 25.81 |
| 12 | 13177 | NCU01838 | jgi|Trire2|65106| | Nitrilase | 722.38 ± 13.8 | 25.71 |
| 13 | 11468 | NCU04142 | jgi|Trire2|123114| | Heat shock protein 80 | 717.96 ± 59.51 | 24.68 |
| 14 | 12062 | NCU07498 | jgi|Trire2|79304| | DNA excision repair protein Rad2 | 715.8 ± 57.68 | 24.32 |
| 15 | 16319 | NCU02842 | jgi|Trire2|121087| | Hypothetical protein | 712.55 ± 32.87 | 23.89 |
| 16 | 18830 | NCU03811 | jgi|Trire2|121453| | Hypothetical protein | 710.24 ± 91.52 | 23.16 |
| 17 | 19831 | NCU03545 | jgi|Trire2|82153| | Hypothetical protein | 706.55 ± 32.91 | 22.85 |
| 18 | 11956 | NCU08809 | jgi|Trire2|74765| | Hypothetical protein | 704.12 ± 19.44 | 22.5 |
| 19 | 16594 | NCU03914 | jgi|Trire2|64375| | Glucan 1,3-beta-glucosidase | 704.88 ± 64.43 | 22.38 |
| 20 | 20296 | NCU08364 | jgi|Trire2|78320| | Choline sulfatase | 702.59 ± 19.06 | 22.23 |
| 21 | 11040 | NCU05411 | jgi|Trire2|70071| | Pathway-specific nitrogen regulator | 701.46 ± 65.9 | 21.78 |
| 22 | 19539 | NCU07334 | jgi|Trire2|68425| | Uracil permease | 698.2 ± 25.84 | 21.72 |
| 23 | 18971 | NCU00503 | jgi|Trire2|71037| | Nonselective cation channel protein | 700.38 ± 66.13 | 21.59 |
| 24 | 13289 | NCU00427 | jgi|Trire2|80691| | Hypothetical protein | 698.5 ± 19.9 | 21.52 |
| 25 | 15939 | NCU00754 | jgi|Trire2|58561| | Multidrug resistant protein | 697.71 ± 79.14 | 21.05 |
| 26 | 13436 | NCU05089 | jgi|Trire2|64882| | MFS monocarboxylate transporter | 692.03 ± 30.99 | 20.33 |
| 27 | 16570 | NCU04886 | jgi|Trire2|28409| | MFS multidrug transporter | 693.08 ± 69.06 | 20.3 |
| 28 | 16331 | NCU03068 | jgi|Trire2|58790| | Glycerol-3-phosphate phosphatase 1 | 690.98 ± 32.01 | 20.14 |
| 29 | 20407 | NCU11050 | jgi|Trire2|75105| | DUF455 domain-containing protein | 689.63 ± 40.55 | 19.86 |
| 30 | 11084 | NCU09549 | jgi|Trire2|26255| | C6 zinc finger domain-containing protein | 683.56 ± 79.47 | 18.58 |
| 31 | 18230 | NCU09887 | jgi|Trire2|67030| | Drp1p | 678.63 ± 42.93 | 17.93 |
| 32 | 19296 | NCU07276 | jgi|Trire2|67732| | ABC bile acid transporter | 677.02 ± 69.64 | 17.5 |
| 33 | 15452 | NCU06309 | jgi|Trire2|22841| | Hypothetical protein | 674.61 ± 18.59 | 17.36 |
| 34 | 15930 | NCU00648 | jgi|Trire2|59952| | Choline transporter | 675.67 ± 114.38 | 17.02 |
| 35 | 13757 | NCU01420 | jgi|Trire2|54157| | Hypothetical protein | 672.9 ± 36.3 | 16.97 |
| 36 | 15627 | NCU04626 | jgi|Trire2|28731| | G-protein coupled receptor | 667.67 ± 37.79 | 16.05 |
| 37 | 15627 | NCU04626 | jgi|Trire2|123806| | G-protein coupled receptor | 667.67 ± 37.79 | 16.05 |
| 38 | 16748 | NCU00799 | jgi|Trire2|40758| | Homocysteine S-methyltransferase | 669.17 ± 128.93 | 15.8 |
| 39 | 14141 | NCU08499 | jgi|Trire2|58161| | GTPase-activating protein GYP5 | 663.7 ± 13.31 | 15.49 |
| 40 | 13568 | NCU04521 | jgi|Trire2|54511| | Hypothetical protein | 665.59 ± 127.62 | 15.19 |
| 41 | 20195 | NCU06578 | jgi|Trire2|104161| | KapG | 662.02 ± 104.79 | 14.69 |
| 42 | 19233 | NCU01997 | jgi|Trire2|74570| | ABC transporter | 662.83 ± 141.67 | 14.63 |
| 43 | 12060 | NCU07381 | jgi|Trire2|3027| | DNA cross-link repair protein pso2/snm1 | 659.15 ± 93.95 | 14.25 |
| 44 | 18123 | NCU07703 | jgi|Trire2|55887| | Hypothetical protein | 655.25 ± 49.89 | 13.82 |
| 45 | 13910 | NCU05195 | jgi|Trire2|75072| | Hypothetical protein | 653.76 ± 52.14 | 13.55 |
| 46 | 11279 | NCU03125 | jgi|Trire2|79405| | NIMA-interacting protein TinC | 651.64 ± 26.12 | 13.32 |
| 47 | 12569 | NCU00321 | jgi|Trire2|67024| | Hypothetical protein | 652.72 ± 75.61 | 13.23 |
| 48 | 19403 | NCU07564 | jgi|Trire2|78465| | Siderophore iron transporter mirC | 648.04 ± 5.97 | 12.87 |
| 49 | 15566 | NCU01961 | jgi|Trire2|59147| | DNA lyase Apn2 | 647.69 ± 69.51 | 12.39 |
| 50 | 19712 | NCU00497 | jgi|Trire2|58391| | Hypothetical protein | 643.14 ± 129.41 | 11.27 |
| 51 | 11150 | NCU00278 | jgi|Trire2|73912| | Hypothetical protein | 643.22 ± 144.06 | 11.2 |
| 52 | 11459 | NCU00340 | jgi|Trire2|36543| | Transcription factor steA | 637.2 ± 73.32 | 10.54 |
| 53 | 13984 | NCU05837 | jgi|Trire2|65104| | Vacuolar protein sorting-associated protein 13a | 635.14 ± 16.93 | 10.5 |
| 54 | 19183 | NCU05477 | jgi|Trire2|102776| | Hypothetical protein | 632.43 ± 18.1 | 10.02 |
| 55 | 11061 | NCU08443 | jgi|Trire2|77513| | Transcription factor ace3 | 635.36 ± 137.21 | 9.87 |
| 56 | 13086 | NCU00541 | jgi|Trire2|80592| | Hypothetical protein | 627.2 ± 73.28 | 8.8 |
| 57 | 16803 | NCU04809 | jgi|Trire2|82037| | MFS phospholipid transporter | 630.47 ± 248.43 | 8.4 |
| 58 | 15478 | NCU08642 | jgi|Trire2|78268| | Cyclic nucleotide-binding domain-containing protein | 624.52 ± 83.42 | 8.28 |
| 59 | 15647 | NCU00025 | jgi|Trire2|82499| | Integral membrane protein | 623.56 ± 73.21 | 8.17 |
| 60 | 16280 | NCU04108 | jgi|Trire2|105874| | Isoamyl alcohol oxidase | 616.8 ± 25.92 | 7.26 |
| 61 | 15869 | NCU00335 | jgi|Trire2|5140| | Pre-mRNA-splicing factor cwc15 | 612.18 ± 15.31 | 6.51 |
| 62 | 13160 | NCU01633 | jgi|Trire2|62380| | Hexose transporter HXT13 | 611.29 ± 19.85 | 6.33 |
| 63 | 19733 | NCU06832 | jgi|Trire2|112231| | Kinesin | 609.8 ± 45.91 | 5.93 |
| 64 | 12341 | NCU09864 | jgi|Trire2|56726| | 2-Oxoisovalerate dehydrogenase alpha subunit | 608.18 ± 4.15 | 5.88 |
| 65 | 11030 | NCU07788 | jgi|Trire2|52368| | Fungal specific transcription factor | 606.86 ± 36.13 | 5.47 |
| 66 | 12282 | NCU06647 | jgi|Trire2|5403| | Enoyl-CoA hydratase/isomerase | 605.74 ± 7.76 | 5.43 |
| 67 | 12018 | NCU02751 | jgi|Trire2|120806| | Serine/threonine-protein kinase | 605.22 ± 59.32 | 5.05 |
