Abstract
Cancer, developmental biology and tissue injury present multiple examples where groups of cells residing in close proximity communicate via paracrine factors. It is nearly impossible to dissect such cellular interactions in vivo and is quite challenging in vitro. The goal of this study is to utilize a reconfigurable microfluidic device in order to study paracrine signal exchange between groups of primary hepatocytes in vitro. Previously, we demonstrated that hepatocytes residing on protein spots containing collagen/HGF spots expressed epithelial (hepatic) phenotype and also rescued epithelial phenotype in neighboring hepatocytes on collagen spots that did not receive direct HGF stimulus. Herein, we designed a microfluidic device with parallel fluidic channels separated by retractable (reconfigurable) walls and employed this device to investigate interactions between groups of hepatocytes, some stimulated with HGF others not stimulated. Using novel reconfigurable microfluidic device, we demonstrate that cultivation on HGF-containing protein spots upregulates production of endogenous HGF in hepatocytes and that these HGF molecules diffuse over, causing phenotype enhancement in the recipient cells. We also show that selective treatment of the recipient hepatocytes with c-met inhibitor (SU11274) diminishes the rescue effect, as gauged by the down-regulation of albumin and HGF expression. Our study is one of the first to demonstrate paracrine signaling via HGF in primary hepatocytes. More broadly, tools and methods described here may be used to study paracrine signaling in other types of cells and will have relevance for various fields of biomedical research from cancer to immunology.
Introduction
The liver is a metabolically complex organ responsible for synthesis of serum proteins, production of urea and metabolism of xenobiotics. It is typically subdivided into three zones as a function of proximity to the portal vein – a major blood conduit in the liver that brings nutrients to be consumed and chemicals/toxins to be metabolized. Different zones are exposed to varying oxygen concentrations and have different metabolic capacities. For example, the region near the central vein (perivenous) expresses higher amounts of alcohol dehydrogenase and CYP2E1 and is better-abled to metabolize hepatotoxicants such as alcohol.1 However, this zone is also exposed to lower oxygen concentrations making it more vulnerable to reactive oxidative species and is the region where alcoholic liver disease originates. It is likely that upon injury, liver cells residing in neighboring zones communicate with each other via soluble factors.2, 3
A traditional view of hepatocytes as parenchymal cells that receive paracrine signals from neighboring nonparenchymal cells (e.g. stellate cells) without firing signals back is changing, with hepatocytes recognized as active participants in the cross-talk.4, 5 After all, hepatocytes do express genes encoding for growth factors (GFs) TGF-α, IGF, IL-1, EGF, HGF and their receptors.5-7 Among these GFs, hepatocyte growth factor (HGF) has been identified as a key signal contributing to liver development, regeneration and repair after injury.8, 9 Improved understanding of paracrine signal exchange in the context of liver injury and regeneration will be of benefit to the development of novel therapeutic strategies.
Microfluidic devices allow for cells to be seeded into desired channels, selectively stimulated and retrieved, making such devices particularly well suited for investigating interactions between cells.10-17 Because convection and diffusion can be controlled precisely, microfluidic devices are being used to study cellular interactions via secreted signals to model injury, immune response or tissue regeneration.18-21
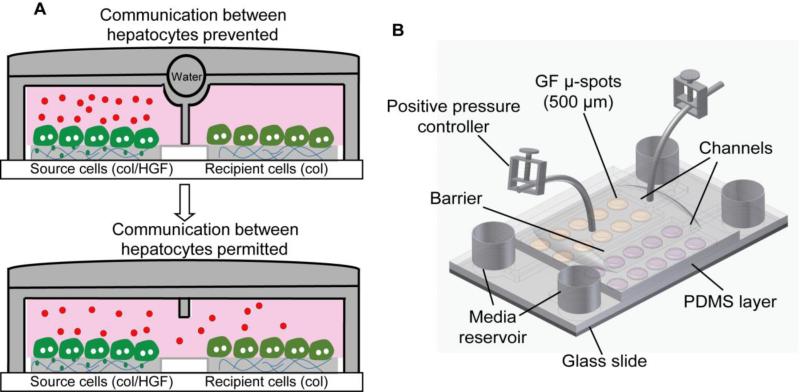
In a series of papers we demonstrated that hepatocytes cultured atop protein spots containing HGF and matrix protein like collagen I (col) retain differentiated epithelial phenotype.22, 23 In the most recent study we printed collagen spots with and without HGF in alternating columns and observed that hepatocytes receiving HGF signals from the spots appeared to rescue phenotype of hepatocytes residing on collagen alone.24 However, because cells resided in the open culture system it was not possible to conclusively determine whether groups of cells were communicating by paracrine factors. In the present study, a reconfigurable microfluidic device was used to place groups of hepatocytes in neighboring chambers separated by a retractable barrier (valve) that permitted or prevented cellular communication. The diagram describing biological questions investigated in this paper is shown in Fig. 1A. Using this microfluidic system we demonstrated that primary hepatocytes cultured atop HGF spots upregulate production of endogenous HGF, which then diffuses to neighboring hepatocytes, and protects these neighboring cells against de-differentiation and epithelial-to-mesenchymal transition (EMT). A reconfigurable microfluidic device described here offers new ways to study cross-talk between groups of cells residing in close proximity to each other. Biologically, our findings are quite interesting since growth factor production and release is normally attributed to to nonparenchymal cells of the liver (e.g. stellate cells). We demonstrate that primary hepatocytes produce sufficient levels of HGF to affect phenotype of neighboring cells.
Fig. 1.
Reconfigurable microfluidic device. (A) Schematic showing the communication between two groups of hepatocytes through endogenous signals. Microfluidic chambers in its close state allow accumulation of secreted signals produced by differentiated hepatocytes (source cells) on col/HGF spots in small volume. The accumulated signals can diffuse into next chamber with deactivation of barrier and may control the cellular behavior of neighboring cells (recipient cells). (B) Schematic illustration of a two-layer reconfigurable microfluidic device. The bottom PDMS layer binds to glass and defines the dimensions of the microfluidic channels. The top layer binds to the bottom layer and contains a water chamber that is used to switch between open and closed states of the device. Col, collagen type I.
