Abstract
Mulberry vein banding associated virus (MVBaV) that infects mulberry plants with typical vein banding symptoms had been identified as a tentative species of the genus Tospovirus based on the homology of N gene sequence to those of tospoviruses. In this study, the complete sequence of the tripartite RNA genome of MVBaV was determined and analyzed. The L RNA has 8905 nucleotides (nt) and encodes the putative RNA-dependent RNA polymerase (RdRp) of 2877 aa amino acids (aa) in the viral complementary (vc) strand. The RdRp of MVBaV shares the highest aa sequence identity (85.9%) with that of Watermelon silver mottle virus (WSMoV), and contains conserved motifs shared with those of the species of the genus Tospovirus. The M RNA contains 4731 nt and codes in ambisense arrangement for the NSm protein of 309 aa in the sense strand and the Gn/Gc glycoprotein precursor (GP) of 1,124 aa in the vc strand. The NSm and GP of MVBaV share the highest aa sequence identities with those of Capsicum chlorosis virus (CaCV) and Groundnut bud necrosis virus (GBNV) (83.2% and 84.3%, respectively). The S RNA is 3294 nt in length and contains two open reading frames (ORFs) in an ambisense coding strategy, encoding a 439-aa non-structural protein (NSs) and the 277-aa nucleocapsid protein (N), respectively. The NSs and N also share the highest aa sequence identity (71.1% and 74.4%, respectively) with those of CaCV. Phylogenetic analysis of the RdRp, NSm, GP, NSs, and N proteins showed that MVBaV is most closely related to CaCV and GBNV and that these proteins cluster with those of the WSMoV serogroup, and that MVBaV seems to be a species bridging the two subgroups within the WSMoV serogroup of tospoviruses in evolutionary aspect, suggesting that MVBaV represents a distinct tospovirus. Analysis of S RNA sequence uncovered the highly conserved 5’-/3’-ends and the coding regions, and the variable region of IGR with divergent patterns among MVBaV isolates.
Introduction
The mulberry (Morus spp.) is an economically important plant grown widely throughout Asia, for the cultivation of the silkworms (Bombyx mori Linn.) and for the sericulture industry. Diverse virus-like symptoms, including mosaic, vein banding, vein necrosis, chlorotic ringspots, and leaf deformation were frequently observed on mulberry. The viral diseases have been a major factor restricting yield and quality of mulberry. Two mulberry-infecting viruses, Mulberry latent virus (Genus Carlavirus) [1] and the Mulberry ringspot virus (Genus Nepovirus) [2], have been partly characterized in Japan. Recently, a mulberry-infecting Tospovirus, temporarily named Mulberry vein banding virus (MuVBV), was found in China and identified to be a new species of Tospovirus based on the homology of N protein sequence to other Tospovirus species [3].
Tospoviruses cause significant loss of yield and quality to vegetables, legumes, and ornamental crops worldwide and are transmitted by thrips in a circulative and propagative manner [4, 5]. Typical symptoms induced by members of the Tospovirus genus include foliar necrotic spots, necrotic stems, bronzing, wilting, and ring spots in leaves and fruits [4, 6]. Tospoviruses are characterized with enveloped quasi-spherical particles of approximately 80–120 nm in size and a tripartite negative and ambisense RNA genome, i.e. small RNA (S RNA), medium RNA (M RMA), and large RNA (L RNA) [4]. The 3' termini of all RNA segments are highly conserved in the first 9 nucleotides and show inverted complementarity to the 5' ends [7].
The L RNA is with a negative polarity and encodes a putative RNA-dependent RNA polymerase (RdRp), which is also called L protein, in the viral complementary (vc) strand for virus replication [4]. The other two genomic RNAs use an ambisense coding strategy. The M RNA encodes a cell-to-cell movement protein (NSm) and the envelope glycoprotein precursor (GP), whereas the S RNA encodes a nonstructural RNA-silencing suppressor protein (NSs) and the nucleocapsid protein (N) [7, 8]. The open reading frames (ORFs) in the M and S RNA segments are separated by large AU-rich intergenic regions (IGR), which forms a stable hairpin structure and is assumed to be involved in transcription termination [4, 9].
