Abstract
Background
Plants are continuously challenged by different environment stresses, and they vary widely in their adjustability. NAC (NAM, ATAF and CUC) transcription factors are known to be crucial in plants tolerance response to abiotic stresses, such as drought and salinity. ANAC019, ANAC055, and ANAC072, belong to the stress-NAC TFs, confer the Arabidopsis abiotic stress tolerance.
Results
Here we isolated two stress-responsive NACs, CiNAC3 and CiNAC4, from Caragana intermedia, which were induced by ABA and various abiotic stresses. Localization assays revealed that CiNAC3 and CiNAC4 localized in the nuclei, consistent with their roles as transcription factors. Histochemistry assay using ProCiNAC4::GUS transgenic Arabidopsis showed that the expression of the GUS reporter was observed in many tissues of the transgenic plants, especially in the root vascular system. Overexpression of CiNAC3 and CiNAC4 reduced ABA sensitivity during seed germination, and enhanced salt tolerance of the transgenic Arabidopsis.
Conclusions
We characterised CiNAC3 and CiNAC4 and found that they were induced by numerous abiotic stresses and ABA. GUS histochemical assay of CiNAC4 promoter suggested that root, flower and local damaged tissues were the strongest stained tissues. Overexpression assay revealed that CiNAC4 play essential roles not only in promoting lateral roots formation, but also in responding to salinity and ABA treatment of Arabidopsis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0591-5) contains supplementary material, which is available to authorized users.
Background
Many adverse environmental conditions, such as drought, high salinity, extreme temperature, have severe effects on the vegetative growth and development of plants. The decrease of the productivity caused by these abiotic stresses is major challenges for modern agriculture. Plants have evolved various tiers of adaptation mechanisms to unfavorable conditions, including strategies at molecular, cellular, physiological, and biochemical level. The transcriptional activation of a large number of genes upon perception of external stresses include function proteins such as chaperones, the ion transporters, LEA (late embryogenesis abundant) proteins, osmotin, regulatory proteins such as the transcription factors (TFs) [1, 2]. The interactions between transcription factors (TFs) and their corresponding cis-acting elements act as molecular switches for gene expression, directing their temporal and spatial expression [3].
The plant-specific transcription factor NAC family (NAM, ATAF and CUC) shares a conserved NAC domain in the N terminus responsible for DNA binding and diversified C terminal domains for transcription activation [4]. Investigations among several plant species with complete genome sequences have identified 117 NACs in Arabidopsis (Arabidopsis thaliana), 151 in rice (Oryza sativa) [5], 74 in grape (Vitis vinifera) [6], and 152 in soybean (Glycine max) [7], which makes the NAC family one of the largest of TFs in plants. Tran et al. revealed that ANAC019, ANAC055, and ANAC072 responded to abiotic stress and their over-expression conferred improved drought tolerance in Arabidopsis [8]. Thus the roles of NAC family in various abiotic stresses had been noticed as compared to their role in plant development. Puranik et al. listed many NAC TFs that influenced plant stress tolerance along with their target genes [9]. Dimerization of TFs can function in modulating the DNA-binding specificity [10]. The NAC domain of approximately 150 amino acids was sufficient to form a homodimer or heterodimer [11]. The consensus sequences that NAC family members were distinguished in most cases with weak base requirements [8, 9, 12–14]. ANAC019, ANAC055, and ANAC072, belong to the stress-NAC TFs [15], and could bind to the promoter of ERD1 (EARLY RESPONSIVE TO DEHYDRATION STRESS 1) by the CATGTG motif and activate its expression [8].
In soybean, Tran et al. analyzed 31 full-length NAC genes and found that nine were induced by dehydration and exhibited a large diversity in response to high salinity, cold and ABA treatment [16]. Among these 31 NACs, GmNAC2 [17], GmNAC3, GmNAC4 [18], GmNAC5 [19], GmNAC6 [20], GmNAC11 and GmNAC20 [21] were reported either to be induced by abiotic stresses or to participate in stress tolerance. Caragana intermedia Kuang & H.C. Fu, a leguminous plant, is a native desert shrub with strong drought, salinity, cold resistance, sand-fixing capacity and high forage value. It is extensively distributed in Inner Mongolia, Ningxia Autonomous Regions and Shanxi Province of China [22]. Due to its tolerance to various stresses, C. intermedia is an ideal material for studying the mechanism of stress tolerance and can offer effective opportunities for genetic engineering.
In this study, we identified two stress-NAC TFs encoding genes, CiNAC3 and CiNAC4, which were responsive to various abiotic stresses such as dehydtration, drought, salt, cold, heat and wounding. GFP-tagged CiNAC3 and CiNAC4 revealed their nuclear localization. To investigate the in vivo functions of CiNAC3 and CiNAC4, we generated transgenic Arabidopsis plants overexpressed CiNAC3 and CiNAC4 driven by the CaMV35S promoter. The ectopic expression of CiNAC3 and CiNAC4 altered ABA sensitivity during seed germination and salt tolerance of the transgenic plants.
Result
Identification and cloning of CiNAC3 and CiNAC4
The cDNA and gDNA sequences of CiNAC3 and CiNAC4 were isolated by RACE from C. intermedia and the intermediate fragments were acquired from the C. korshinskii (a very closer species of C. intermedia) SSH library under dehydration treatment [23]. Both genes contain three exons and two introns. The putative amino acid sequences were compared with those in the GenBank database by BLAST. CiNAC3 shares identities of 61 % to ANAC072 (RD26) [GenBank: NP_567773.1], and 76 % to GmNAC3 [GenBank: NP_001238234.1], while CiNAC4 has an identities of 59 % to ANAC072, and 80 % to GmNAC4 [GenBank: NP_001238424.1]. A multiple alignment of some stress-NAC TFs [15] by DNAMAN revealed that CiNAC3 and CiNAC4 shared a highly conserved N terminal DNA binding domain, named NAC domain which consists of 5 consensus sub-domains, and a highly variable C-terminal transcriptional regulation domain (Fig. 1). To investigate the divergence of CiNAC3 and CiNAC4 proteins with other NAC proteins during evolution, we analyzed the phylogenetic relationship of CiNAC3 and CiNAC4 with some stress-NAC proteins from A. thaliana, Brassica napus, Ricinus communis, V. vinifera and the leguminous plants by MEGA5.1 (Fig. 2). It suggested that CiNAC3 and CiNAC4 belong to the stress-NAC proteins and indicated a regulatory role in stress responses.