Materials and methods
Chemicals and materials
Glass slides (75 × 25 mm2) were obtained from VWR (West Chester, PA). Thiol-silane was purchased from Gelest, Inc. (Morrisville, PA). Poly(dimethylsiloxane) (PDMS) was acquired from Dow Corning (Midland, MI). Collagenase, collagen from rat tail (type I), AlexaFluor 488 anti-sheep IgG and AlexaFluor 546 anti-mouse IgG were purchased from Invitrogen (Carlsbad, CA). Hepatocyte growth factor (HGF) and transforming growth factor-β1 (TGF-β1) were obtained from Sigma– Aldrich (St. Louis, MO). Sheep anti-rat albumin antibody was purchased from Bethyl Laboratories (Montgomery, TX). Phosphate-buffered saline (1× PBS), Duelbecco's modified eagle medium (DMEM), sodium pyruvate, DAPI stain and fetal bovine serum (FBS) were purchased from Life technologies. Paraformaldehyde was purchased from Electron Microscopy Sciences (Hatfield, PA). SU11274 was purchased from Selleckchem.
Fabrication of microfluidic devices
The two-layer microfluidic cell culture platform was fabricated using standard soft-lithography techniques and replica molding of PDMS (Ellsworth Adhesives, Germantown, WI) (Fig. 1B). The bottom layer was comprised of two adjacent microchambers (of the dimension 8×1.8×0.075 mm (length×width×height)), separated by a narrow wall of PDMS (width of 200 μm). The mold for the first layer of PDMS was made by a two-layer fabrication technique.25 Briefly SU8 2010 first formed a 10 μm high rectangular slab on a silicon wafer. SU8 2050 was then spin-coated to define the mold for the 75 μm high chambers and channels. PDMS base and its curing agent were mixed in a ratio of 10:1 and poured onto the molds in a petri dish. The dish was degassed for 1 hour before being incubated for 2 hours at 70 °C for curing. After the solidified PDMS layers were peeled from its mold, a sharp metal puncher was used to generate holes for inlets and outlets. The first and second PDMS layers were bonded together after both surfaces were treated with oxygen plasma. Subsequently, the PDMS piece was placed on top of a glass slide with imprinted protein spots to complete the device. Four cloning cylinders (Fisher Scientific, Pittsburgh, PA) were attached to the loading wells as reservoirs for culture media and two microbore tubes (Cole-Parmer, Vermon Hills, IL) were attached to the control chamber by gluing with liquid PDMS. The microfluidic device was then sterilized under UV light in a tissue culture hood for 1-2 h before it was ready to culture cells.
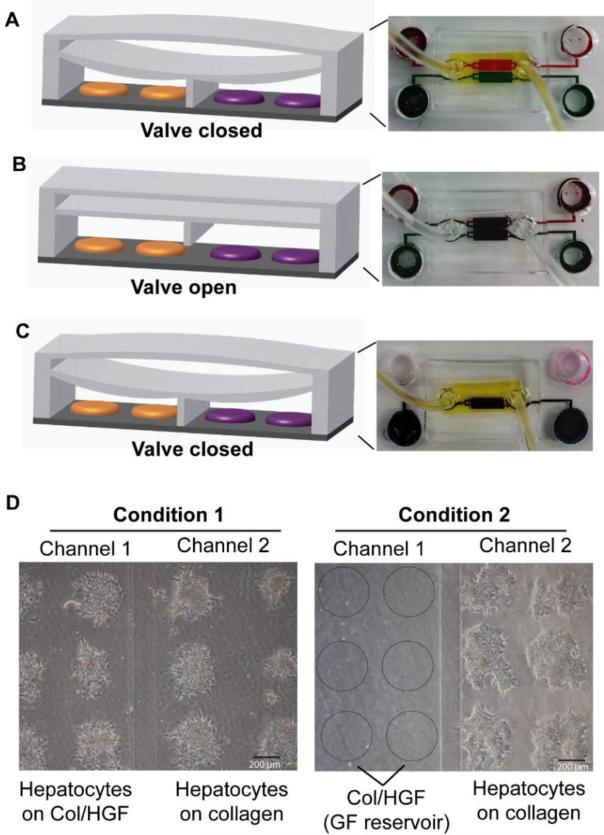
The second PDMS layer was used to separate the cell culture chambers by creating a pressure chamber on the top that can be filled with 0.2–0.3 ml of water. A microbore tube attached with a syringe was used to inject water in order to create a hydraulic pressure on the chambers when separation between two chambers was needed (Fig. 2A). The tubes were clamped in order to retain fluid within the pressure chamber. The valve barrier was opened by removing water from the pressure chamber (Fig. 2B).
Fig. 2.
Operation of reconfiguration microfluidic platform. (A-C) Demonstration of closed and open states with different color dyes. (A) The top chamber is filled with yellow dye and the valve barrier pushes down to completely isolate the two chambers that are then infused with red and green dyes. (B) The yellow dye is flowed out, which reverses the device in the open state, where the gap under the PDMS valve barrier allows red and green dyes mix together. (C) Yellow dye is infused again to seal off chambers. In this closed state, the dye only from one chamber is selectively replaced with water. (D) Demonstration of selective cell seeding. In condition 1, the open valve allowed seeding of cells in both chambers. In condition 2, the closed valve physically separated two chambers and allowed seeding of cells only in one chamber leaving the neighboring chamber pre-coated with collage/HGF mixture (col/HGF) as growth factor (GF) reservoir. Bright filed images shows primary hepatocytes cultured on micropatterned surfaces for 24 h.
Printing collagen and GF spots on glass substrates
Glass slides were modified with thiol-silane using protocols described previously 26. HGF was mixed with collagen and printed onto the silane-modified substrates. Briefly, collagen was dissolved in 1× PBS + 0.005% Tween-20 at 0.2 mg mL−1 concentration. HGF was mixed with the ECM solution to the desired final concentration of 500 ng mL−1 and allowed to bind to the ECM protein for 30 min at room temperature prior to printing. Protein microarrays were contact-printed under ambient conditions on silane-modified 75 × 25 mm2 glass slides using a spotbot arrayer. Pins collected protein from a 384-well plate, dispensing 20–70 nL of solution onto the glass slide and forming circle spots 500 μm in diameter. Pins were cleaned in acetone via sonication for 5 min and subsequently washed with pin-cleaning solution (DI water and isopropanol) between each protein pick-up. Protein arrays were kept in a refrigerator overnight prior to cell cultivation.