| 68 | 11677 | NCU01868 | jgi|Trire2|59388| | MFS maltose permease MalP | 595.74 ± 3.43 | 3.71 |
| 69 | 18917 | NCU05459 | jgi|Trire2|65773| | Mitochondrial AAA ATPase | 595.8 ± 64.3 | 3.39 |
| 70 | 19405 | NCU07574 | jgi|Trire2|22294| | Hypothetical protein | 591.48 ± 5.41 | 2.96 |
| 71 | 17946 | NCU04755 | jgi|Trire2|45456| | Protein kinase domain-containing protein ppk32 | 581.14 ± 12.76 | 1.12 |
| 72 | 17081 | NCU01044 | jgi|Trire2|63464| | Hypothetical protein | 580.08 ± 10.41 | 0.95 |
| 73 | 19059 | NCU08307 | jgi|Trire2|56077| | Hypothetical protein | 579.21 ± 100.55 | 0.3 |
| 74 | 17389 | NCU02337 | jgi|Trire2|80332| | Mitochondrial carrier protein | 576.12 ± 32.03 | 0.14 |
| 75 | 2489 | ##### | ##### | Wild type | 575.45 ± 36.81 | 0 |
| 76 | 16836 | NCU04847 | jgi|Trire2|52520| | cyclin | 573.97 ± 15.22 | −0.13 |
| 77 | 20073 | NCU06341 | jgi|Trire2|44956| | MFS transporter | 573.58 ± 9.57 | −0.17 |
| 78 | 19245 | NCU02220 | jgi|Trire2|64866| | hypothetical protein | 563.98 ± 19.59 | −1.9 |
| 79 | 12078 | NCU00523 | jgi|Trire2|50268| | NAD-dependent deacetylase sirtuin-2 | 551.74 ± 23.25 | −4.05 |
| 80 | 12072 | NCU04203 | jgi|Trire2|121351| | Glucosidase II alpha subunit | 508.03 ± 35.04 | −11.73 |
| 81 | 13475 | NCU07119 | jgi|Trire2|60458| | Nonribosomal peptide synthase 2 | 503.65 ± 12.98 | −12.37 |
| 82 | 19165 | NCU05213 | jgi|Trire2|75074| | Hypothetical protein | 449.4 ± 86.39 | −22.22 |
| 83 | 18185 | NCU08452 | jgi|Trire2|110570| | Hypothetical protein | 439.95 ± 23.92 | −23.52 |
| 84 | 16956 | NCU02152 | jgi|Trire2|3400| | RRM domain-containing protein | 430.62 ± 18.69 | −25.11 |
| 85 | 11357 | NCU03184 | jgi|Trire2|4921| | C2H2 conidiation transcription factor FlbC | 419.08 ± 52.77 | −27.31 |
| 86 | 20306 | NCU08380 | jgi|Trire2|122050| | Plasma membrane phosphatase | 416.5 ± 18.07 | −27.57 |
| 87 | 11360 | NCU03244 | jgi|Trire2|62053| | WD repeat protein | 346.47 ± 27.22 | −39.81 |
a N. crassa orthologs: locus selected according to the N. crassa database (v7) (https://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
bRUTC30 targets: locus selected according to the T. reesei genome sequence web site (http://genome.jgi-psf.org/Trire2/Trire2.home.html).
cThe increased percentage of secreted protein relative to the WT. Strains more than 25% increased or decreased compared with the WT are shown in italics.
Ncap3m encodes the μ subunit of the AP-3 complex in N. crassa
The AP-3 complex is well conserved in various eukaryotes [25, 26]. We were able to identify NcAP3m sequence homologs in Saccharomyces cerevisiae (identity: 25%, E value: 4 × 10−15), Arabidopsis thaliana (identity: 32%, E value: 2 × 10−27), Drosophila melanogaster (identity: 34%, E value: 9 × 10−33), Mus musculus (identity: 29%, E value: 2 × 10−32), and even Homo sapiens (identity: 29%, E value: 2 × 10−32). To reveal the phylogenetic position of Ncap3m within eukaryotes, we generated the phylogenetic tree of AP-3 complex μ-subunit proteins shown in Fig. 2. Proteins in this tree were significantly divided into three major groups corresponding to fungi, plants, and metazoans. Ncap3m was clustered along with its ortholog tre53811 from T. reesei (identity: 57%, E value: 0.00) in a well-supported clade within the fungal group.
Fig. 2.
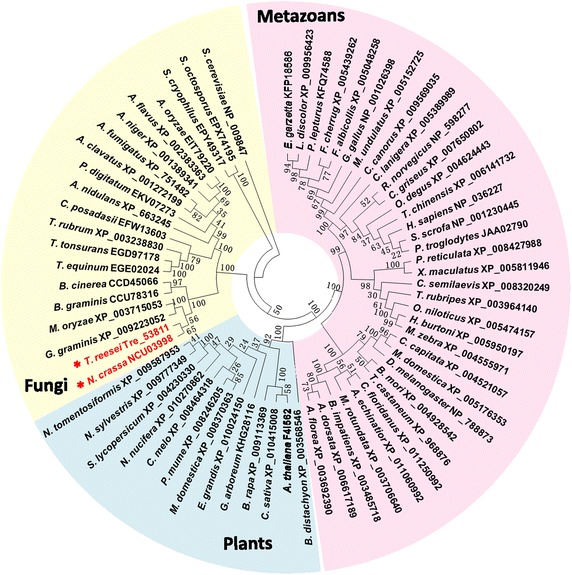
Phylogenetic tree of adaptor protein 3 (AP-3) complex μ-subunit proteins. Amino acid sequences were obtained from the NCBI database based on ortholog calling using the local BLASTp program. The phylogenetic analysis was performed by MEGA6 using the maximum likelihood method with 1,000 bootstrap replicates.
Ncap3m is involved in lignocellulase production in N. crassa
To further determine how Ncap3m impacts lignocellulase production in N. crassa, we performed batch culturing of ΔNcap3m using Avicel or xylan as the sole carbon source, with the WT used as the positive control. In batch cultures with 2% (w/v) Avicel, the protein secretion titer of ΔNcap3m increased by more than 80% compared with the WT. Filter paper activity (FPA) and xylanase activities were, respectively, 44 and 80% higher in the mutant than in the WT, with endoglucanase and exoglucanase activities showing respective increases of approximately 24 and 20% (Fig. 3a). When we changed the culture conditions to 2% xylan as the sole carbon source, thereby allowing xylanase to be specifically induced and secreted [27], the extracellular protein concentration of the ΔNcap3m mutant was improved by approximately 50% and xylanase activity increased by about 100% (Fig. 3c). All of these results were additionally confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) profiling of the extracellular secretome (Fig. 3b, d), with known cellulase proteins from a previously reported LC–MS analysis used as a reference [23].
Fig. 3.
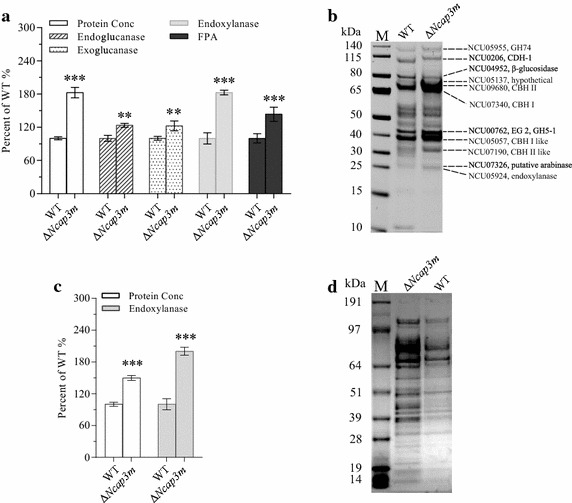
Hypersecretion of lignocellulases by Neurospora crassa due to deletion of Ncap3m. Typical secretomes of the wild type (WT) and ΔNcap3m are shown on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (a, c) after 7 days of culturing in Avicel (a, b) and xylan (c, d) media. Total extracellular protein concentration and enzyme activity (b, d) were measured and evaluated after normalization to the WT control according to percentage (standard error of the mean, n = 3). Asterisks indicate significant differences from the WT (**P < 0.01; ***P < 0.001) based on one-way analysis of variance.