Fig. 1.
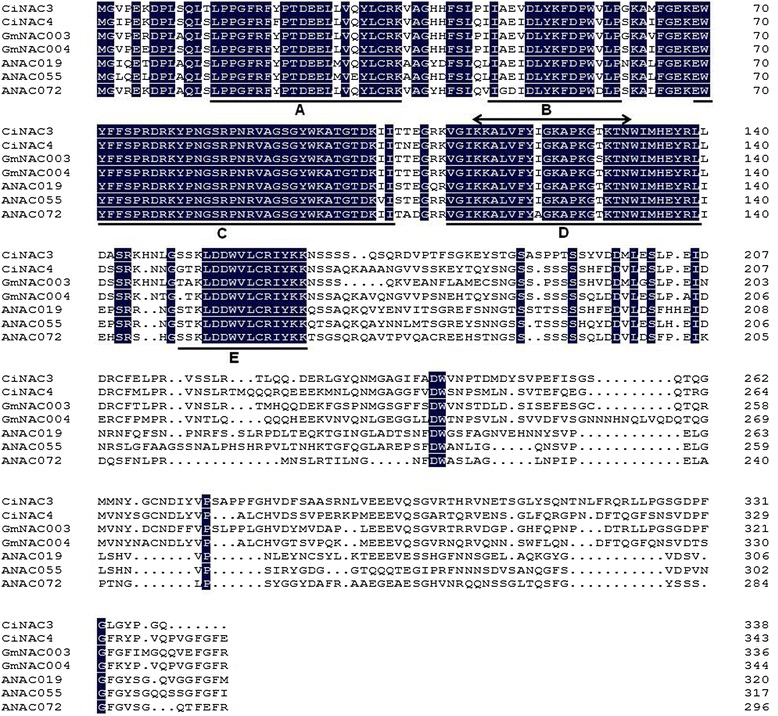
Alignment of the amino acid sequences of some stress-NAC TFs. The putative nuclear localization signal is shown by a double-headed arrow above the sequence. The consensus sub-domains (a-e) in the NAC binding domain are indicated by underlines. Identical amino acids are indicated by white letters on a black background
Fig. 2.
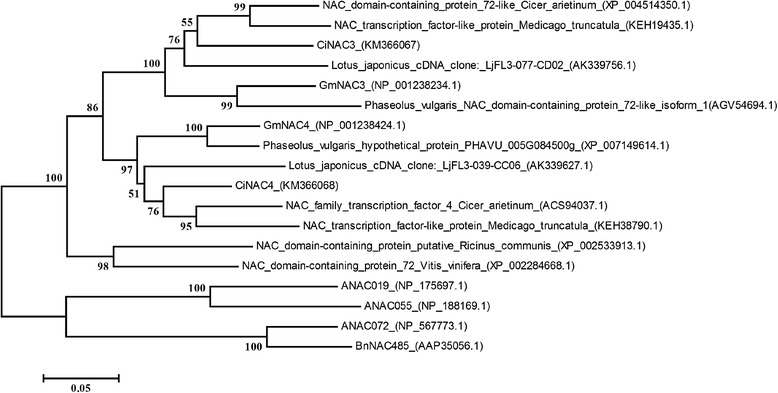
The phylogenetic relationship of the deduced CiNAC3 and CiNAC4 proteins. Phylogenetic analyses were conducted by MEGA5 using the neighbor-joining method. All nodes have 0.80 or greater posterior possibilities. The numbers beside each node represent bootstrap values based on 1,000 replications. The scale bar indicates the relative amount of change along branches
The expression profiles of CiNAC3 and CiNAC4 under various stresses
To investigate the response of CiNAC3 and CiNAC4 to various abiotic stress, quantitative real time RT-PCR analysis was performed. Both CiNAC3 and CiNAC4 were induced by abiotic stresses, such as osmotic stress, salt, wounding, high and low temperature (Fig. 3 and Additional file 1). Particularly, there were hundreds or even thousands fold of changes under drought condition. There exists a distinction between ABA-dependent and ABA-independent pathways for regulating gene expression in response to abiotic stress [1, 9]. We further examined whether CiNAC3 and CiNAC4 responded to ABA treatment or not, and found that these two genes were induced within 3 h after application of ABA (Fig. 3 and Additional file 1). These results suggested that CiNAC3 and CiNAC4 might be involved in osmotic stresses and ABA signaling.
Fig. 3.
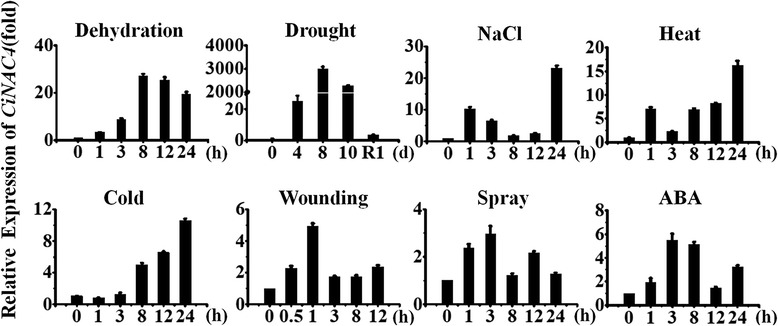
CiNAC4 were induced by abiotic stress and ABA. One-month-old C. intermedia seedlings treated with exogenous ABA (sprayed with 200 μM ABA), cold stress (put into 4 °C incubator), heat stress (put into 42 °C incubator), NaCl (watered with 200 mM NaCl), wounding (2/3 of the total leaves were pierced with tweezers), spray (sprayed with water, used as the control of ABA treatment), dehydration stress (cleaned the soil on the root and put on the filter paper), or drought stress (withholding water) were harvested at the indicated time points. Expression values were calculated using 2-ΔΔCT method and CiEF1a as endogenous control. Two independent biological replicates were performed with similar result. Three technical replicates of each biological replicate were analyzed in quantitative real-time PCR analysis
Subcellular localization of CiNAC3 and CiNAC4
To confirm the subcellular localization of CiNAC3 and CiNAC4, the coding region of CiNAC3 and CiNAC4 were fused to the C-terminal of GFP marker gene, and the fusion genes were driven by the cauliflower mosaic virus (CaMV) 35S promoter. Root tips of the transgenic seedlings were examined for GFP fluorescence. A strong fluorescence signal was predominately observed in the nuclei (Fig. 4). In contrast, the GFP signal distributed throughout the cell in the 35S::GFP transgenic lines. These results were consistent with the role of CiNAC3 and CiNAC4 as TFs.