Cultivation of primary hepatocytes in reconfigurable microfluidic devices
Adult female Lewis rats weighing 125–200 g were purchased from Charles River Laboratories (Boston, MA) and fed with a commercial diet and water. All animal experiments were performed according to the National Institutes of Health (NIH) guidelines for the ethical care and use of laboratory animals. The experimental protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of California, Davis. Primary hepatocytes were isolated from adult female Lewis rats using a standard two-step collagenase perfusion procedure.27 Typically, 100 to 200 million hepatocytes were obtained with viability >90% as determined by trypan blue exclusion. Primary hepatocytes were maintained in DMEM supplemented with epidermal growth factor (EGF), glucagon, hydrocortisone sodium succinate, recombinant human insulin, 200 units per mL penicillin, 200 mg mL−1 streptomycin and 10% FBS. For cell seeding experiments, water was first injected in the top PDMS layer to isolate the two cell chambers. Hepatocytes were suspended in culture medium at a concentration of 20 × 106 cells mL−1 and introduced to the inlet of a microfluidic chamber (50 μL per inlet) to induce passive flow. After 30 min of incubation at 37 °C, hepatocytes bound of ECM/GF domains, but did not attach on the surrounding silane-modified surface. Subsequently, the inlets were washed twice with media to remove unbound hepatocytes. Fresh media (450 μL/inlet) was then added and changed daily subsequently. Water in the top PDMS layer was removed to allow communication between the two chambers. For culturing samples in 6-well plates, a glass slide containing printed arrays of ECM/GF was cut into (1 × 1) squares to fit into a 6-well plate. The distance between rows was 250 μm and the center-to-center spacing of the spots was 1 mm. Hepatocytes were seeded to form cellular arrays using protocols described elsewhere.28 In brief, glass slides containing printed ECM/GF spots were first exposed to 3 mL of hepatocytes suspended in culture medium at a concentration of 1 × 106 cells per mL. After 1 h of incubation at 37 °C, hepatocytes bound on ECM/GF domains, but did not attach on the surrounding silane-modified surface. The samples were then washed twice in PBS to remove unbound hepatocytes and fresh media was added to the sample well.
Molecular and biochemical assays for hepatic functionality
Quantitative real-time PCR
Two groups of hepatocytes residing in neighboring chambers were recovered selectively by lowering the valve that separates the two chambers (Fig. 2C). Cells within the chamber in its closed state were treated for 10 min with trypsin-EDTA, followed by aspirating a volume of 500 μL of fresh media using a pipette and recovery of cells from outlet. The cells were then resuspended in 200 mL of lysis buffer (Roche) and then stored at −80 °C. PCR primers, listed below in Table 1, were designed for rat gene analysis. Total RNA extraction and cDNA preparation was performed according to the manufacturer's instructions (Roche). All PCR reactions were done in duplicate. The relative expression level of each gene was calculated using the comparative threshold cycle (Ct) method with GAPDH as a housekeeping gene. The number of samples (culture surfaces) used for statistical analysis of PCR data was n = 3 for all conditions.
Table 1.
Primer sequences for real-time PCR
| Gene | Sequence 5’ to 3’ |
|---|---|
| Housekeeping - GAPDH | F: AGACAGCCGCATCTTCTTGT |
| R: CTTGCCGTGGGTAGAGTCAT | |
| Albumin | F: CATCCTGAACCGTCTGTGTG |
| R: TTTCCACCAAGGACCCACTA | |
| HGF | F: CTT CTG CCG GTC CTG TTG |
| R: TCT TCT CTT CTT CTG TCC TTC TGC |
Albumin staining
For albumin immunostaining, cells were fixed in 4% paraformaldehyde + 0.3% Triton-X100 in PBS for 15 min. The cells were then incubated in blocking solution (1% bovine serum albumin (BSA) in PBS) for 1 h and exposed to sheep anti-albumin antibodies for 90 min. The samples were washed three times with PBS, and incubated with a mixture of secondary antibodies and DAPI (Invitrogen) for 1 h. Goat anti-sheep IgG antibodies conjugated with alexa-488 was used at 1:1000 dilutions. After three times washing, slides were mounted onto cover slips using DAPI stain mounting media. All incubations were performed at room temperature unless otherwise mentioned. Fluorescent images for at least three independent experiments of each condition were captured by confocal microscope (Zeiss LSM Pascal).
Albumin ELISA
The cell media was collected from microfluidic channel outlets every 24 h. After 5 days of culture, media from all the time points were mixed together and the cumulative albumin released by cells was quantified as per the manufacturer's instructions (Bethyl labs). HGF released by the cells was quantified by a mouse duoset HGF ELISA kit (R&D Systems) as per the manufacturer's instructions. For each condition, samples were acquired from three independent microfluidic devices. Each sample was run in triplicate on an ELISA plate. The antibodies used in this ELISA are cross-reactive with rat HGF but not human HGF and thus they could not detect the human HGF imprinted with the ECM mixture onto the glass slide.
Modeling secretion and diffusion of HGF in microchambers
Using geometry of the actual device, we set up a finite element model combining growth factor secretion, diffusion and convection (COMSOL Inc., Los Angeles, CA). The secretion rates of stimulated and recipient hepatocytes were derived from HGF ELISA. Cells were assumed to be uniformly distributed across ECM spots and secreting HGF at a constant rate. The main parameters used for modeling are listed in Table 2.
Table 2.
Main Parameters for modeling autocrine and paracrine signaling
| Parameters | Values |
|---|---|
| Diffusion coeff. HGF | 0.95 × 10−6 cm2/s |
| Cells per spot | 200 |
| Secretion rate of “stimulated” cells | 16.5 ng/106 cells/24 hrs |
| Secretion rate of “recipient” cells | 5.0 ng/106 cells/24 hrs |
| Media flow rate | 0.1 μL/hr |
Inhibition of HGF signaling in microfluidic devices
In order to study the role of HGF in rescuing neighboring hepatocytes in paracrine fashion, we administered c-met inhibitor (SU11274, 5 μM). Following cell seeding step, actuating the PDMS layer sealed off communication between the two chambers and the cells residing on collagen spots were exposed to c-met inhibitor for two days. The SU11274 was then washed off and the barrier separating the two groups of cells was raised to allow communication between the cells. Hepatocyte cultures without inhibitors were maintained inside microfluidic devices as controls. Media was changed daily in all the experiments described here.