To further show that the lignocellulase hypersecretion phenotype was caused by the loss of Ncap3m in N. crassa rather than other unknown genetic mutations, additional gene complementation assays were performed. We separately introduced enhanced green fluorescent protein (EGFP)-labeled Ncap3m under the control of ccg-1 [28] or native promoters into ΔNcap3m. Batch culturing under either Avicel or xylan utilization conditions revealed that both complemented isolates restored the altered secretion phenotype to levels similar to that of the WT. These results were verified by enzyme activity measurements (Fig. 4).
Fig. 4.
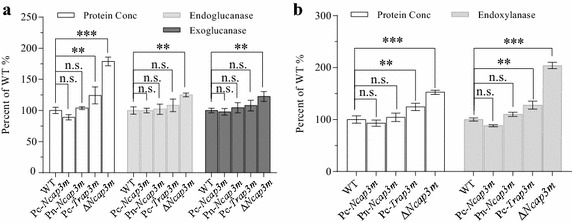
Restoration of the ΔNcap3m hypersecretion phenotype to normal wild-type (WT) levels by Ncap3m or Trap3m. The following strains were grown in 2% (w/v) Avicel (a) or xylan (b) media: the WT, the Ncap3m gene knockout (KO) mutant (ΔNcap3m), the Ncap3m-complemented strain under either the control of the ccg-1 strong promoter (Pc-Ncap3m) or the native promoter (Pn-Ncap3m), and a Trap3m-complemented strain introduced into ΔNcap3m under the control of the ccg-1 strong promoter (Pc-Trap3m). After 7 days of culturing, the total extracellular protein concentration and endoglucanase (a) or endoxylanase (b) activity were measured. Data were normalized to the WT control according to percentage (standard error of the mean, n = 3); asterisks indicate significant differences from the WT (**P < 0.01; ***P < 0.001; ns not significant) based on one-way analysis of variance.
To investigate whether the hypersecretion observed in N. crassa due to the loss of the ap3m gene is conserved in T. reesei, we conducted an inter-complementation assay by introducing the ap3m gene of T. reesei into ΔNcap3m. We first cloned the 1,608-bp open reading frame (ORF) of Trap3m (tre53811) in the T. reesei QM6a cDNA (in-house annotated; see “Methods”; Additional file 1: Figure S1, Additional file 2: Figure S2 for details) and placed it under the control of the N. crassa ccg-1 promoter. The plasmid was then transformed into ΔNcap3m:his3−N. crassa to form the strain Pc-Trap3m. When compared with its parental strain ΔNcap3m:his3−, protein secretion and enzyme activities in Pc-Trap3m were restored to WT levels, similar to the complemented strain Pc-ap3m (Fig. 4). This result implies that Trap3m can genetically complement the ap3m deletion phenotype in N. crassa. Taken together, these results indicate that the function of ap3m is evolutionarily conserved between the two lignocellulolytic ascomycetes.
Sub-cellular localization of the NcAP3m protein in N. crassa
To assess NcAP3m protein sub-cellular localization, we observed the EGFP signal of NcAP3m–EGFP recombinant protein in young hyphae of the complemented strain ΔNcap3m::Ncap3m–EGFP (Fig. 5). After pre-growth in minimal medium for 16 h followed by culturing in Avicel (2% w/v) medium for another 4 h, we found that NcAP3m–EGFP proteins were unevenly distributed in cytosol; they were primarily located in the extreme tip region of young hyphae, where they accumulated in a ring-like structure. Co-staining with membrane dye FM4-64 showed that NcAP3m–EGFP proteins overlapped with a structure known as the Spitzenkörper that has been reported to serve as a vesicle trafficking center in filamentous fungal cells [29]. This overlap implies that NcAP3m may be located in small secretory vesicles. When the culture time was extended to 48 h, we found that NcAP3m–EGFP proteins no longer accumulated in tip regions. Instead, they were spotted around large vacuole-like structures near the tip region. This observed location is consistent with previous reports showing that the AP-3 complex in higher eukaryotes is primarily located in vacuoles and lysosomes and plays an important role in protein sorting and trafficking between the trans-Golgi network and vacuoles/lysosomes or endosomes [30, 31].
Fig. 5.
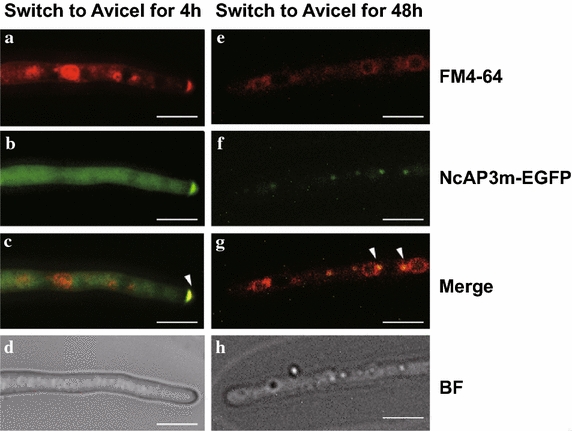
Sub-cellular localization of the adaptor protein 3 (AP-3) complex in Neurospora crassa. Wild-type (WT) and ΔNcap3m strains were pre-grown in minimal medium with 2% (w/v) sucrose as the sole carbon source for 16 h and then switched to Avicel medium to elicit lignocellulase production for another 4 h (initial stage; a–d) or 48 h (logarithmic stage; e–h). Extreme tip regions of mycelia of the WT and ΔNcap3m were stained with 5 µg/mL of the membrane dye FM4-64 for 30 min to label membrane structures such as the Spitzenkörper or vacuoles (a, e). Locations of NcAP3m proteins were monitored by recording enhanced green fluorescent protein (EGFP) signal (b, f). Merged yellow fluorescence signal from FM4-64 and EGFP (c, g) are denoted by white arrows in the photos. Each scale bar represents 10 µm.
NCU06569 encoding the β subunit of the AP-3 complex can also affect protein secretion in N. crassa
Whether other subunits of the AP-3 complex besides NcAP3m can affect protein secretion remain to be explored. Based on genome annotation, three other subunits of the AP-3 complex were found to exist in N. crassa; these were separately encoded by NCU06569 (β subunit, Ncap3b), NCU04652 (δ subunit, Ncap3δ) and NCU09461 (σ subunit, Ncap3σ). Among the three genes, the KO mutant was available for Ncap3b (FGSC#11856, ΔNcap3b) and we therefore investigated its lignocellulase secretion capacity (Fig. 6a, b). When grown on 2% (w/v) Avicel, the extracellular protein secreted by the ΔNcap3b mutant was about 63% higher than that in the WT; FPA increased by approximately 37%, while endoglucanase, exoglucanase, and xylanase activities increased by about 20% compared with the WT. These results demonstrate that Ncap3b also markedly affected protein secretion. We next constructed a double mutant of two subunit-encoding genes of the AP-3 complex (ΔNcap3mΔNcap3b). This double mutant had an extracellular protein secretion titer and related enzyme activities similar to those of the ΔNcap3m single mutant (Fig. 6a, b), suggesting that NcAP3m is the critical subunit for full functionality of the AP-3 complex. Taking all these results into consideration, we deduced that the AP-3 complex has a significant impact on the secretion of proteins such as lignocellulases.
Fig. 6.