Fig. 4.
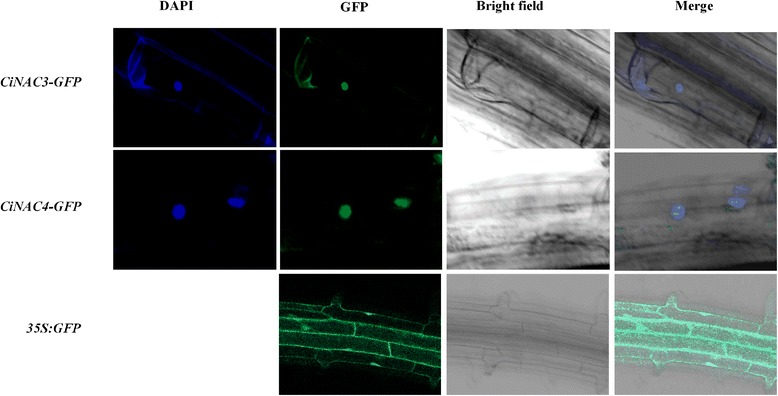
Subcellular localization of CiNAC3 and CiNAC4. Root of the transgenic plants containing the 35S:CiNAC3-GFP (upper panel), 35S:CiNAC4-GFP (middle panel), and 35S:GFP (bottom panel) fusion genes were observed. DAPI was used to visualize the nucleus
Stage and tissue specificity of CiNAC4 expression
To investigate the temporal and spatial expression patterns of these two genes in more details, we cloned the 1,143 bp promoter region (the promoter fragment and 5′-untranslated region) of CiNAC4 using Genome Walking Kit. We failed to clone the promoter of CiNAC3. The transgenic lines containing the ProCiNAC4::GUS construct was used to determine the GUS expression. Histochemical GUS staining was detected, with varying intensity, in many tissues of the transgenic plants (Fig. 5). High GUS expression was limited to the vascular system of root (b, f, g), sepals (c), and in the filament of the stamen (d), but not in root tips (g). The staining was also observed in the cotyledons (b) and both ends of siliques (e) and stigma (d). However, true leaves among the different transgenic lines showed a different pattern. Four out of 19 positive lines showed a strong vein staining (h), while others just showed a slight and smear staining among whole leaves but had a strong GUS activity in the local damaged tissues after wounding treatment (i). The phenotype of wounding induced expression was in accordance with the qPCR result (Fig. 3).
Fig. 5.
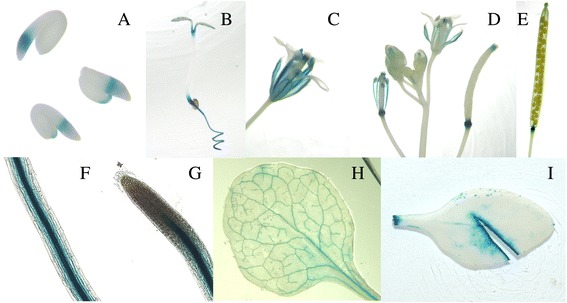
Tissue-specific expression patterns of CiNAC4. a, Seeds imbibed for 36-h. b, Five-day seedlings. c, Flowers. d, Inflorescences. e, Mature siliques. f, Roots. g, Root tips. h and i, Leaves
Overexpression of CiNAC3 and CiNAC4 altered ABA sensitivity during seed germination
To explore the function of CiNAC3 and CiNAC4 in planta, we developed transgenic Arabidopsis constitutively expressing CiNAC3 or CiNAC4 genes under control of the 35S promoter. Real-time RT-qPCR was used to detect the transcripts of CiNAC3 and CiNAC4 in their overexpression homozygous plants (Fig. 6). Four representative homozygote lines (NAC3-51, NAC3-60, NAC4-45and NAC4-76) with different expression levels were used in the following experiments. No notable morphological differences were observed between wild-type and the transgenic plants throughout their life cycle.
Fig. 6.
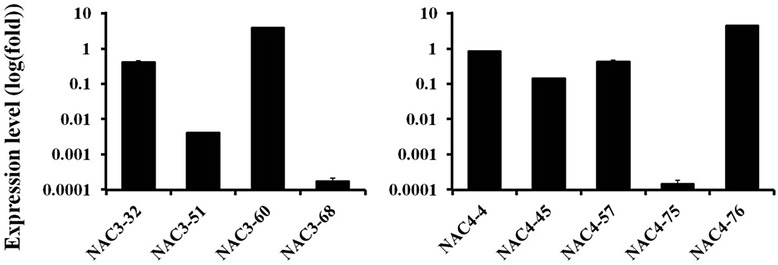
Expression level of CiNAC3 or CiNAC4 in their overexpression lines. The T3 transgenic plants growing under normal condition was detected by relative quantitative real-time PCR. There was no amplification in Col-0. Expression values were calculated using 2-ΔCT method and AtEF1α as endogenous control. Two independent biological replicates were performed with similar result.Three technical replicates of each biological replicate were analyzed in quantitative real-time PCR analysis
ABA has been shown to regulate many aspects of plant growth and development, and its role in seed germination has been illustrated [24]. In order to understand whether the ectopic expression of CiNAC3 or CiNAC4 altered seed germination phenotype, the germination rates were analyzed under different ABA concentrations (Fig. 7 and Additional file 2). CiNAC4 transgenic lines could germinate even on the medium with 6ìM ABA (Fig. 7b), while the wild-type was seriously inhibited. The germination rate was comparable in ABA-free medium between the two genotypes (Fig. 7c). Without stratification, the CiNAC4 overexpression line also exhibited a higher germination rate (Fig. 7d). CiNAC3 transgenic lines showed a similar phenotype (Additional file 2). These results suggested that CiNAC3 and CiNAC4 counteract the ABA-induced inhibition of seed germination.