In recent years, new tospoviruses such as Pepper necrotic spot virus (PNSV) [10], Soybean vein necrosis associated virus (SVNaV) [11], Hippeastratum chlorotic ring virus (HCRV) [12–14], Bean necrotic mosaic virus (BeNMV) [15], Tomato necrotic spot virus (TNSV) [16], MuVBV [3], and Lisianthus necrotic ringspot virus (LNRV) [17] have been identified.
Tospoviruses are classified into four major serogroups, designated by their serological relatedness to the type viruses Watermelon silver mottle virus (WSMoV), Tomato spotted wilt virus (TSWV), Groundnut yellow spot virus (GYSV), and Iris yellow spot virus (IYSV) [18,19]. Impatiens necrotic spot virus (INSV) was classified together as a distinct serotype [18]. BeNMV and SVNaV, phylogenetically clustered into a new branch and exhibited low cross-reactivity with other species, may represent a new evolutionary lineage within the genus Tospovirus [15, 20]. As of to date, the full genome sequences were available for sixteen species in the genus Tospovirus [8, 11, 13, 21–24].
In our previous survey, a new tospovirus tentatively named Mulberry vein banding virus (MuVBV) with highest N protein homology of 74.4% to Capsicum chlorosis virus (CaCV) was identified from mulberry with vein-banding symptom in Guangxi Province of China by RT-PCR and sequencing of a fragment of the S RNA of the viral genome [3]. Since this virus has not been fully characterized, particularly due to the lack of the vector information, we rename this virus Mulberry vein banding associated virus (MVBaV), to more precisely reflect its current taxonomic status. To better understand the nature of this virus, we cloned and sequenced the whole genome of MVBaV, and characterized its histopathology and serology properties. Our results show that MVBaV is a distinct member of Tospovirus belonging to the WSMoV serogroup.
Material and Methods
Ethics Statement
This work was supported by the government of Guangxi province and the sampling activities were conducted with the permission of the agricultural authority of local counties or authorities in charged. The mulberry orchards where the plant samples were collected were in public domain and in regular agricultural farm lands where no endangered or protected species were involved. The sampling locations of the isolates of MVBaV in this study and the authorities who issued the permission were listed in S1 Table.
Virus sources, electron microscopy and indirect enyzme-linked immunosorbent assay (ELISA)
During 2011 to 2012, ten isolates of MVBaV designated XCSY-3, XCBY-1, XZDL-1, SL-3, HX-2, NN-5, NN-10, NN-16, YZ-3, YZ-4, were collected from mulberry plants showing characteristic symptoms including vein banding, mosaic, chlorotic ringspots, necrotic ringspots or vein necrosis on leaves in mulberry orchards around Guangxi province, China. Due to difficulty of sap transmission, the virus was maintained on mulberry by grafting to the virus-free mulberry seedlings. The leaf samples were stored at −80°C until total RNA was extracted. Ultrathin sections of diseased tissues were prepared as described [25] and examined with the H-7650 transmission electron microscope (Hitachi High-Technologies, Tokyo, Japan).
Mixed antisera of WSMoV/Groundnut bud necrosis virus (GBNV) for WSMoV serogroup, Groundnut ringspot virus (GRSV)/Tomato chlorotic spot virus (TCSV) for TSWV serotype, and antisera against TSWV for TSWV serogroup, IYSV for IYSV serogroup, and INSV for the distinct serotype, were purchased from Agdia Inc. (Elkhart, IN, USA). Indirect ELISA was performed as described Chen et al. (2010) with minor modifications [18]. Crude extracts from infected and healthy mulberry plants were diluted 1:30 and used as the antigen source and negative control, respectively.
RNA extraction and rapid amplification of cDNA ends (RACE)
Total RNA was extracted from the infected mulberry leaves using RNAplant plus reagent (TIANGEN, Beijing, China) according to the manufacturer's instruction and the quality of the purified RNA was evaluated by gel electrophoresis on 0.8% (w/v) agarose gels. The 5′ and 3′-terminal sequences of the viral genomic RNA were determined by using the RACE cDNA Amplification Kit (BD Biosciences, Franklin Lakes, NJ, USA) coupling with Sanger sequencing. Primers were designed according to the available sequences obtained through viral small RNA deep sequencing. The RACE products were purified using DNA gel recovery kit (Sangon, Shanghai, China) and cloned into the pCR2.1-TOPO vector (Thermo Fisher Scientific, Waltham, MA, USA) before sequencing.