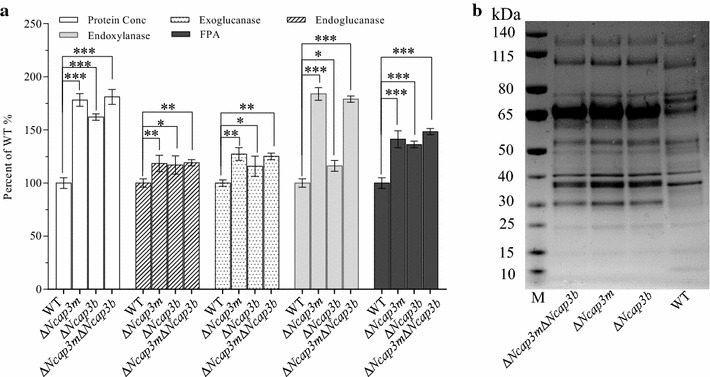
Involvement of the adaptor protein 3 (AP-3) complex in lignocellulase secretion in Neurospora crassa. Conidia from the wild type (WT) and Ncap3m and Ncap3b single (ΔNcap3m, ΔNcap3b) and double mutants (ΔNcap3mΔNcap3b) were separately inoculated into Avicel medium and batch cultured for 7 days. The typical secretome of each strain was then observed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (b), while total extracellular protein concentration and enzyme activity (a) were measured and displayed after normalization to the WT control according to percentage (standard error of the mean, n = 3). Asterisks indicate significant differences from the WT (*P < 0.05; **P < 0.01; ***P < 0.001) based on one-way analysis of variance.
Lignocellulase gene transcriptional abundance is maintained at relatively higher levels during the late fermentation phase in AP-3 complex mutants
To further elucidate the hypersecretion phenotype of the AP-3 complex KO mutants, especially those of the key subunit NcAP3m, we monitored changes in the expressions of lignocellulase genes by quantitative real-time PCR (qPCR) during batch culturing of ΔNcap3m. To avoid potential differences induced by variable growth rates of isolates, shift cultures were used. All strains were pre-grown in 2% (w/v) sucrose until the formation of young hyphae and then switched to Vogel’s medium containing 2% (w/v) Avicel to induce lignocellulase gene expression. We monitored the expression patterns of three genes, namely, cbh-1 (NCU07340), cbh-2 (NCU09680), and gh5-1 (NCU00762), which respectively encode the major exoglucanase, cellobiohydrolase, and endoglucanase proteins in N. crassa [23, 24] (Fig. 7). In WT, all three genes had their highest expression values 4 h after induction; expression then rapidly decreased, normally within 48 h, as culturing continued [32]. This phenomenon is known as repressed expression of secreted sequences (RESS), a feedback mechanism that selectively down-regulates transcription of genes encoding extracellular enzymes upon increased protein flux, thereby helping to reduce ER load [33–35]. In ΔNcap3m, however, lignocellulase gene expression levels had not obviously decreased after 48 h and remained at a relatively high level compared with the WT until day 7 (Fig. 7). All of these results indicate that the RESS mechanism might be compromised in ΔNcap3m, even though we still cannot interpret the detailed mechanism at present. This consistently higher induction might be an additional reason for the higher secretion of lignocellulase proteins in AP-3 complex defect mutants.
Fig. 7.
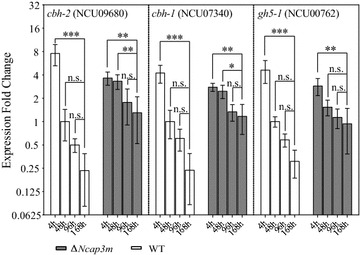
Maintenance of high lignocellulase gene expression levels in ΔNcap3m relative to the wild-type strain (WT) at the late fermentation stage. After ΔNcap3m and WT conidia were grown on Avicel for 4, 48, 96, or 168 h, the transcriptional abundance of three major lignocellulase genes was evaluated by quantitative real-time PCR (qPCR). The data are normalized to the expression of the WT strain at 48 h for each tested gene, with actin (NCU04173) gene expression levels used as an endogenous control in all samples (standard error of the mean, n = 3). Asterisks indicate significant differences from the control (*P < 0.05; **P < 0.01; ***P < 0.001; ns not significant) based on one-way analysis of variance.
Alkaline phosphatase influences lignocellulase secretion
In S. cerevisiae, the AP-3 complex has been shown to be specifically involved in the transport of alkaline phosphatase (ALP) from the Golgi apparatus to vacuoles/lysosomes [30]. Whether ALP affects lignocellulase secretion remains unclear. To address this question, we used the protein sequence of Pho8p, the sole vacuolar ALP in S. cerevisiae, as a query in BLASTp searches against the NCBI database (E value <1 × 10−5), thereby identifying two homologs in the N. crassa genome: NCU08997 (identity: 51%, E value: 1 × 10−135) and NCU01376 (identity: 26%, E value: 2 × 10−17). Further phylogenetic analysis revealed that the two ALP proteins were also well conserved in other filamentous fungal genomes, including that of T. reesei (Additional file 3: Figure S4). When we monitored the lignocellulase secretion capacity of gene KO mutants for the two targets using batch culturing, we discovered that the deletion of NCU08997 had a markedly positive effect on lignocellulase secretion and the deletion of NCU01376 yielded no obvious phenotypes (Fig. 8). This result suggests that NCU08997 serves as the main functional vacuolar ALP in N. crassa. Our findings also indicate that ALP involved in efficient protein degradation may be responsible for the observed induction of lignocellulase hypersecretion caused by the AP-3 complex malfunction.
Fig. 8.
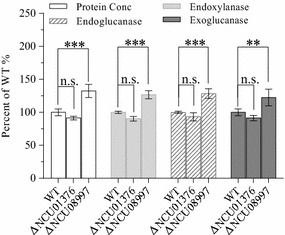
Screenings of two putative alkaline phosphatase knockout (KO) mutants in Neurospora crassa to reveal their potential functional association with lignocellulase secretion. Conidia of ΔNCU08997, ΔNCU01376, and wild-type (WT) strains were separately inoculated into Avicel medium and batch cultured for 7 days; total extracellular protein concentration and enzyme activity were then measured and normalized to the WT control according to percentage (standard error of the mean, n = 3). Asterisks indicate significant differences from the control (**P < 0.01; ***P < 0.001; ns not significant) based on one-way analysis of variance.
Discussion
Because lignocellulases are a pivotal factor in the biorefinery process, improvement of their production titers could effectively reduce the cost of biofuel production. Most currently used industrial hosts, such as T. reesei (Hypocrea jecorina) and Aspergillus spp., can achieve an outstanding output; because most of these strains were generated by random mutagenesis, however, a clear genetic basis underpinning the requirements of effective secretion has not yet been elucidated [11]. Previous work comparing the genomic data of two T. reesei hyperproducing mutants, NG14 and RUT-C30, with their reference isolate QM6a, uncovered more than 200 mutagenic including many single-nucleotide substitutions, some small insertions/deletions, and more than 100 kb of larger genomic DNA deletions [19]. Some of these mutagenic events have affected genes engaged in functions such as secretion/vacuolar targeting, mRNA stability, and transcription, and thus might contribute to hypersecretion phenotypes. Meanwhile, independent work using high-resolution array comparative genomic hybridization analysis of T. reesei NG14 and RUT-C30 uncovered an additional 17 previously undocumented mutation sites. Importantly, two deletions identified in RUT-C30, of a large 85-kb genomic DNA segment and a transcription factor, were both determined to not be involved in cellulase production [18]; this raises the question of whether other mutagenic events have potential roles in boosting secretion capacity. A large-scale screening of each mutation in the T. reesei system seems unreachable; however, N. crassa is genetically very similar to T. reesei and possesses a nearly complete genomic KO strain set, which provides an alternative opportunity to test the possible contribution of chromosomal mutations to effective secretion. In this work, we systematically screened deletion mutants of 86 N. crassa genes that were orthologs of T. reesei genes having mutations in RUT-C30 relative to its parental strain QM6a. Further examination of the lignocellulase production capacities of these corresponding KO strains by batch culturing revealed 12 strains, including well-known mutant Δcre-1 [16], which were able to promote protein secretion by more than 25% in comparison with the WT. Among these strains, we found several targets involved in cell cycle-related functions, such as NCU07880, the homolog of the Aspergillus nidulans Cdc2-related kinase gene npkA [36]; NCU01242, which encodes G2/mitotic-specific cyclin; and NCU06992, which encodes a homolog of the fission yeast DNA repair protein Nse1 possibly contributing to genome stability [37]. Previous work on T. reesei has demonstrated that effective secretion is related to low growth rate [38], implying that interruption of the cell cycle may contribute to enhanced lignocellulase secretion. Moreover, NCU01161 encodes an actin polymerization protein that acts as the homolog of S. cerevisiae BZZ1 [39]. In filamentous fungi, actin cytoskeleton organization is commonly linked with endo- or exocytic pathways [40]; thus, hypersecretion induced by a defect in NCU01161 might result from alteration of such pathways. In addition, our screening identified several deletion strains, namely, ΔNCU02152, ΔNCU03184, ΔNCU08380, and ΔNCU03244, with decreased secretion capacities, which suggests that they play positive roles in regulating lignocellulase secretion. All four strains were found to have secretion that was decreased by more than 25% compared with WT N. crassa, although the detailed mechanisms remain to be elucidated by further studies. Unlike previous observations, we found that deletion of NCU04203, which encodes the alpha subunit of glucosidase II, decreased lignocellulase production by approximately 10% rather than promoting its secretion [17]. We speculate that improperly glycosylated enzyme protein might be degraded intracellularly rather than being secreted to the extracellular matrix. Despite the identification of dozens of mutations in many interesting genes associated with protein synthesis and secretion, it should be noted that our screenings found that most are likely non-functional mutations—at least, no obvious phenotype could be seen at single-gene disruption levels. These results clearly agree with the idea that the hypersecretion phenotype of T. reesei RUT C30 is not caused by any single-gene mutation. Because only single-gene KO stocks were screened in this study, the interactions among these mutant alleles could not be elucidated by the present work. However, N. crassa can be easily sexually crossed to generate double-, triple-, and multiple-gene mutants, which would help to further investigations of the mechanism of hypercellulase production in RUT C30 in future.