Results
Operation of the reconfigurable microfluidic device
The reconfigurable microfluidic device was used to create cell culture systems where two groups of hepatocytes were placed in proximal but distinct compartments, thus allowing communication between the groups of cells and molecular biology analysis of each cell compartment separately. The concept of this experiment is shown in Fig. 1A. Groups of cells are seeded into adjacent microfluidic channels separated by a retractable wall. Raising or lowering this wall (valve) may be used to permit or prevent cellular communications by secreted factors. As highlighted in Fig. 1B, each channel contained 20 spots with 500 μm diameter spots, each spot supporting attachment of ~200 hepatocytes. Spots in one of the channels contained collagen/HGF while spots in the other channel were composed of collagen only. The adjacent cell culture chambers had individual inlets and outlets and could be perfused separately. Experiments with food dye shown in Fig. 2C highlight the possibility of controlling composition of media in each chamber individually. In addition to selective treatment of cells, this microfluidic platform allowed for selectively retrieval of cells after interactions have taken place for downstream analysis, for example by RT-PCR. Fig. 2D provides view from the bottom of hepatocytes residing on protein spots inside the microfluidic devices. Note the 200 μm wide lane running through the middle of the chamber – this is the retractable wall in the lowered position. This retractable wall allows for two populations of cells to be in close proximity without interactions, until, at a chosen time, the device is actuated and the wall is raised. One of the key questions addressed in this study is whether hepatocytes residing atop HGF-containing spots produce factors that affect neighboring hepatocytes in paracrine fashion or whether HGF imprinted into protein spots diffuses over and influences neighboring cells. The use of reconfigurable microfluidics allowed to ask these questions by creating cultures where recipient cells (on collagen) were exposed to source cells (on HGF) (left panel Fig. 2D) or to HGF spots without cells (right panel Fig. 2D).
Effects of HGF-stimulated hepatocytes on neighboring cells
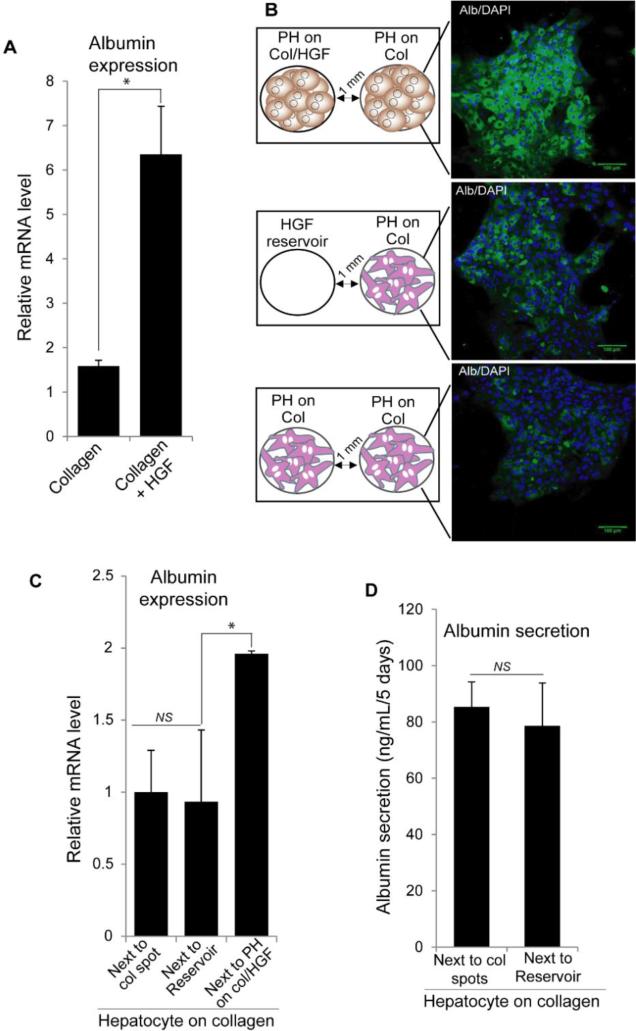
Similar to our previous reports,23, 24 hepatocytes residing on HGF-carrying protein spots cultured in microfluidic chambers for 5 days expressed higher levels of albumin compared to cells on collagen spots without HGF (Fig. 3A). In fact, hepatocytes on collagen only protein spots were expected to de-differentiate, losing epithelial phenotype and becoming more mesenchymal. We hypothesized that EMT in these cells could be prevented by HGF-producing cells placed in the neighboring microfluidic chamber.
Fig. 3.
Hepatocytes atop col/HGF spots (HGF-stimulated) rescue hepatic phenotype in neighboring cells on collagen (recipients) via cell secreted factors. (A) Expression of albumin in primary hepatocytes (PH) cultured on collagen and col/HGF spots inside microfluidic chambers analyzed by qPCR. (B) Immunofluorescence staining reveals high levels of albumin in recipient cells in three different conditions: recipient cells next to HGF-stimulated hepatocytes, col/HGF spots (reservoir), and another group of recipient cells. (C) Albumin expression analysis by qPCR in recipient hepatocytes cultured next to collagen black spots, HGF reservoirs, and HGF-stimulated hepatocytes. (D) Albumin production by recipient hepatocytes cultured next to collagen black spots and HGF reservoirs, showing there not influence on rescuing functionality by HGF released from reservoir. Results are represented as mean±SD (n=3). *: p-value < 0.05. NS: Non-significant.