Amplification and cloning of viral genomic cDNAs
MVBaV-specific primer pairs were designed based on the sequence information obtained by 5′ and 3′ RACE, and used for the amplification of complete fragments of the MVBaV genome. The primers LRNAF1/LRNAR1 and LRNAF2/LRNAR2 were used for the PCR amplification of MVBaV L RNA, MRNAF/MRNAR for MVBaV M RNA, and SRNAF/SRMAR for S RNA, respectively. The reverse transcription reaction was carried out using the Reverse Transcriptase Kit (Thermo Fisher Scientific) and the PCR was carried out in a total reaction volume of 25 μl. After denaturizing at 95°C for 2 min, the PCR were performed with the following parameters: 35 cycles with each cycle at 94°C for 30 s, 55–58°C for 30 s (the temperature varied with the length of template fragments), 72°C for 45 s, and final extension at 72°C for 10 min. The PCR products were analyzed by electrophoresis on a 0.8% (w/v) agarose gel. The PCR products of interest were recovered from the gel and cloned into pCR2.1-TOPO vector plasmid. The primers used for RACE and amplifying the MVBaV Genome are shown in Table 1.
Table 1. Primers used for amplification of the MVBaV genome.
| Primer | Sequence (5' to 3') | Location and orientation |
|---|---|---|
| Tos-UPA | GACCACGCGTATCGATGTCGACAGAGCAATC | 3' termination of S, M and L RNA |
| SRNA 5–1 | GCTGGGAACTGTGCTGTCAGAAAGGC | S 670-695(vc) |
| SRNA 5–2 | ATGTTCTCTCTCCAGTAAGACGTTG | S 610-634(vc) |
| S RNA 3 | ATTGCTCTCGCTCAACATCTTAAC | S 2547-2570(v) |
| S RNAF | AGAGCAATCAGGGTATTAATT | S 1-21(v) |
| S RNAR | AGAGCAATCGAGGTATTAATAAAAACA | S 3268-3294(vc) |
| MRNA 5–1 | CACTTCTAGAGACTTGTCCATATC | M 961-984(vc) |
| MRNA 5–2 | CTATATCAATGTCAATCTCAACCTC | M 937-961(vc) |
| M RNA 3 | GCTATGCTTGATGGTATGTATGAAATAAG | M3378-3406(v) |
| MRNAF | AGAGCAATCGGTGCACGAATTCACAGAATC | M 1-30(v) |
| M RNAR | AGAGCAATCAGTGCAACAATTAAAGA | M4706-4731(vc) |
| LRNA 5–1 | GGCCTGAATCAAGAATAGACAGAG | L1402-1425(vc) |
| LRNA 5–2 | TCTTCTAGTGAGATAATGACCTCG | L 1241-1264(vc) |
| LRNA 3 | TCAGTTATAGCATCTCTGAATGACTG | L 7380-7405(v) |
| LRNAF1 | AGAGCAATCGAGCAACATATTAAAGTA | L 1-27(v) |
| LRNAR1 | GCTACTATCCGACTTCAACTGC | L 4328-4349(vc) |
| LRNAF2 | TTAGGGTTCAGGGTTATACAATAGC | L 4254-4278(v) |
| LRNAR2 | AGAGCAATCGTGCAACAATAAAATATCAG | L 8877-8905(vc) |
The cloned PCR products were sequenced with sequencer ABI 3700. At least three independent clones were sequenced for a target PCR product. The assembled full-length sequence data of MVBaV genome were deposited in the GenBank with accession numbers KM819698-KM819709.
Analysis of viral RNA sequences
Sequence analysis was carried out using programs within the software VECTOR NTI V11.0 (Thermo Fisher Scientific). Phylogenetic trees were constructed by the neighbor-joining method with 1000 bootstrap replications using a program included in MEGA 6.06 [26]. The putative peptide cleavage sites and the N- and O-linked glycosylation sites of the GP of MVBaV were predicted using the software programs SignalP 4.1, NetNGlyc 1.0, and NetOGlyc 3.1, respectively. For transmembrane domain prediction, the TMHMM Server 2.0 program was used. The sequences of Tospoviruses were down loaded from the GenBank and listed in Table 2.