Notably, we found that deletion of Ncap3m (NCU03998) in N. crassa achieved the highest extracellular protein yield among all screened mutants, with amounts of secreted proteins comparable to Δcre-1. Furthermore, the results of FPA and xylanase activity measurements confirm the observed enhancement of cellulase and hemicellulase production in this strain, suggesting that NCU03998 functions in the regulation of secretion without enzyme specificity. Protein sequence alignment revealed that NCU03998 encodes a protein homologous to the AP-3 complex subunit μ3a in mammals and Amp3p in S. cerevisiae. Adaptor protein complexes (AP complexes) mediate the formation of vesicles and participate in intra-organelle membrane trafficking in eukaryotes [25, 41, 42]. To date, five AP complexes (AP-1 to AP-5) have been identified [43], and AP-3 has been extensively studied in mammals, flies, Arabidopsis, and yeast [25, 41, 42, 44]. In mammalian cells, the AP-3 complex is a four-subunit heterotetrameric complex consisting of two large subunits (β3a and δ), a medium subunit (μ3a) and a small subunit (σ3) [45, 46]. In addition to Ncap3m, we also tested the secretion capacity of another AP-3 complex large-subunit-β3a mutant (ΔNCU06569, ΔNcap3b) in N. crassa; this deletion mutant also displayed an enhanced secretion phenotype similar to that of ΔNcap3m, suggesting that the AP-3 complex is indeed involved in lignocellulase secretion. Moreover, we successfully complemented Trap3m in ΔNcap3m, implying that the AP-3 complex, and Trap3m in particular, may contribute to the hypersecretion phenotype of the T. reesei RUT-C30 strain.
Previous work on S. cerevisiae has demonstrated that the AP-3 complex is required for the intracellular retention of the major chitin synthase Chs3p located in the plasma membrane [47], suggesting that the AP-3 complex can influence secretory protein secretion. Details of the process remain to be elucidated, however, especially in filamentous fungal systems. In addition, the S. cerevisiae AP-3 complex has been shown to be specifically involved in the transport of ALP from the Golgi apparatus to vacuoles/lysosomes. Deletion of Amp3p resulted in a pronounced accumulation of ALP in cytoplasm rather than at the vacuolar membrane [30], suggesting that Amp3p plays an important role in ALP location. PHO8 encodes the sole vacuolar integral membrane ALP in S. cerevisiae [48] and is commonly used as a marker protein for monitoring autophagy [49, 50]. ALP can efficiently dephosphorylate a variety of phosphopeptides, thereby releasing phosphate groups [51]. Although the exact physiological function of ALP remains unclear, research using animal models has demonstrated that phosphate can mediate autophagy stimulation [52], which has been partially interpreted to be the potential link between ALP and autophagy. Notably, a previous study of fibroblasts, which are secretory cells that can secrete large quantities of collagen for immunity functions, found that ALP activity can contribute to collagen degradation [53]. That observation raises the possibility that ALP may also contribute to the degradation of other secretory proteins such as lignocellulases in filamentous fungi. In the present study, we found that NcAP3m protein following brief exposure (4 h) of N. crassa cells to microcellulose was not located at vacuole/lysosome-like structures as previously reported in eukaryotes. Instead, NcAP3m was located in the tip region of fungal hyphae and likely overlapped with the Spitzenkörper comprising numerous small secretory vesicles. This localization implies that NcAP3m is a critical component of the secretory vesicle membrane and mediates extracellular enzyme secretion. As the culture time was increased to 48 h, rapid lignocellulase synthesis was observed, protein secretion flux increased exponentially, and NcAP3m protein was found to have relocated to large vacuole/lysosome-like structures. As vacuole/lysosome function is usually linked to protein degradation and nitrogen source recycling, we speculate that high rates of secretory protein synthesis and secretion should cause fungal cells to temporarily experience nitrogen source starvation; this in turn would activate the recycling of nutrients via partial lignocellulase degradation. The AP-3 complex consequently seems to balance the secretion and degradation of secretory proteins in fungal cells. Malfunction of the AP-3 complex attenuates the degradation process, which in turn enhances secretion. To check whether ALP contributes to lignocellulase degradation, we also tested lignocellulase secretion capacity in two putative ALP gene KO mutants and found that the deletion of the ALP encoded by NCU08997 led to a hypersecretion phenotype in N. crassa. Because the level of hypersecretion was lower than that expected based on the disruption of the AP-3 complex, we hypothesize that other classes of vacuolar enzymes involved in the degradation process may also require the AP-3 complex for correct sorting. Nevertheless, the means by which the AP-3 complex mediates these degradation enzymes locations in N. crassa remains unknown and requires further investigation. We found that the deletion of NCU03145, which encodes a vacuolar membrane zinc transporter in N. crassa, also enhances lignocellulase secretion. In S. cerevisiae, the counterpart of NCU03145 is ZRC1, which functions in transporting zinc from cytosol to vacuole for storage [54]. Vacuolar zinc transporters have been shown to contribute to the maintenance of ALP accumulation and activity [55], implying that the hypersecretion phenotype of ΔNCU03145 might also result from disruption of ALP function. Considering all of these results, we propose a novel model, illustrated in Fig. 9, to explain how the AP-3 complex determines the ultimate fate of lignocellulase.
Fig. 9.
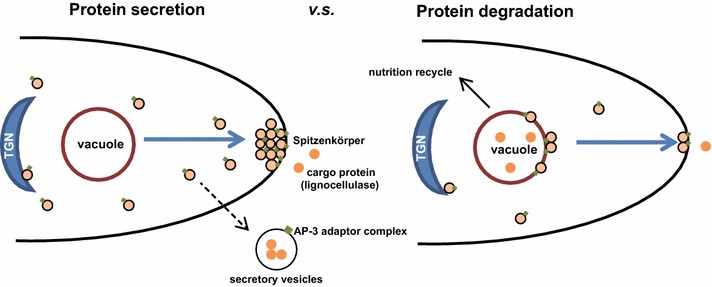
Schematic model illustrating a proposed mechanism for adaptor protein 3 (AP-3) complex-mediated lignocellulase fate decision in Neurospora crassa. When N. crassa cells are exposed to Avicel at an early stage (4 h), lignocellulases are synthesized and secreted into the extracellular matrix for nutrient acquisition. During this period, the AP-3 complex was observed to accumulate in the tip region—considered the main region for cellulolytic enzyme secretion. As high protein flux tends to temporarily lead to intracellular nitrogen source depletion, partial enzyme molecules at the logarithmic stage of fermentation (48 h) might suffer degradation so that the cell can obtain amino acids through nutrient recycling to maintain normal cell physiological activities such as somatic growth. At this time, the AP-3 complex was found to have re-located to vacuolar membranes, likely mediating delivery of cargo protein (lignocellulases) to secretory vesicles for degradation. Under this scenario, disruption of AP-3 complex function would lead to continued protein secretion, and the hypersecretion phenotype of ΔNcap3m can be reasonably explained.