To test this hypothesis, two groups of hepatocytes were placed in close proximity inside microfluidic chambers to evaluate the role of hepatocytes atop HGF spots in rescuing the neighboring hepatocytes atop collagen spots. In addition to paracrine interactions, it was also possible that HGF imprinted into proteins spots leached out affected neighboring cells. In order to elucidate these possibilities, we tested three different conditions: (i) recipient cells next to hepatocytes on col/HGF, (ii) recipient cells next to col/HGF spots without cells, and (iii) recipient cells residing in both chambers (Fig. 3B) – meaning hepatocytes in both channels were on collagen spots without HGF. The center-to-center spacing between spots was 750 μm in each chamber. Hepatocytes were first seeded in parallel chambers with valve closed and cultured for 24 h. The valves were then raised to commence communication and allow for secreted factors to diffuse over. After 5 days of culture, brightfield images revealed that there were no significant differences in morphology between recipient cells next to either stimulated cells or col/HGF spots (Supp. Fig. 1). The cells were polygonal shaped and formed tight junctions with other cells. Immunofluorescence staining for albumin, a marker for hepatocyte functionality, showed that recipient hepatocytes (on collagen) continued to express highest levels of albumin after 5 days of being cultured next to HGF-stimulated hepatocytes (Fig. 3B). On the other hand, recipient hepatocytes cultured next to col/HGF spots or cultured next hepatocytes residing on collagen were synthesizing less albumin, indicating dedifferentiation and loss epithelial phenotype. RT-PCR analysis of albumin gene expression carried out by reconfiguring the device and trypsinizing the recipient cells demonstrated a 2-fold higher expression in recipient cells cultured next to HGF-stimulated hepatocytes compared to recipient cells cultured alone or next to the col/HGF spots without cells (Fig. 3C). This observation was further validated by albumin ELISA, where the level of albumin secretion over 5 days of culture showed no significant difference between recipient cells residing next to a collagen spots and next to col/HGF spots (Fig. 3D). These results suggest that HGF leaching from protein spots does not have significant effect on phenotype of neighboring recipient cells and that hepatocytes residing atop HGF spots likely secrete signals that act in paracrine manner on the neighboring cells.
Hepatocytes cultured on top of HGF-containing substrates produce HGF in autocrine manner
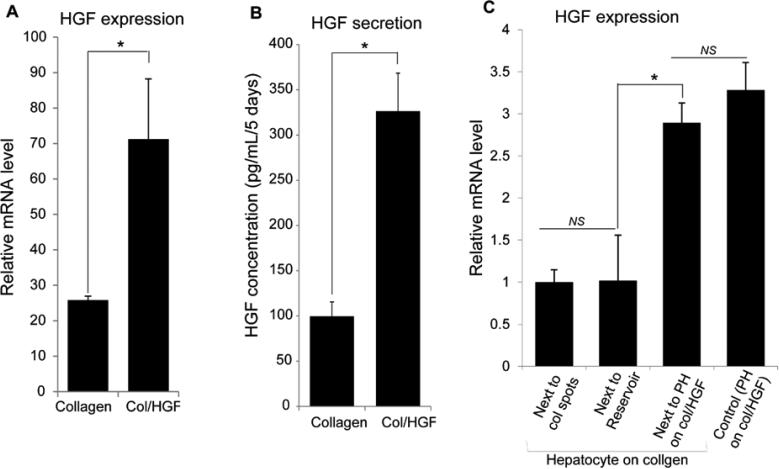
We hypothesized that cultivation of hepatocytes on top of HGF-containing surfaces triggers an autocrine loop, causing these cells to produce even more HGF. To test this hypothesis, we first compared HGF expression in hepatocytes residing on collagen only vs. collagen/HGF protein spots. Strikingly, hepatocytes cultured on col/HGF spots showed 2-fold increase in HGF expression than those hepatocytes cultured on collagen, as gauged by PCR (Fig. 4A). We also determined cumulative amounts of HGF secreted by HGF-stimulated and unstimulated hepatocytes over 5 days of culture (Fig. 4B). These ELISA results revealed that HGF-stimulated cells secreted three times higher levels of HGF relative to the cells cultured on collagen alone. These results support our hypothesis that solid phase presentation of HGF on protein spots induces hepatocytes to upregulate production of HGF in an autocrine manner. This in turn led us to hypothesize that HGF may be the key paracrine signal being exchanged between the source and recipient cells residing on protein spots in our culture system.
Fig. 4.
Hepatocytes atop HGF containing spots stimulated to secrete more HGF in an autocrine and a paracrine fashion. (A) Expression of HGF in primary hepatocytes (PH) cultured on collagen and col/HGF spots inside microfluidic chambers analyzed by qPCR. (B) Cumulative production of HGF by hepatocytes cultured for 5 days on collagen and col/HGF spots inside microfluidic chambers analyzed by ELISA. (C) Gene expression of HGF by qPCR reveal a ~3 fold higher expression in recipient hepatocytes than cells cultured next to collagen or col/HGF spots without cells. The expression of HGF transcript in recipient hepatocytes was comparable to that of hepatocytes cultured on col/HGF spots. Results are represented as mean±SD (n=3). *: p-value < 0.05. NS: Non-significant.
HGF acts in a paracrine fashion to rescue epithelial phenotype in neighboring hepatocytes
In the next set of experiments we wanted to test our hypothesis of HGF being the key paracrine factor involved in cell-cell communication and phenotype rescue in our system. Leveraging our reconfigurable microfluidic platform, we analyzed HGF gene expression in recipient cells cultured under three conditions mentioned before: (i) next to hepatocytes on col/HGF, (ii) next to col/HGF spots without cells, and (iii) recipient cells on collagen in both chambers. Hepatocytes residing on collagen/HGF were used as a positive control. As highlighted by data in Fig. 4C, recipient cells cultured next to hepatocytes on col/HGF spots expressed 3 fold higher levels of HGF compared to two other conditions tested. Strikingly, the level of HGF gene expression in these cells was similar to HGF expression in hepatocytes receiving direct stimulation from HGF-containing protein spots.