Fig. 7.
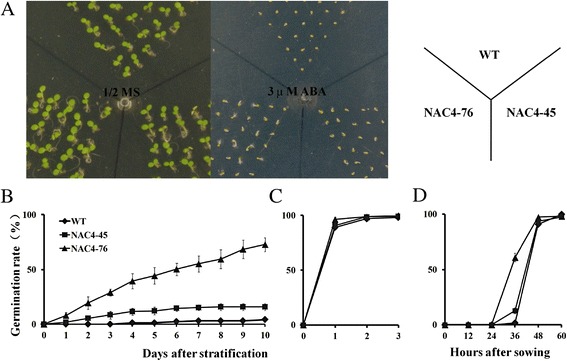
Germination of CiNAC4 transgenic seeds under ABA treatment. (a) The transgenic seeds showed a higher germination rate on 3 μM ABA medium compared with the wild-type. The picture was taken 7 d (3 d for control) after imbibition. The germination rate of wild-type and two overexpression lines on medium with (b) or without (c) 6 μM ABA. (d) Germination of transgenic seeds without stratification. Error bars are standard errors of the means from three replications. Three independent biological replicates have been performed
Overexpression of CiNAC3 and CiNAC4 increased the expression of AtMYB2
Because the CiNAC3 and CiNAC4 transgenic plants showed a high germination rate on the medium containing ABA (Fig. 7 and Additional file 2). We detected the downstream genes of ABA biosynthesis and signaling, and found that AtMYB2, a positive regulator in ABA signaling [25], was increased in transgenic plants, especially the two lines NAC3-60 and NAC4-76 with high expression level (Fig. 8). We searched the microarray data, and the similar result was found that MYB2 had an expression level of 1.6-fold in the 35S::ANAC055 transgenic plants [8]. ADH1, a downstream gene of AtMYB2, was also up-regulated in CiNAC3 and CiNAC4 transgenic plants (Fig. 8). This result further confirmed CiNAC3 and CiNAC4 have a function in ABA signaling.
Fig. 8.
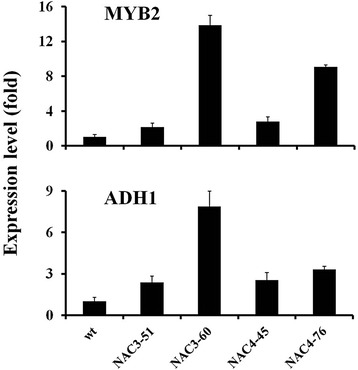
Transcription of MYB2 and ADH1 were increased in the transgenic lines. Two-week old seedlings without treatment were harvested. Expression values were calculated using 2-ΔΔCT method and AtEF1a as endogenous control. Three independent biological replicates were performed with similar result, and the representative data from one repetition are presented. Three technical replicates of each biological replicate were analyzed in quantitative real-time PCR analysis
Overexpression of CiNAC3 and CiNAC4 enhanced salt tolerance of the transgenic Arabidopsis
To examine whether the transgenic plants confer tolerance to salt conditions, wild- type and transgenic plants under salt stress were compared. Under normal growth conditions, four transgenic lines showed no obvious abnormal morphological phenotype compared with the wild-type. Five-day old CiNAC4 transgenic seedlings kept a continuous growth comparing to the wild-type after transferred to half strength MS medium containing 80 mM NaCl (Fig. 9a). The fresh weight was examined and showed a significant difference (Fig. 9b). In addition, CiNAC4 transgenic plants were allowed to grown in pots for 4 weeks under normal condition, then were watered with 200 mM NaCl and continuous growth was also observed (Fig. 9c). CiNAC3 transgenic lines also showed great tolerance under watering with 200 mM NaCl (Additional file 3), but the phenotype was not that obvious on half strength MS medium (data not show). These results showed that overexpression of CiNAC3 and CiNAC4 in Arabidopsis enhanced tolerance to salt stress.
Fig. 9.
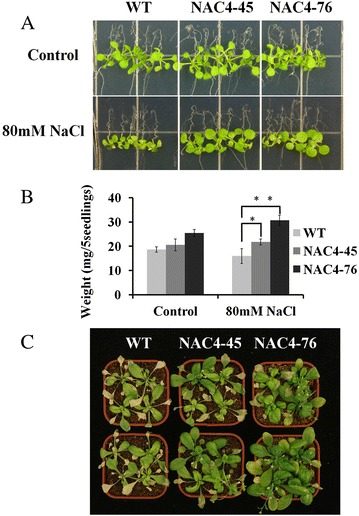
CiNAC4 overexpression enhanced salt tolerance of the transgenic plants. Five-day old seedlings were transferred to half strength MS medium with or without 80 mM NaCl, and photographed (a) and weighed (b) after 5 days. (c) Four-week old plants were watered with 200 mM NaCl twice, photo was taken after one week
Overexpression of CiNAC4 increased lateral root numbers
According to the histochemical assay, CiNAC4 has a high expression in the vascular system of root. We examined whether CiNAC3 and CiNAC4 could affect the root development. Compared to the wild-type, the CiNAC4 transgenic plants showed no significant difference in primary root length. However, the lateral root numbers significantly increased in the CiNAC4 transgenic plants (Fig. 10). This result showed that CiNAC4 promoted lateral root development, and this is consistent with the function of GmNAC4 [18].
Fig. 10.