According to our results, the expression of lignocellulase genes at the transcriptional level had an effect in ΔNcap3m and ΔNcap3b. The typical RESS phenomenon attenuated in these mutants. Although we could not determine the precise mechanism, we speculate that this change may be an indirect consequence of lignocellulase secretion induced by AP-3 complex malfunction, which could yield more efficient degradation of cellulose to produce biopolymer inducers to maintain lignocellulase gene expression. Although the xylanase activity change was proportionate to the protein enhancement level, we additionally noticed that the activity increase of cellulases such as endoglucanase under Avicel conditions, approximately 20%, was not as high as the enhancement of secreted enzymes protein titers (80%). We currently have no good explanation for this observation. As mentioned above, deletion of S. cerevisiae amp3 resulted in a pronounced accumulation of ALP in the cytoplasm, raising the possibility that ALPs in ΔNcap3m or ΔNcap3b may be secreted in the extracellular matrix, thus modifying lignocellulases and interfering with the enzyme activity.
Conclusions
In this study, we used gene deletion mutant stocks of N. crassa to investigate the function of orthologs of T. reesei genes whose mutations might contribute to the hypersecretion phenotype of the RUT-C30 strain. We identified several potential targets, especially the AP-3 complex, involved in the lignocellulase secretion pathway. This study provides a novel view of lignocellulase secretion and suggests a new strategy for future improvement of fungal strains.
Methods
Strains
Neurosporacrassa strains, including the WT (FGSC#2489) as well as gene KO mutants, were obtained from the Fungal Genetics Stock Center (FGSC; http://www.fgsc.net/) [56]. Double deletion strains (such as ΔNcap3mΔNcap3b) were generated by performing sexual crosses following previously described protocols [57]. The genotypes of single and double deletion strains were confirmed by PCR as described in Wu et al. [58] using the primers listed in Additional file 4: Table S1. The QM6a strain of T. reesei was kindly donated by Dr. Monika Schmoll (Department of Health and Environment-Bioresources, Austrian Institute of Technology).
Culture conditions and screens for N. crassa KO mutants
Neurospora crassa stock cultures were maintained on minimal medium agar slants containing 1× Vogel’s salt with 2% (w/v) sucrose [59] at 25°C. For enzyme production, 10-day-old conidia of N. crassa strains were suspended in sterile water and inoculated using a final concentration of 1 × 106 per mL into a 250-mL Erlenmeyer flask containing 100 mL medium [60] [1× Vogel’s salt, 2% w/v crystalline cellulose (Avicel PH-101; Sigma-Aldrich, St. Louis, MO, USA), and 0.75% w/v yeast extract (Sigma-Aldrich)]. Flask batch culturing was performed at 25°C under constant light and with agitation (200 rpm) for 7 days; extracellular protein titers were then measured based on the Bradford method. For secreted protein assays, two biological replicates were cultured per strain and three technical replicates were conducted per culture.
Identification of orthologs and phylogenetic analysis
The protein sequences of genes with potential malfunctions due to various mutations of T. reesei RUTC30 were extracted from the latest genome data maintained by the DOE Joint Genome Institute (http://genome.jgi.doe.gov/TrireRUTC30_1/TrireRUTC30_1.home.html) using Perl scripts. Homologs of N. crassa proteins were identified using local BLASTp (version Blast+ 2.2.28) with an E value <10−5 applied as a cutoff. Phylogenetic analysis of Ncap3m was carried out in MEGA6 using the maximum likelihood method with 1,000 bootstrap replicates. Database homology searching was performed with the local BLAST program as described above.
Complementation of ΔNcap3m in N. crassa
Complementary plasmids containing the Ncap3m coding region under the control of native or strong promoters were constructed separately. For the former case, the 1,566-bp full-length ORF of Ncap3m without a stop codon and with a 1,000-bp upstream putative native promoter region was PCR-amplified from WT N. crassa genomic DNA using the primers Pnative-F and ap3m-R (Additional file 4: Table S1). After digestion with NotI and PacI, the fragment was inserted into plasmid pMF272 [61] and the resulting plasmid was designated as Pn-ap3m. For the latter case, the 1,566-bp ORF region of Ncap3m was PCR amplified from WT N. crassa genomic DNA using the primer set ap3m-F/R (Additional file 4: Table S1), digested with XbaI and PacI, and then cloned into the downstream ccg-1 promoter within plasmid pMF272. The resulting plasmid was designated as Pc-ap3m. Complementary plasmids were transformed into the host strain with a double deletion (ΔNcap3m:his3-) according to the method described in Margolin et al. (http://www.fgsc.net/fgn44/margol.html). Transformants were screened by selection for histidine prototrophy and green fluorescent protein fluorescence of conidia.
T. reesei ap3m gene re-annotation and cloning
For a heterologous functional complementation analysis, we conducted an inter-complementation assay by introducing the orthologs of NCU03998 in T. reesei into the ap3m mutant. The current ORF of tre53811 (the predicted ortholog of NCU03998) contained 3,658 bp with three introns and encoded a polypeptide of 1,007 amino acids containing three domains: a clathrin adaptor complex small chain, an AP-3 complex medium Mu3, and a malonyl CoA-acyl carrier protein transacylase (Additional file 1: Figure S1). We cloned the homologous gene cDNA of ap3m in T. reesei QM6a (in-house annotation, modified from jgi_Trire2_53811), consisting of 1,611 bp encoding a polypeptide of 536 amino acids containing the first two domains (amplified with primers Trap3m-F and Trap3m-R) (Additional file 4: Table S1, Additional file 2: Figure S2, Additional file 5: Figure S3). This gene under the control of the ccg1 promoter was named Pc-Trap3m and then transformed into the host strain as described above to yield the selected transformant Pc-Trap3m.
RNA extraction
Ten-day-old conidia of N. crassa strains were collected and inoculated into 1× Vogel’s salt solution with 2% sucrose and grown for 16 h. Mycelia were collected, washed several times with 1× Vogel’s salt solution, transferred into medium with 2% Avicel or xylan as the carbon source, and grown to different time points (4, 48, 96, or 168 h). Mycelia were harvested by vacuum filtration, frozen immediately in liquid nitrogen, and stored at −80°C for RNA extraction. Total RNA from frozen samples was isolated using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) in accordance with the manufacturer’s protocol. An additional clean-up was performed using an RNeasy mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s RNA Clear Up instructions. RNA integrity and concentration were checked on a Nanodrop instrument and by agarose gel electrophoresis.
qPCR
Quantitative real-time PCR was performed on a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) using reagents supplied with a Toyobo One-Step qPCR kit (Toyobo, Osaka, Japan). The 20-µL reaction mixtures, with three replicates, included 75-ng template RNA, 0.4 µM primers, and 10-µL RNA-direct SYBR Green Real-Time PCR Master Mix. The relative transcript level of each gene was calculated by the method, with its expression in the WT strain used as the control and the expression of the actin gene (NCU04173) used as an internal standard for all experiments. Specific primers for qPCR are listed in Additional file 4: Table S1.
Protein and enzyme activity measurements
Total extracellular protein content in supernatants was measured using a Bio-Rad DC protein assay kit (Bio-Rad) based on absorbance at 595 nm, with bovine serum albumin used as the standard. FPA was measured by the 3,5-dinitrosalicylic acid method [62]. Exoglucanase activity was measured at 50°C using 1.0 mg/mL p-nitrophenyl-β-d-cellobioside (Sigma-Aldrich) in 50 mM citrate buffer (pH 4.8) as the substrate. The reaction mixture containing 250 µL of properly diluted enzyme and 250 µL of 1.0 mg/mL substrate in 50 mM citrate buffer (pH 4.8) was incubated for 10 min at 50°C, and the reaction was terminated by the addition of 500 µL of 1 M Na2CO3. The release of p-nitrophenol (pNP) was monitored at an absorbance at 420 nm. The control was the inactivated enzyme, which was boiled at 100°C for 10 min. pNP was used to generate a standard curve. In exoglucanase activity analyses, one unit of enzymatic activity was defined as the amount of pNP released from the substrate per minute using 1 mL enzyme under the standard assay conditions. Endoglucanase activity in culture supernatants was determined using an azo-cm-cellulose assay kit (Megazyme, Wicklow, Ireland) as described by the manufacturer. Endo-1,4-β-xylanase activities were assayed using an azo-xylan kit (Megazyme) according to the manufacturer’s instructions.