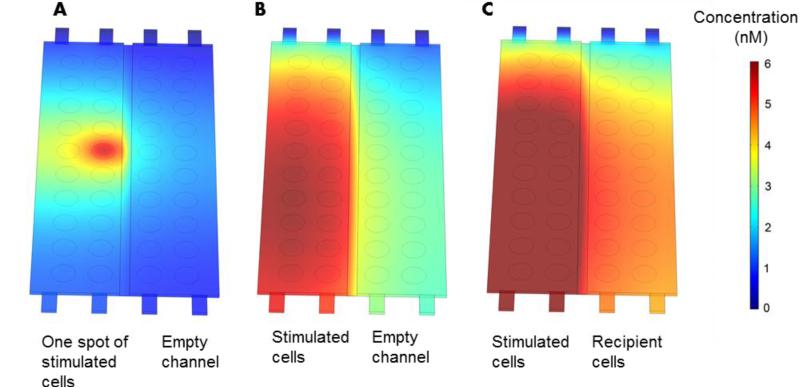
One may wonder about the distances between HGF producing cells and recipient cells as well as about GF concentration gradients that may be present inside the microfluidic devices. To answer this question we set-up a diffusion model that utilized device geometry and cell numbers. HGF secretion rates for source and recipient hepatocytes were determined from ELISA data described in Fig. 4B. The summary of parameters used for constructing this model is provided in Table 2. First, modeling was used to visualize HGF concentration field around one cluster of hepatocytes - 500 μm island containing ~200 cells. As seen from Fig. 5A, after 24 h HGF diffusion field spread beyond the source spot and into the neighboring groups of hepatocytes. Next we modeled secretion form 20 such hepatocytes islands assuming secretion rate for hepatocytes residing on HGF spots (stimulated cells). As seen from Fig. 5B, after 24 h, HGF secretions from individual hepatocyte islands combined to form a uniform HGF field with concentration ranging from 4 to 6 nM. One should note that HGF levels inside the microchambers are 20 to 30 fold higher than typical levels of exogenous HGF added into culture media (20 ng/ml or 0.25 nM). Thus, our modeling demonstrates that hepatocyte secreting HGF established a high local concentration of this hepato-inductive signal. The model described in Fig. 5B also incorporates slow flow (convection) which explains asymmetric concentration profile with lower concentration in the top of the device. This model also reveals that HGF secreted by hepatocytes in one chamber diffuses readily into the adjacent cell chamber. The final model, presented in Fig. 5C, was used to visualize HGF concentrations in the scenario with source (stimulated) and recipient hepatocytes being present in the adjacent microchambers. This model, highlights the fact that after 24h the recipient cells experience concentration of HGF similar to that of source of cells in the adjacent chamber. Overall, modeling in Fig. 5 underscores that HGF produced by the cells diffused rapidly across adjacent microchambers and supports the feasibility of secreted HGF affecting phenotype of recipient hepatocytes in paracrine manner.
Fig. 5.
COMSOL simulation of HGF secretion, diffusion and convection in the microchambers. (A) One group of stimulated hepatocytes secreting HGF. (B) 20 islands containing hepatocytes secreting HGF. Note adjacent chamber is assumed to be free of cells in this model. (C) A scenario where source and recipient hepatocytes are secreting HGF. Source hepatocytes reside on top of HGF spots and become stimulated to produce HGF at 3 times the rate of recipient cells. Secretion rates and other parameters relevant to modeling are described in Table 2.
The findings described in Fig. 4 and Fig. 5 clearly demonstrate that recipient cells in our system expressed HGF at elevated levels and that these recipient cells needed to reside next to HGF-producing hepatocytes. However, these results did not conclusively prove that HGF was the key paracrine factor delivered from the source cells to the recipient cells.
Blocking HGF receptors in the recipient cells eliminates rescue effect
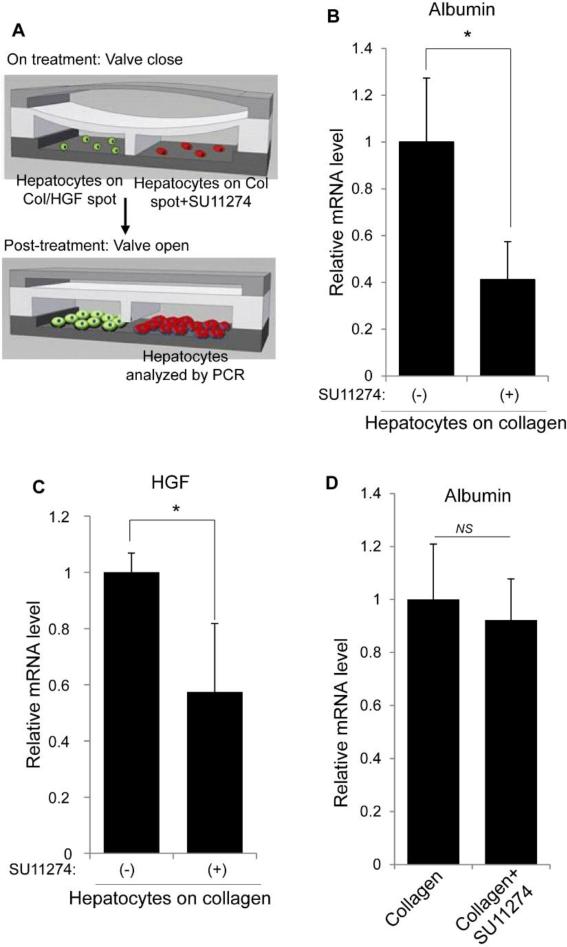
To verify the role of HGF as a paracrine factor, hepatocytes atop collagen were selectively treated with HGF receptor (c-met) inhibitor SU11274. Fig. 6A shows a schematic of how the reconfigurable microfluidic device was used to selectively expose recipient hepatocytes to SU11274. Briefly, both recipient and HGF-stimulated hepatocytes were cultured for 24 h in separate channels with the valve in closed state. The recipient hepatocytes were then exposed to SU11274 for 48 h while the HGF-stimulated hepatocytes were cultured with regular hepatocyte culture media. After 48 h of incubation, the SU11274 was washed off and the barrier separating the two groups of cells was raised to allow communication between recipient and producer hepatocytes. A control experiment was carried out in parallel where recipient cells were maintained in the same condition without SU11274. The cells were then incubated for an additional 2 days (total 5 days of culture) before being harvested for PCR analysis. As shown in Fig. 6B and 6C, c-met inhibition caused a 2.5-fold decrease in albumin expression and a 1.7-fold decrease in HGF expression compared to control experiment without c-met inhibitor. Before raising the valve to initiate communication, cells at day 3 were analyzed for albumin expression to ensure that the two groups of recipient hepatocytes were functionally indistinguishable (Fig. 6D). These findings indicate that recipient cells treated with c-met inhibitor were not able to respond to paracrine HGF and as a result expressed lower levels of albumin and HGF. Fig. 6D demonstrates that c-met inhibition made HGF and albumin expression in hepatocytes residing next to paracrine source of HGF indistinguishable from hepatocytes alone. These results underscore central role played by paracrine HGF signals in our culture system.