Table 2. List of accession number and abbreviation of Tospoviruses used in this study.
| Virus | Abbreviation | S RNA | M RNA | L RNA |
|---|---|---|---|---|
| Mulberry vein banding associated virus | MVBaV | KM819701.1 | KM819699.2 | KM819698.1 |
| Alstromeria necrotic streak virus | ANSV | GQ478668.1(N) | – | – |
| Bean necrotic mosaic virus | BeNMV | JN587268.1 | JN587269.1 | JF417980.1 |
| Calla lily chlorotic spot virus | CCSV | AY867502.1 | FJ822961.1 | FJ822962.1 |
| Capsicum chlorosis virus AIT isolate | CaCV-AIT | NC_008301.1 | NC_008303.1 | NC_008302.1 |
| Capsicum chlorosis virus HT-1 isolate (synonyms: Gloxinia tospovirus) | CaCV-HT-1 | AF059578 (N) AF059577(NSs) | AF023172.1 | – |
| Chrysanthemum stem necrosis virus | CSNV | KM114548 | KM114547 | KM114546 |
| Groundnut bud necrosis virus | GBNV | AY871098.2 | U42555.1 | AF025538.1 |
| Groundnut chlorotic fan- spot virus | GCFSV | AF080526.1 | – | – |
| Groundnut ringspot virus (South African isolate) | GRSV-SA | AF487516.1 for N, JN571117.1 for NSs | AY574055.2 for GPs, AF213673.1 for NSm | - |
| GRSV (south Florida isolate from the United State) | GRSV-US | HQ644140.1 | HQ644141.1 | HQ644142.1 |
| Groundnut yellow spot virus | GYSV | AF013994.1 | – | – |
| Hippeastrum chlorotic ringspot virus | HCRV | JX833564 | JX833565 | HG763861 |
| Impatiens necrotic spot virus | INSV | X66972.1 | DQ425095.1 | DQ425094.1 |
| Iris yellow spot virus | IYSV | AF001387.1 | FJ361359.1 | FJ623474.1 |
| Lisianthus necrotic ringspot virus | LNRV | AB852525 | – | – |
| Melon severe mosaic virus | MSMV | EU275149.1 | – | – |
| Melon yellow spot virus | MYSV | AB038343.1 | AB061773.1 | AB061774.1 |
| Pepper necrotic spot virus | PNSV | HE584762.1 | – | – |
| Physalis silver mottle virus | PhySMV | AF067151 | – | – |
| Polygonum ringspot virus | PolRSV | KJ541744 | KJ541745 | KJ541746 |
| Soybean vein necrosis associated virus | SVNaV | HQ728387.1 | HQ728386.1 | HQ728385.1 |
| Tomato chlorotic spot virus | TCSV | AF282982.1 (N) | AY574054.2 for GPs, AF213674.1 for NSm | HQ700667.1 |
| Tomato necrosis virus | TNeV | AY647437 (N) | – | – |
| Tomato necrotic ringspot virus | TNRV | FJ489600.2 | FJ947152.1 | – |
| Tomato necrotic spot virus | TNSV | KM355773.1 | – | – |
| Tomato spotted wilt virus | TSWV | AF020660.1 | JN664253.1 | NC_002052.1 |
| Tomato yellow ring virus | TYRV | AY686718.1 | JN560177 | JN560178 |
| Tomato zonate spot virus | TZSV | EF552433.1 | EF552434.1 | EF552435.1 |
| Watermelon bud necrosis virus | WBNV | GU584184.1 | GU584185.1 | GU735408.1 |
| Watermelon silver mottle virus | WSMoV | AB042650.1 | DQ157768.1 | AY863200.1 |
| Zucchini lethal chlorosis virus | ZLCV | AF067069.1(N) | AB274027.1for GPs, AF213676.1 for NSm | – |
Results
Symptomatology and virion morphology
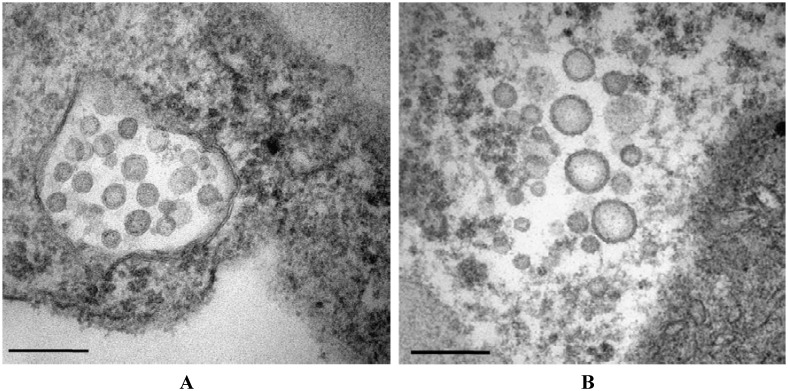