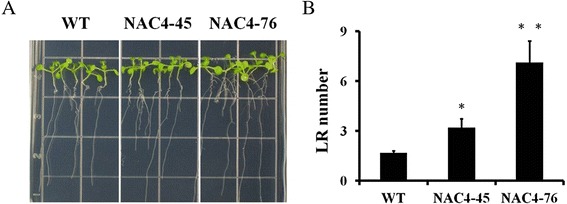
CiNAC4 overexpression increased lateral root number. The lateral roots of ten-day old seedlings grown on half strength MS media were photographed (a) and the number of the lateral roots were counted (b). Each data point represents the mean of 15 seedlings. Error bars indicate SD, and asterisks indicate a significant difference (P < 0.05) compared to the control
Discussion
To overcome the unfavourable effects of different stresses on crops in agriculture, efforts have been made to enhance the tolerance of crop through both plant breeding and genetic engineering approaches. Ectopic expression of several genes in stress signaling pathways, including NAC TFs, had resulted in different stress tolerance in model plants [9]. The distribution of Caragana species was in the arid and semi-arid area figured their plasticity to the stress conditions. We demonstrated the function of two stress NAC TFs named CiNAC3 and CiNAC4 from C. intermedia, and their phylogenetic relationship with other stress NAC TFs from A. thaliana, B. napus, R. communis, V. vinifera and leguminous plants have been compared (Fig. 2). ANAC019, ANAC055, ANAC072 and BnNAC485 were in one clade, and the left NACs were organized in the other clade. In the leguminous clade grouped by G. max, Medicago truncatula, Cicer arietinum, Phaseolus vulgaris, Lotus japonicas and C. intermedia, there is a distinction between the NAC3 and the NAC4, indicating they might have a different role in certain process. The three ANAC TFs play a key role in the stress-specific gene regulatory network [26]. Then a presumption is that NAC3 and NAC4 may play a comparative role in leguminous plants as the three ANAC TFs in Arabidopsis. It should be noticed that CiNAC3 and CiNAC4 had the closest relationship with the NACs from C. arietinum and M. truncatula, respectively, and these two species had been known to be tolerant to drought. But further evidence is required.
CiNAC3 and CiNAC4 can be induced under abiotic stresses including wounding (Fig. 3). The stress-NAC TFs ANAC019, ANAC055 and ANAC072 were induced by dehydration and salt, this is in accordance with their regulatory role in abiotic stresses [8, 26, 27]. Here, we also found CiNAC3 and CiNAC4 had an accumulation and peaked at 1 h after wound treatment (Fig. 3 and Additional file 1). GUS staining of transgenic plants that harbored a ProCiNAC4::GUS construct showed an strong activity in the local damaged tissues (Fig. 5). GUS expression pattern of ATAF1 and ATAF2, which belongs to the ATAF subgroup and is a relative closer subgroup with stress-NAC TFs [15], also showed an strong GUS activity in the local damaged tissues [28, 29]. Other ATAFs, such as OsNAC6 [30] and StNAC [31] were also wounding-induced. It has been found that the ATAFs participated in both abiotic and biotic stresses [9, 32]. ATAF1 and its homolog HvNAC6 in barley positively regulated penetration resistance towards the biotrophic fungus Blumeria graminis f.sp. hordei (Bgh) [33, 34]. We also checked the expression of defense genes PR1 and PDF1.2 in the transgenic Arabidopsis, and found that CiNAC3 and CiNAC4 down-regulated PDF1.2 and slightly up-regulated PR1 (Additional file 4). This suggests that CiNAC3 and CiNAC4 may have a role in plant defense responses. We did not have the opportunity to check the disease tolerance phenotype yet.
CiNAC3 and CiNAC4 could be induced under ABA treatment (Fig. 3 and Additional file 1). The expression of CiNAC3 and CiNAC4 under JA, SA, IAA and NAA treatments was comparable with that in the control (Additional file 5). The CiNAC3 and CiNAC4 ectopic expression lines also exhibited a high germination rate on the ABA medium (Fig. 7 and Additional file 2). ABA plays an important role in many plant processes such as formation and dormancy of seeds, inhibition of germination, stress response and stomata regulation [24]. A previous study had shown that ANAC072 functioned as a transcriptional activator and played a positive role in ABA sensitivity of seedlings and ABA inducible gene expression under abiotic stress [27]. ANAC019 also showed a positive role in ABA signaling of seed germination and early seedling development [35]. BnNAC485, the homologous gene of ANAC072, conferred ABA hypersensitivity in transgenic Arabidopsis [36]. Interestingly, GmNAC3 and GmNAC4 play an inconsistent role in seed germination [18]. Our results showed that both CiNAC3 and CiNAC4 could counteract ABA inhibition of seed germination with a dose-dependent manner (Fig. 7 and Additional file 2).
Previous study had shown that AtMYB2 acted as transcriptional activator in ABA signaling [25]. High expression level of AtMYB2 was found in the CiNAC3 and CiNAC4 transgenic plants, especially the high expression lines NAC3-60 and NAC4-76 (Fig. 8), showed a dose-dependent pattern. Abe et al. found that overexpression of AtMYB2 in Arabidopsis resulted in ABA sensitive phenotype during seed germination [25]. One explanation is that the high expression level of AtMYB2 is not the reason of the high germination rate of CiNAC3 and CiNAC4 transgenic plants under ABA treatment. We also assayed the expression of other representative genes in ABA synthesis, catabolism and signaling pathways, such as ABA1, ABA2, AAO3, ABI1, ABI3, ABI5, ABF1, ABF2, ABF3, ABF4, and RGL2, a DELLA that negatively regulated the seed germination, most of the genes checked showed an increase within one fold in the transgenic plants compare to wild type (Additional file 6A). It has been reported that GmNAC3 and GmNAC4 regulated ABA signaling genes [18]. This suggests that CiNAC3 and CiNAC4 may also function in a dose-dependent manner and play a role in ABA signaling of seed germination.
CiNAC3 and CiNAC4 can be induced by hundreds or even thousands fold under drought treatment, while only over ten-fold elevation under NaCl treatment (Fig. 3 and Additional file 1). Interestingly, CiNAC3 and CiNAC4 transgenic plants showed tolerance to NaCl but not to drought (data not shown), this is different from the drought tolerance phenotype of the ANAC019, ANAC055, and ANAC072 overexpression lines [8]. One reason might be the signaling network mediated by NACs in Arabidopsis was distinct from that in C. intermedia, although many researchers used Arabidopsis as the model organism to validate the function of their interested genes from various plant species. Currently, we did have trouble in transformation with C. intermedia. And M. truncatula was also a ideal plants that have close relationship with C. intermedia. We are now trying to set up the successful transformantion system of M. truncatula.