Fig. 6.
Effects of abrogating HGF signals received by hepatocytes on collagen (recipient). (A) Illustration of selective application of c-met receptor inhibitor (SU11274) to one chamber that contained recipient hepatocytes. The other chamber contained hepatocytes on col/HGF (HGF-stimulated). (B-C) PCR results show a ~2 fold decrease in both albumin (B) and HGF (C) expression, suggesting the important role HGF signaling in maintaining hepatocyte functionality. (D) Primary hepatocytes cultured inside microfluidic chambers in presence and absence of SU11274 for 48 h (from 2 to 3 days of culture) were analyzed for albumin expression to ensure that the two groups of recipient hepatocytes were functionally indistinguishable. Results are represented as mean±SD (n=3). *: p-value < 0.05. NS: Non-significant.
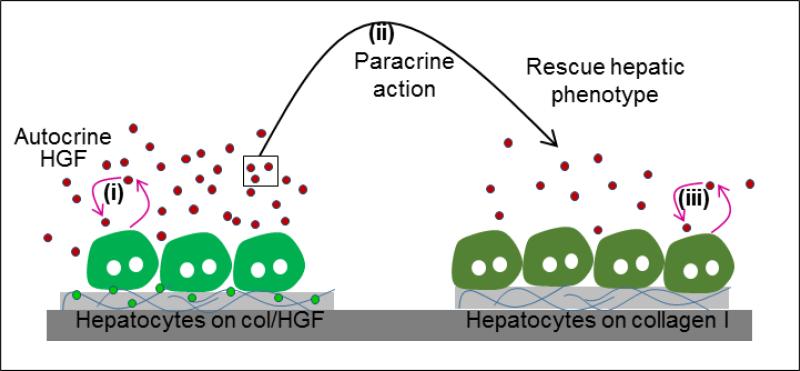
Overall, we demonstrate that reconfigurable microfluidic devices may be used to parse out role of paracrine and autocrine signaling in cell communication. From a liver biology standpoint, we demonstrate that primary hepatocytes are able to respond to surface-immobilized HGF in autocrine fashion by producing more HGF. While we did not measure local HGF concentration, our studies revealed that this concentration was high enough to rescue epithelial phenotype of neighboring hepatocytes that did not receive direct HGF stimulus from the surface. In fact, as highlighted in Fig. 4C, paracrine HGF signal caused recipient cells to upregulate production of their own HGF, possibly triggering an autocrine loop. The mechanism of interaction is summarized in Fig. 7. One group of hepatocytes receives HGF signal from the surface, produces autocrine HGF, which then diffuses over to the neighboring group of cells and triggers autocrine signaling there.
Fig. 7.
Schematic showing the proposed mechanism involve in the rescue of liver-specific functionality in hepatocytes. (i) Hepatocytes on HGF spots are stimulated to produce more HGF. (ii) These secreted HGF molecules diffuses to neighboring hepatocytes atop collagen spots in close proximity, and (iii) rescue epithelial phenotype by activating autocrine production of HGF. Abbreviation: col/HGF, mixture of collagen type I and HGF.
Discussion
In this study, we sought to develop a novel culture system for addressing a scenario where groups of cells of the same type communicate with each other via similar secreted factors. This scenario is commonly encountered during tissue development and injury; therefore, while our study focused on the liver cells, the tools and methods described here are broadly applicable to other cellular systems. The biological goal of this study was to investigate a hypothesis that hepatocytes are able to secrete hepato-inductive signals that can act in paracrine and autocrine manner to rescue phenotype of neighboring cells. To prove this point, we set up an experiment where HGF was mixed with collagen I and was printed in the form of protein spots onto cell culture surfaces. Hepatocytes cultured on top of these spots upregulated expression and synthesis of HGF, suggesting that printed GF spots served as triggers of autocrine signaling. Furthermore, we deployed reconfigurable microfluidic devices to conclusively prove that hepatic HGF diffused from the source cells and rescued hepatic phenotype of neighboring cells that did not receive direct HGF signals. To put our study in a perspective, hepatocytes are not typically thought of as growth factor-producing cells in the liver. This role is typically assigned to hepatic stellate cells – resident stromal cells in the liver.29 Our study is one of the first to demonstrate that hepatocytes may constitute a significant source of HGF and that hepatic HGF may influence epithelial phenotype of neighboring cells. Our study supports the notion that hepatocyte (parenchymal cells of the liver) are active participants in the induction and maintenance of phenotype in the healthy and diseased liver.30
Conclusion
Liver is functionally heterogeneous with phenotypically different hepatocytes residing in different zones. During injury some hepatocytes may be more affected by the injurious agent, becoming more mesenchymal while neighboring hepatocytes may retain epithelial phenotype. In the present study, we recreated such a scenario by delivering hepato-inductive signals to select groups of primary hepatocytes. We then employed reconfigurable microfluidic devices to study interactions between those hepatocytes that received direct inductive signals (in the form of HGF) and the neighboring hepatocytes that did not receive such signals. We observed that cells residing atop HGF-containing spots affected phenotype of neighboring cells that did not receive HGF stimulation. We demonstrated that hepatocytes atop HGF-containing surfaces produced HGF in autocrine manner and that these secreted GF molecules likely diffused across to rescue phenotype of neighboring recipient cells. Selective inhibition of c-met in recipient cells abrogated rescue effect despite the fact that “source” hepatocytes and signals secreted by them were still present in the microfluidic device.
Beyond liver-related applications, this study demonstrates the utility of reconfigurable microfluidic devices to study paracrine interactions between very similar groups of cells residing in close proximity to each other. The need to study such interactions arises in cancer biology, neuroscience, immunology and developmental biology; therefore, we envision broad applications for this technology.
Supplementary Material
Insight, innovation, integration.
This paper describes the use of reconfigurable microfluidic devices to dissect signaling of HGF between two groups of hepatocytes. We demonstrate that hepatocytes, that are typically not known as growth factor-producing cells, can be induced to secrete large amount of important factors like HGF that can diffuse to neighboring hepatocytes and trigger autocrine signaling. The concept of studying interactions between similar cells in close proximity has broad implications in tissue engineering, cancer biology and among other fields.