Fluorescence microscopy and image processing
To localize NcAP3m–EGFP fusion protein using microscopy, the complemented ΔNcap3m::Ncap3m–EGFP strain was inoculated into liquid minimal medium and grown for 16 h. The hyphae were harvested, washed with Vogel’s salt solution, and transferred into inducing medium containing 0.5% (w/v) Avicel for another 4 or 48 h. EGFP fluorescence observations were performed on an Olympus BX51 fluorescence microscopy system. To co-localize the Spitzenkörper, ΔNcap3m::Ncap3m–EGFP cells were also stained with red-fluorescent FM4-64 dye (Invitrogen) at a final concentration of 10 µM in Hanks’ balanced salt solution buffer following the manufacturer’s instructions.
Statistical analyses
Unless otherwise noted, all experiments were performed in triplicate and statistical tests for significance were determined via one-way analysis of variance using R (version 3.1.1).
Authors’ contributions
XP, FF, LL, SZ, and CT designed the study and XP, FF, LL, WS, and YC performed the research. XP, FF, and LL analyzed the data and XP, FF, SZ, and CT wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by a Grant from the 973 Program of China (2011CB707403 and 2011CBA00803) and the 863 Project (2012AA022203D and SS2014AA021300). The authors wish to thank Zhiyong Sun, Guoli Ma, and Huiyan Li for help with the experiments and for critical reading of the manuscript.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Abbreviations
- ALP
alkaline phosphatase
- AP-3
adaptor protein complex 3
- bp
base pair
- CCR
carbon catabolite repression
- EGFP
enhanced green fluorescent protein
- FM4-64
N-(3-triethylammoniumpropyl)-4-(6-(4(diethylamino) phenyl) hexatrienyl) pyridinium dibromide
- FPA
filter paper activity
- KO
knockout
- ORF
open reading frame
- pNP
p-nitrophenol
- qPCR
quantitative real-time PCR
- RESS
repression under secretion stress
- WT
wild type
Additional files
Protein sequence of tre53811 from Trichoderma reesei QM6a.
Diagram of tre2_53811 protein from Trichoderma reesei QM6a.
Phylogenetic tree of alkaline phosphatase proteins.
Primers used in this study.
Protein sequence of the in-house-annotated AP-3 μ subunit from Trichoderma reesei QM6a.
Footnotes
Xue Pei and Feiyu Fan contributed equally to this work
Contributor Information
Xue Pei, Email: pei_x@tib.cas.cn.
Feiyu Fan, Email: fan_fy@tib.cas.cn.
Liangcai Lin, Email: lin_lc@tib.cas.cn.
Yong Chen, Email: chen_y@tib.cas.cn.
Wenliang Sun, Email: sun_wl@tib.cas.cn.
Shihong Zhang, Email: zhang_sh@jlu.edu.cn.
Chaoguang Tian, Email: tian_cg@tib.cas.cn.
References
- 1.Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, et al. Toward a systems approach to understanding plant-cell walls. Science. 2004;306(5705):2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 2.Glass NL, Schmoll M, Cate JHD, Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 3.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y, Song X, Qin Y, Qu Y. Genome shuffling improves production of cellulase by Penicillium decumbens JU-A10. J Appl Microbiol. 2009;107(6):1837–1846. doi: 10.1111/j.1365-2672.2009.04362.x. [DOI] [PubMed] [Google Scholar]
- 5.Glenn M, Ghosh A, Ghosh BK. Subcellular fractionation of a hypercellulolytic mutant, Trichoderma reesei Rut-C30—localization of endoglucanase in microsomal fraction. Appl Environ Microbiol. 1985;50(5):1137–1143. doi: 10.1128/aem.50.5.1137-1143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan SM, Wu G. Secretory pathway of cellulase: a mini-review. Biotechnol Biofuels. 2013;6:177. doi: 10.1186/1754-6834-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mcgavin M, Lam J, Forsberg CW. Regulation and distribution of Fibrobacter succinogenes subsp. succinogenes S85 endoglucanases. Appl Environ Microbiol. 1990;56(5):1235–1244. doi: 10.1128/aem.56.5.1235-1244.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conesa A, Punt PJ, van Luijk N, van den Hondel CAMJJ. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet Biol. 2001;33(3):155–171. doi: 10.1006/fgbi.2001.1276. [DOI] [PubMed] [Google Scholar]
- 9.Zhang FX, Paterson AJ, Huang P, Wang K, Kudlow JE. Metabolic control of proteasome function. Physiology. 2007;22(6):373–379. doi: 10.1152/physiol.00026.2007. [DOI] [PubMed] [Google Scholar]
- 10.Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. BBA Mol Cell Res. 2009;1793(4):650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson R, Nevalainen H. Trichoderma reesei RUT-C30—thirty years of strain improvement. Microbiology. 2012;158(Pt 1):58–68. doi: 10.1099/mic.0.054031-0. [DOI] [PubMed] [Google Scholar]
- 12.Bisaria VS, Ghose TK. Biodegradation of cellulosic materials—substrates, microorganisms, enzymes and products. Enzyme Microb Technol. 1981;3(2):90–104. doi: 10.1016/0141-0229(81)90066-1. [DOI] [Google Scholar]
- 13.Montenecourt BS, Eveleigh DE. Semiquantitative plate assay for determination of cellulase production by Trichoderma viride. Appl Environ Microbiol. 1977;33(1):178–183. doi: 10.1128/aem.33.1.178-183.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montenecourt BS, Eveleigh DE. Preparation of mutants of Trichoderma reesei with enhanced cellulase production. Appl Environ Microbiol. 1977;34(6):777–782. doi: 10.1128/aem.34.6.777-782.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherry JR, Fidantsef AL. Directed evolution of industrial enzymes: an update. Curr Opin Biotech. 2003;14(4):438–443. doi: 10.1016/S0958-1669(03)00099-5. [DOI] [PubMed] [Google Scholar]
- 16.Ilmén M, Thrane C, Penttilä M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet. 1996;251(4):451–461. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 17.Geysens S, Pakula T, Uusitalo J, Dewerte I, Penttila M, Contreras R. Cloning and characterization of the glucosidase II alpha subunit gene of Trichoderma reesei: a frameshift mutation results in the aberrant glycosylation profile of the hypercellulolytic strain Rut-C30. Appl Environ Microbiol. 2005;71(6):2910–2924. doi: 10.1128/AEM.71.6.2910-2924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vitikainen M, Arvas M, Pakula T, Oja M, Penttila M, Saloheimo M. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genom. 2010;11:441. doi: 10.1186/1471-2164-11-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Crom S, Schackwitz W, Pennacchio L, Magnuson JK, Culley DE, Collett JR, et al. Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106(38):16151–16156. doi: 10.1073/pnas.0905848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poggi-Parodi D, Bidard F, Pirayre A, Portnoy T, Blugeon C, Seiboth B, et al. Kinetic transcriptome analysis reveals an essentially intact induction system in a cellulase hyper-producer Trichoderma reesei strain. Biotechnol Biofuels. 2014;7:173. doi: 10.1186/s13068-014-0173-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Znameroski EA, Glass NL. Using a model filamentous fungus to unravel mechanisms of lignocellulose deconstruction. Biotechnol Biofuels. 2013;6(1):6. doi: 10.1186/1754-6834-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J, Glass NL. Identification of the CRE-1 Cellulolytic Regulon in Neurospora crassa. PLoS One. 2011;6(9):e25654. doi: 10.1371/journal.pone.0025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, et al. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA. 2009;106(52):22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips CM, Iavarone AT, Marletta MA. Quantitative proteomic approach for cellulose degradation by Neurospora crassa. J Proteome Res. 2011;10(9):4177–4185. doi: 10.1021/pr200329b. [DOI] [PubMed] [Google Scholar]