Osmotic stress such as dehydration, drought, salt, and cold has a critical effect on plant growth and development. For survival, plant evolved some adaptive mechanism that included accumulation of osmolytes, maintenance of the ion homeostasis and detoxification [1, 37]. Salt stress causes more toxic ions entering the plants. We also detected some ion channels encoding genes including HKT, NHX, SOS1, and SOS2, which have been proved to be involved in balancing the ion homeostasis, and no more than 2.5 fold changes were found (Additional file 6B). Some reproducible stress responsive marker genes include COR15A, COR47, RD22, ERD1, and ERA1, also had no remarkable changes in the transgenic plants (Additional file 6C).
The promotion of numbers and length of lateral roots in response to water deficit, is considered as an avoidance mechanism of soybean plants to water stress [38]. Several NAC TFs have been found to respond to environmental stresses and promote lateral root development [21, 39, 40]. We found that the lateral root number of the CiNAC4 transgenic lines were significantly enhanced compared to wild-type (Fig. 10). This result was in accordance with Quach et al. [18], that GmNAC4 significantly promoted LR number under non-stress and mild water deficit conditions. CiNAC3 and GmNAC3 did not promote LR number [18]. This suggests that CiNAC3 and CiNAC4 play a partially redundant role in plant stress responsive signaling, and their roles in lateral root development process are different.
Conclusions
We identified two stress-NAC TFs, CiNAC3 and CiNAC4, and found that their transcripts accumulated under various abiotic stresses and ABA treatment. Both TFs were localized in nuclei. The GUS staining experiments indicated that the CiNAC4 promoter have a high activity in root, flower and local damaged tissues of Arabidopsis. CiNAC3 and CiNAC4 altered the ABA signaling and NaCl tolerance of the transgenic Arabidopsis, while only CiNAC4 overexpression increased the lateral root number.
Methods
Growth conditions and treatments
The Arabidopsis wild-type (Columbia-0) and the transgenic lines were grown on half strength MS medium or a 1:1 mixture of peat soil and vermiculite under long-day conditions (16-h-light/8-h-dark cycle) at 22 °C. To determine the germination rate, seeds were surface sterilized and sown on half strength MS containing 0.65 % agar powder and 3 % sucrose with different concentrations of NaCl or ABA. Germination rates were scored based on radicle protrusion after three days of stratification. Seeds of C. intermedia were collected from Hohhot, Inner Mongolia, China. One-month-old seedlings which were sown in pots containing a soil mixture were used to detect the transcript level of CiNAC3 and CiNAC4 under various treatments.
For NaCl treatment, the seedlings growing under normal conditions were watered with 200 mM NaCl. For cold and heat treatments, the seedlings were transferred to the 4 °C or 42 °C incubator. For ABA treatment, the seedlings were sprayed with 200 μM ABA plus 0.05 % Tween. For the “spray” treatment which was used as the control of ABA treatment, the seedlings were sprayed with only water containing 0.05 % Tween. For wounding treatment, 2/3 of the total leaves of each seedling was pierced with tweezers. For dehydration treatment, soil was removed and the seedlings were cleaned with tap water, then placed on the filter paper at room temperature. For drought treatment, the seedlings were subjected to drought conditions by withholding water for ten days and then re-watered. Each sample contains three seedlings and the shoots (including stems and leaves) were taken as samples.
RNA extraction and real-time RT-qPCR analysis
Total RNA was isolated according to manufactures’ instructions (Invitrogen) using Trizol reagent. After DNase I (Ambion Cat# AM2224) treatment, 500 ng (for Arabidopsis) or 1ìg (for Caragana) of RNA was used for reverse transcription (TaKaRa, Dalian, China Cat# D2640A). The cDNA was diluted 40 times (for Arabidopsis) or 16 times (for Caragana), and 5 μL was used as a template in a 20-μL PCR reaction. Real-time PCR analysis was performed using SYBR Green Perfect mix (TaKaRa, Cat# DRR041A) on a LightCycler 480 system (Roche), with the program of 40 cycles under the following conditions: 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 15 s. AtEF1α and CiEF1α [GenBank: KC679842] was used to normalize the Arabidopsis or Caragana samples respectively [41]. The primers used in this study are listed in Additional file 7.
Identification of CiNAC3 and CiNAC4
The cDNA intermediate fragments of CiNAC3 and CiNAC4 were acquired from a Suppression Subtractive Hybridization (SSH) library of C. korshinskii (a very closer species of C. intermedia) under dehydration stress which was constructed by Yang et al. [23]. Full length cDNA was obtained by Rapid Amplification of cDNA Ends (RACE) using the mRNA extracted from C. intermedia as template (TaKaRa, Dalian, China Cat#6107 and Cat#6106). The promoter of CiNAC4 was cloned using Genome Walking Kit (TaKaRa, Dalian, China Cat#6108). The primers used in this study are listed in Additional file 7: Table S1. PCR for the cloning of CiNAC3 and CiNAC4 was performed with the following cycling profile: 98 °C for 2 min; 35 cycles at 98 °C for 10 s, 57 °C for 15 s, and 72 °C for 1.5 min; and a final extension for 10 min at 72 °C. PCR for the cloning of the CiNAC4 promoter was : 98 °C for 2 min; 35 cycles at 98 °C for 10 s, 59 °C for 15 s, and 72 °C for 1.5 min; and a final extension for 10 min at 72 °C.
Protein sequences were aligned using the DANMAN. Phylogenetic tree was conducted by MEGA5 using the Neighbor-Joining (NJ) method and bootstrap analysis of 1000 replications.
Construction of CiNAC3 and CiNAC4 transgenic plants
The open reading frames (without the termination codon) of CiNAC3 and CiNAC4 were cloned into pENTR/D-TOPO (Invitrogen, Cat.#K2420-20). The primer were designed following the direction of pENTR™ Directional TOPO® Cloning Kits, and “CACC” was added to the 5′ end of the sense primer to serve as a recombination site for introducing the PCR product into the entry plasmid, pENTR/D-TOPO. After sequencing and validation of the entry plasmid, the LR recombination reaction was performed between the entry plasmid and the gateway destination vector (Invitrogen Cat.#11791-020). The vector pMDC32 was used for overexpression construct, while pMDC43 was used for C-terminal GFP fused construct to detect the subcellular localization.