Acknowledgements
Financial support for this project was provided by NIH (R01DK079977) and NSF. Additional funding came from “Research Investments in Science and Engineering from UC Davis”
References
- 1.Jungermann K, Kietzmann T. Hepatology. 2000;31:255–260. doi: 10.1002/hep.510310201. [DOI] [PubMed] [Google Scholar]
- 2.Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Xu J, Yarborough KM, Hengstler JG. Arch Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka M, Itoh T, Tanimizu N, Miyajima A. J Biochem. 2011;149:231–239. doi: 10.1093/jb/mvr001. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Lamouille S, Derynck R. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosgrove BD, Cheng C, Pritchard JR, Stolz DB, Lauffenburger DA, Griffith LG. Hepatology. 2008;48:276–288. doi: 10.1002/hep.22335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams CM, Mehta G, Peyton SR, Zeiger AS, Van Vliet KJ, Griffith LG. Tissue Eng Part A. 2011;17:1055–1068. doi: 10.1089/ten.tea.2010.0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desbois-Mouthon C, Wendum D, Cadoret A, Rey C, Leneuve P, Blaise A, Housset C, Tronche F, Le Bouc Y, Holzenberger M. Faseb j. 2006;20:773–775. doi: 10.1096/fj.05-4704fje. [DOI] [PubMed] [Google Scholar]
- 8.Sun S, Chen G, Xu M, Qiao Y, Zheng S. PLoS One. 2013;8:e83523. doi: 10.1371/journal.pone.0083523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue F, Takahara T, Yata Y, Minemura M, Morioka CY, Takahara S, Yamato E, Dono K, Watanabe A. Gut. 2002;50:558–562. doi: 10.1136/gut.50.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giridharan GA, Nguyen MD, Estrada R, Parichehreh V, Hamid T, Ismahil MA, Prabhu SD, Sethu P. Anal Chem. 2010;82:7581–7587. doi: 10.1021/ac1012893. [DOI] [PubMed] [Google Scholar]
- 11.Hsu WM, Carraro A, Kulig KM, Miller ML, Kaazempur-Mofrad M, Weinberg E, Entabi F, Albadawi H, Watkins MT, Borenstein JT, Vacanti JP, Neville C. Ann Surg. 2010;252:351–357. doi: 10.1097/SLA.0b013e3181e982ba. [DOI] [PubMed] [Google Scholar]
- 12.Huang SB, Wang SS, Hsieh CH, Lin YC, Lai CS, Wu MH. Lab Chip. 2013;13:1133–1143. doi: 10.1039/c2lc41264k. [DOI] [PubMed] [Google Scholar]
- 13.Jang KJ, Suh KY. Lab Chip. 2010;10:36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 14.Schutte J, Hagmeyer B, Holzner F, Kubon M, Werner S, Freudigmann C, Benz K, Bottger J, Gebhardt R, Becker H, Stelzle M. Biomed Microdevices. 2011;13:493–501. doi: 10.1007/s10544-011-9517-7. [DOI] [PubMed] [Google Scholar]
- 15.Guo F, French JB, Li P, Zhao H, Chan CY, Fick JR, Benkovic SJ, Huang TJ. Lab Chip. 2013;13:3152–3162. doi: 10.1039/c3lc90067c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang CP, Lu J, Seon H, Lee AP, Flanagan LA, Kim HY, Putnam AJ, Jeon NL. Lab Chip. 2009;9:1740–1748. doi: 10.1039/b818401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu T, Yue W, Li CW, Yao X, Cai G, Yang M. Lab Chip. 2010;10:2271–2278. doi: 10.1039/c004844e. [DOI] [PubMed] [Google Scholar]
- 18.Byrne MB, Trump L, Desai AV, Schook LB, Gaskins HR, Kenis PJ. Biomicrofluidics. 2014;8:044104. doi: 10.1063/1.4887098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domenech M, Yu H, Warrick J, Badders NM, Meyvantsson I, Alexander CM, Beebe DJ. Integr Biol (Camb) 2009;1:267–274. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SA, No da Y, Kang E, Ju J, Kim DS, Lee SH. Lab Chip. 2013;13:3529–3537. doi: 10.1039/c3lc50197c. [DOI] [PubMed] [Google Scholar]
- 21.Przybyla L, Voldman J. Annu Rev Anal Chem (Palo Alto Calif) 2012;5:293–315. doi: 10.1146/annurev-anchem-062011-143122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones CN, Tuleuova N, Lee JY, Ramanculov E, Reddi AH, Zern MA, Revzin A. Biomaterials. 2009;30:3733–3741. doi: 10.1016/j.biomaterials.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 23.Jones CN, Tuleuova N, Lee JY, Ramanculov E, Reddi AH, Zern MA, Revzin A. Biomaterials. 2010;31:5936–5944. doi: 10.1016/j.biomaterials.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel D, Haque A, Jones CN, Tuleouva N, Foster E, Vu T, Reddi AH, Revzin A. Integr Biol (Camb) 2014;6:44–52. doi: 10.1039/c3ib40140e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y, Majumdar D, Jovanovic B, Shaifer C, Lin PC, Zijlstra A, Webb DJ, Li D. Biomed Microdevices. 2011;13:539–548. doi: 10.1007/s10544-011-9523-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo JH, Shin DS, Mukundan P, Revzin A. Colloids Surf B Biointerfaces. 2012;98:1–6. doi: 10.1016/j.colsurfb.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 27.Koebe HG, Dunn JC, Toner M, Sterling LM, Hubel A, Cravalho EG, Yarmush ML, Tompkins RG. Cryobiology. 1990;27:576–584. doi: 10.1016/0011-2240(90)90045-6. [DOI] [PubMed] [Google Scholar]
- 28.Revzin A, Rajagopalan P, Tilles AW, Berthiaume F, Yarmush ML, Toner M. Langmuir. 2004;20:2999–3005. doi: 10.1021/la035827w. [DOI] [PubMed] [Google Scholar]
- 29.Friedman SL. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meindl-Beinker NM, Dooley S. J. Gastro. Hepatol. 2008;S1:S122–S127. doi: 10.1111/j.1440-1746.2007.05297.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.