- 25.Feraru E, Paciorek T, Feraru MI, Zwiewka M, De Groodt R, De Rycke R, et al. The AP-3 β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell. 2010;22(8):2812–2824. doi: 10.1105/tpc.110.075424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr opin Cell Biol. 2001;13:10. doi: 10.1016/S0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 27.Sun JP, Tian CG, Diamond S, Glass NL. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot Cell. 2012;11(4):482–493. doi: 10.1128/EC.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 2004;41(10):897–910. doi: 10.1016/j.fgb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Harris SD. The Spitzenkorper: a signalling hub for the control of fungal development? Mol Microbiol. 2009;73(5):733–736. doi: 10.1111/j.1365-2958.2009.06803.x. [DOI] [PubMed] [Google Scholar]
- 30.Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91(1):109–118. doi: 10.1016/S0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- 31.Zwiewka M, Feraru E, Moller B, Hwang I, Feraru MI, Kleine-Vehn J, et al. The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 2011;21(12):1711–1722. doi: 10.1038/cr.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan FY, Ma GL, Li JG, Liu Q, Benz JP, Tian CG, et al. Genome-wide analysis of the endoplasmic reticulum stress response during lignocellulase production in Neurospora crassa. Biotechnol Biofuels. 2015;8:66. doi: 10.1186/s13068-015-0248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodriguez-Carmona E, Baumann K, et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez IM, Chrispeels MJ. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15(2):561–576. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pakula TM, Laxell M, Huuskonen A, Uusitalo J, Saloheimo M, Penttila M. The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei—evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J Biol Chem. 2003;278(45):45011–45020. doi: 10.1074/jbc.M302372200. [DOI] [PubMed] [Google Scholar]
- 36.Fagundes MR, Lima JF, Savoldi M, Malavazi I, Larson RE, Goldman MH, et al. The Aspergillus nidulans npkA gene encodes a Cdc2-related kinase that genetically interacts with the UvsBATR kinase. Genetics. 2004;167(4):1629–1641. doi: 10.1534/genetics.103.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pebernard S, Perry JJP, Tainer JA, Boddy MN. Nse1 RING-like domain supports functions of the Smc5–Smc6 holocomplex in genome stability. Mol Biol Cell. 2008;19(10):4099–4109. doi: 10.1091/mbc.E08-02-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pakula TM, Salonen K, Uusitalo J, Penttila M. The effect of specific growth rate on protein synthesis and secretion in the filamentous fungus Trichoderma reesei. Microbiology. 2005;151:135–143. doi: 10.1099/mic.0.27458-0. [DOI] [PubMed] [Google Scholar]
- 39.Soulard A, Lechler T, Spiridonov V, Shevchenko A, Shevchenko A, Li R, et al. Saccharomyces cerevisiae Bzz1p is implicated with type I myosins in actin patch polarization and is able to recruit actin-polymerizing machinery in vitro. Mol Cell Biol. 2002;22(22):7889–7906. doi: 10.1128/MCB.22.22.7889-7906.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wendland J, Walther A. Tip growth and endocytosis in fungi. Plant Endocytosis. 2006;1:8. [Google Scholar]
- 41.Boehm M, Bonifacino JS. Genetic analyses of adaptin function from yeast to mammals. Gene. 2002;286(2):175–186. doi: 10.1016/S0378-1119(02)00422-5. [DOI] [PubMed] [Google Scholar]
- 42.Dell’Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol. 2009;21(4):552–559. doi: 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Hirst J, Barlow LD, Francisco GC, Sahlender DA, Seaman MNJ, Dacks JB, et al. The fifth adaptor protein complex. PLoS Biol. 2011;9(10):e1001170. doi: 10.1371/journal.pbio.1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullins C, Hartnell LM, Bonifacino JS. Distinct requirements for the AP-3 adaptor complex in pigment granule and synaptic vesicle biogenesis in Drosophila melanogaster. Mol Gen Genet. 2000;263(6):1003–1014. doi: 10.1007/PL00008688. [DOI] [PubMed] [Google Scholar]
- 45.DellAngelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor-like protein complex with ubiquitous expression. EMBO J. 1997;16(5):917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson F, Peden AA, Christopoulou L, Robinson MS. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol. 1997;137(4):835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starr TL, Pagant S, Wang CW, Schekman R. Sorting signals that mediate traffic of chitin synthase III between the TGN/endosomes and to the plasma membrane in yeast. PLoS One. 2012;7(10):e46386. doi: 10.1371/journal.pone.0046386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klionsky DJ, Emr SD. Membrane-protein sorting—biosynthesis, transport and processing of yeast vacuolar alkaline-phosphatase. EMBO J. 1989;8(8):2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 50.Klionsky DJ. Monitoring autophagy in yeast: the Pho8Δ60 assay. Methods Mol Biol. 2007;390:363–371. doi: 10.1007/978-1-59745-466-7_24. [DOI] [PubMed] [Google Scholar]
- 51.Donella-Deana A, Ostojic S, Pinna LA, Barbaric S. Specific dephosphorylation of phosphopeptides by the yeast alkaline-phosphatase encoded by Pho8 gene. Biochim Biophys Acta. 1993;1177(2):221–228. doi: 10.1016/0167-4889(93)90044-P. [DOI] [PubMed] [Google Scholar]
- 52.Dai XY, Zhao MM, Cai Y, Guan QC, Zhao Y, Guan YF, et al. Phosphate-induced autophagy counteracts vascular calcification by reducing matrix vesicle release. Kidney Int. 2013;83(6):1042–1051. doi: 10.1038/ki.2012.482. [DOI] [PubMed] [Google Scholar]
- 53.Cate AR, Syrbu S. A relationship between alkaline phosphatase activity and the phagocytosis and degradation of collagen by the fibroblast. J Anat. 1974;117(Pt 2):351–359. [PMC free article] [PubMed] [Google Scholar]
- 54.Miyabe S, Izawa S, Inoue Y. The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem Bioph Res Commun. 2001;282(1):79–83. doi: 10.1006/bbrc.2001.4522. [DOI] [PubMed] [Google Scholar]
- 55.Qiao W, Ellis C, Steffen J, Wu CY, Eide DJ. Zinc status and vacuolar zinc transporters control alkaline phosphatase accumulation and activity in Saccharomyces cerevisiae. Mol Microbiol. 2009;72(2):320–334. doi: 10.1111/j.1365-2958.2009.06644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCluskey K. The fungal genetics stock center: from molds to molecules. Adv Appl Microbiol. 2003;52:245–262. doi: 10.1016/S0065-2164(03)01010-4. [DOI] [PubMed] [Google Scholar]
- 57.Cai PL, Wang B, Ji JX, Jiang YS, Wan L, Tian CG, et al. The putative cellodextrin transporter-like protein CLP1 Is involved in cellulase induction in Neurospora crassa. J Biol Chem. 2015;290(2):788–796. doi: 10.1074/jbc.M114.609875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu W, Hildebrand A, Kasuga T, Xiong X, Fan Z. Direct cellobiose production from cellulose using sextuple β-glucosidase gene deletion Neurospora crassa mutants. Enzyme Microb Technol. 2013;52(3):184–189. doi: 10.1016/j.enzmictec.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 59.Vogel HJ. A convenient growth medium for Neurospora (Medium N) Microbiol Genet Bull. 1956;13:5. [Google Scholar]
- 60.Yazdi MT, Radford A, Keen JN, Woodward JR. Cellulase production by Neurospora Crassa—purification and characterization of cellulolytic enzymes. Enzyme Microb Technol. 1990;12(2):120–123. doi: 10.1016/0141-0229(90)90084-4. [DOI] [PubMed] [Google Scholar]
- 61.Folco HD, Freitag M, Ramon A, Temporini ED, Alvarez ME, Garcia I, et al. Histone H1 is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot Cell. 2003;2(2):341–350. doi: 10.1128/EC.2.2.341-350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghose TK. Measurement of cellulase activities. Pure Appl Chem. 1987;59(2):257–268. doi: 10.1351/pac198759020257. [DOI] [Google Scholar]