The promoter of CiNAC4 was amplified with high fidelity enzyme and cloned into pEASY-Blunt Simple (TransGen, Cat.#CB111-02). After validation, the promoter was fused with the GUS reporter gene of pCAMBIA1305.2 with the CaMV35S promoter removed using HindIII and NcoI restriction sites.
The binary plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation, which was used for floral dip transformation of Arabidopsis. Transformants were selected on half strength MS medium containing the appropriate antibiotics.
Histochemical GUS Staining
The T2 seedlings of ProCiNAC4::GUS transgenic plants were used to determine the GUS activity. The GUS staining protocol was performed as previously described [42]. The images were taken under the dissecting microscope (SMZ800, Nikon).
Subcellular localization
The T2 seeds of 35S::GFP-CiNAC3 or 35S::GFP-CiNAC4 transgenic lines were grown on half strength MS medium containing 13 mg/L hygromycin B. The root tips of 10-day-old positive seedlings were observed under laser scanning confocal microscope (Zeiss LSM 510).
Acknowledgements
We thank Dr. Mark Goettel, the Editor-in-Chief of Biocontrol Science & Technology, for polishing the manuscript carefully. This work was supported by the grants from Chinese National Programs for High Technology Research and Development (No. 2011AA100203), National Natural Science Foundation of China (No. 31360056), Inner Mongolia Science & Technology Plan- Innovation Team (2015-2016), and the Joint Specialized Research Fund for the Doctoral Program of Higher Education, the Ministry of Education (No.20121515110007).
Abbreviations
- NAC
NAM/ATAF/CUC
- ABA
Abscissic acid
- JA
Jasmonic acid
- SA
Salicylic acid
- IAA
Indoleacetic acid
- NAA
Naphthaleneacetic acid
- LEA
Late embryogenesis abundant
- TF
Transcription factor
- ERD1
Early responsive to dehydration stress 1
- Ci
Caragana intermedia
- Gm
Glycine max
- Hv
Hordeum vulgare
- Bn
Brassica napus
- CaMV35S
cauliflower mosaic virus 35S
- SSH
Suppression Subtractive Hybridization
- ABI
ABA insensitive
- ABF
ABRE binding factor
- AAO
Aldehyde oxidase
- RGL
RGA (repressor of ga)-like
- COR
Cold-regulated
- RD
Response to dehydration
- HKT1
High-affinity K+transporter 1
- NHX
Na+/H+exchanger
- SOS
Salt overly sensitive
- RACE
Rapid Amplification of cDNA Ends
- LR
Lateral root
Additional files
CiNAC3 was induced by abiotic stresses and ABA. One-month-old C. intermedia seedlings treated with exogenous ABA (sprayed with 200 μM ABA), cold stress (put into 4 °C incubator), heat stress (put into 42 °C incubator), NaCl (watered with 200 mM NaCl), wounding (2/3 of the total leaves were pierced with tweezers), spray (sprayed with water, used as the control of ABA treatment), dehydration stress (cleaned the soil on the root and put on the filter paper), or drought stress (withholding water) were harvested at the indicated time points. Expression values were calculated using 2-ΔΔCT method and CiEF1a as endogenous control. Two independent biological replicates were performed with similar result. Three technical replicates of each biological replicate were analyzed in quantitative real-time PCR analysis. (TIFF 521 kb)
Germination of the CiNAC3 transgenic seeds under ABA treatment. (A) The transgenic lines showed a higher germination rate on 3 μM ABA medium compared with wild-type. The picture was taken 7 d (3 d for control) after imbibition. The germination rate of wild-type and two overexpression lines on medium with (B) or without (C) 6 μM ABA. (D) Germination of transgenic seeds without stratification. Error bars are standard errors of the means from three replications. Three independent biological replicates have been performed. (TIFF 1072 kb)
CiNAC3 overexpression altered salt tolerance of the transgenic plants. Four-week old wild-type and CiNAC3 overexpression plants were watered with 200 mM NaCl twice, photo was taken after one week. (TIFF 1071 kb)
Expression of some defence genes in the transgenic plants. PDF1.2 was down-regulated and PR1 was slightly up-regulated in transgenic Arabidopsis. Expression values were calculated using 2-ΔΔCT method and AtEF1a as endogenous control. Two independent biological replicates were performed with similar result. Three technical replicates of each biological replicate were analyzed in quantitative real-time PCR analysis. (TIFF 37 kb)
Expression of CiNAC3 and CiNAC4 under various hormones treatment. One-month-old C. intermedia seedlings were sprayed with water, MeJA (100 μM), SA (1 mM), IAA (10 μM), NAA (10 μM). Samples were harvested at the indicated times. Expression values were calculated using 2-ΔΔCT method and CiEF1a as endogenous control. Two independent biological replicates were performed with similar result. Three technical replicates of each biological replicate were analyzed in quantitative real-time PCR analysis. (TIFF 89 kb)
Stress responsive marker genes profile by quantitative real-time PCR analysis in the transgenic plants. ABA signaling genes (A), salt signaling genes and some stress responsive genes (B), (C) were detected. Two-week old seedlings with or without treatment were harvested. Expression values were calculated using 2-ΔΔCT method and AtEF1a as endogenous control. Two independent biological replicates were performed with similar result. Three technical replicates of each biological replicate were analyzed in quantitative real-time PCR analysis. (TIFF 212 kb)
Primers used in this study. (XLSX 15 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XH ang GL designed and drafted the manuscript. XH carried out the experiments, ZF and DX repeated some of the experiments. QY, RW and LQ participated in the design of experiments and edited the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Xiaomin Han, Email: hanxiaominhushi@163.com.
Zongqi Feng, Email: fengzongqier@163.com.
Dandan Xing, Email: dan2008132128@126.com.
Qi Yang, Email: atp_yangqi@163.com.
Ruigang Wang, Email: wangruigang@imau.edu.cn.
Liwang Qi, Email: lwqi@caf.ac.cn.
Guojing Li, Email: liguojing@imau.edu.cn.
References
- 1.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 2.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130(4):2129–41. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10(6):239–47. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 5.Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465(1-2):30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Wang N, Zheng Y, Xin H, Fang L, Li S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013;32(1):61–75. doi: 10.1007/s00299-012-1340-y. [DOI] [PubMed] [Google Scholar]
- 7.Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, et al. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18(4):263–76. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16(9):2481–98. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17(6):369–81. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Muller CW. Transcription factors: global and detailed views. Curr Opin Struct Biol. 2001;11(1):26–32. doi: 10.1016/S0959-440X(00)00163-9. [DOI] [PubMed] [Google Scholar]
- 11.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Olsen AN, Ernst HA, Leggio LL, Skriver K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005;169(4):785–97. doi: 10.1016/j.plantsci.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 13.Zhong R, Lee C, Ye ZH. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant. 2010;3(6):1087–103. doi: 10.1093/mp/ssq062. [DOI] [PubMed] [Google Scholar]
- 14.Ernst HA, Olsen AN, Larsen S, Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5(3):297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinheiro GL, Marques CS, Costa MD, Reis PA, Alves MS, Carvalho CM, et al. Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response. Gene. 2009;444(1-2):10–23. doi: 10.1016/j.gene.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Tran LS, Quach TN, Guttikonda SK, Aldrich DL, Kumar R, Neelakandan A, et al. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genomics. 2009;281(6):647–64. doi: 10.1007/s00438-009-0436-8. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Huang F, Cheng H, Song H, Yu D. Overexpression of the GmNAC2 Gene, an NAC Transcription Factor, Reduces Abiotic Stress Tolerance in Tobacco. Plant Molecular Biology Reporter. 2013;31(2):435–42. doi: 10.1007/s11105-012-0514-7. [DOI] [Google Scholar]
- 18.Quach TN, Tran LS, Valliyodan B, Nguyen HT, Kumar R, Neelakandan AK, et al. Functional Analysis of Water Stress-Responsive Soybean GmNAC003 and GmNAC004 Transcription Factors in Lateral Root Development in Arabidopsis. PLoS One. 2014;9(1):e84886. doi: 10.1371/journal.pone.0084886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin H, Xu G, Meng Q, Huang F, Yu D. GmNAC5, a NAC Transcription Factor Is a Transient Response Regulator Induced by Abiotic Stress in Soybean. Scientific World J. 2013;2013:768972. doi: 10.1155/2013/768972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faria JA, Reis PA, Reis MT, Rosado GL, Pinheiro GL, Mendes GC, et al. The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol. 2011;11:129. doi: 10.1186/1471-2229-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68(2):302–13. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- 22.Xiao CW, Zhou GS, Ceulemans R. Effects of elevated temperature on growth and gas exchange in dominant plant species from Maowusu sandland. China Photosynthetica. 2003;41(4):565–9. doi: 10.1023/B:PHOT.0000027521.86653.07. [DOI] [Google Scholar]
- 23.Yang Q, Zhang T, Wang Y, Li G, Yin JJ, Han XM, et al. Construction of a suppression subtractive hybridization library of Caragana korshinskii under drought stress and cloning of CkWRKY1 Gene. Scientia Silvae Sinicae. 2014;49(7):62–8. [Google Scholar]
- 24.Finkelstein R. Abscisic Acid synthesis and response. Arabidopsis Book. 2013;11:e0166–e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15(1):63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickman R, Hill C, Penfold CA, Breeze E, Bowden L, Moore JD, et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 2013;75(1):26–39. doi: 10.1111/tpj.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39(6):863–76. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 28.Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, et al. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol. 2007;63(2):289–305. doi: 10.1007/s11103-006-9089-8. [DOI] [PubMed] [Google Scholar]
- 29.Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, et al. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005;43(5):745–57. doi: 10.1111/j.1365-313X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- 30.Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51(4):617–30. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 31.Collinge M, Boller T. Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol. 2001;46(5):521–9. doi: 10.1023/A:1010639225091. [DOI] [PubMed] [Google Scholar]
- 32.Christianson JA, Dennis ES, Llewellyn DJ, Wilson IW. ATAF NAC transcription factors: regulators of plant stress signaling. Plant Signal Behav. 2010;5(4):428–32. doi: 10.4161/psb.5.4.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen MK, Rung JH, Gregersen PL, Gjetting T, Fuglsang AT, Hansen M, et al. The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol. 2007;65(1-2):137–50. doi: 10.1007/s11103-007-9204-5. [DOI] [PubMed] [Google Scholar]
- 34.Jensen MK, Hagedorn PH, de Torres-Zabala M, Grant MR, Rung JH, Collinge DB, et al. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008;56(6):867–80. doi: 10.1111/j.1365-313X.2008.03646.x. [DOI] [PubMed] [Google Scholar]
- 35.Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, et al. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J. 2010;426(2):183–96. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- 36.Ying L, Chen HY, Cai WM. BnNAC485 is involved in abiotic stress responses and flowering time in Brassica napus. Plant Physiol Biochem. 2014;79:77–87. doi: 10.1016/j.plaphy.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6(5):441–5. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 38.Manavalan LP, Guttikonda SK, Phan Tran L-S, Nguyen HT. Physiological and Molecular Approaches to Improve Drought Resistance in Soybean. Plant Cell Physiology. 2009;50(7):1260–76. doi: 10.1093/pcp/pcp082. [DOI] [PubMed] [Google Scholar]
- 39.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14(23):3024–36. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005;44(6):903–16. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 41.Yang Q, Yin JJ, Li G, Qi LW, Yang FY, Wang RG, et al. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol Biol Rep. 2014;41(4):2325–34. doi: 10.1007/s11033-014-3086-9. [DOI] [PubMed] [Google Scholar]
- 42.Wan DL, Li RL, Zou B, Zhang X, Cong JY, Wang RG, et al. Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 2012;31(7):1269–81. doi: 10.1007/s00299-012-1247-7. [DOI] [PubMed] [Google Scholar]