The symptoms of MVBaV-infected leaves varied widely, depending on the growing stage of mulberry and the environmental conditions. Initial symptoms of vein banding, mosaic, or chlorotic ringspots (Fig 1A–1C) were observed in early April and leaf deformation could develop later (Fig 1D). In the autumn, necrotic ringspots and vein necrosis on leaves were common (Fig 1E). The diseased mulberry leaf samples were further examined by electron microscopy. Enveloped particles with a quasi-spherical shape of 80–100 nm in diameter were observed to accumulate in the endoplasmic reticulum (Fig 2A) or dispersed in the cytoplasm as single particles (Fig 2B) in leaf cells. These particles had morphological features typical of tospoviruses. The tospovirus-like particles were also present in the symptom-showing leaves of plants infected by grafting inoculation (data not shown), suggesting that the symptoms were associated with MVBaV.
Fig 1. The symptoms of MVBaV-infected mulberry leaves.
(A) vein banding, (B) mosaic, (C) chlorotic spots, (D) leaf deformation and (E) vein necrosis. The photos were taken over a period of eight months from April to November in 2013.
Fig 2. Electron micrograph of a MVBaV-infected mulberry plant leaves showing typical tospovirus-like particle morphology.
Typical spherical, enveloped virions are shown accumulating in the endoplasmic reticulum (A) or dispersing in the cytoplasm as single particles in leaf cells (B). The scale bar represents 200 nm.
Serological characterization of MVBaV
The diseased samples reacted strongly with WSMoV/GBNV mixed antisera, but not with antisera to TSWV, INSV, IYSV, and GRSV/TCSVin ELISA assays (data not shown), indicating that MVBaV is serologically related to the members of the WSMoV serogroup.
Genome organization of the MVBaV
In our previous deep sequencing of small RNAs from MVBaV-infected samples, three incomplete RNA segments of MVBaV responding to the L, M, and S RNA had been assembled (our unpublished data). The complete genome sequence of the MVBaV isolate XCSY-3 was obtained by combining the deep sequencing data, RT-PCR and RACE data (Fig 3, Table 1).
Fig 3. Cloning strategies for L, M, and S RNAs.
Arrows indicate the annealing positions of each primer. To determine the 5′-terminal sequences, total RNAs were denatured by heating at 65°C for 5 min and then mixed with the first primer, LRNA 5–1 for L RNA, MRNA 5–1 for M RNA, and SRNA 5–1 for S RNA, respectively. After removal of template RNAs by RNaseH digestion, PCR amplification of the 5′-cDNAs was performed with Ex Taq DNA polymerase (Takara Bio, Dalian, China) using UPM primer (provided with the kits) and a nested primer, LRNA 5–2 for L RNA, MRNA 5–2 for M RNA, and SRNA 5–2 for S RNA, respectively. To determine the 3′-terminal sequence, first strand cDNA was synthesized by using the primer Tos-UPA. PCR amplification of the 3′-cDNAs was performed with Ex Taq DNA polymerase using the primers Tos-UPA and the LRNA 3 primer for L RNA, MRNA 3 for M RNA, and SRNA 3 for S RNA, respectively. The primers used for RACE and amplifying the MVBaV Genome are shown in Table 1.
L RNA
Two overlapping PCR fragments generated by RT-PCR constitute the complete MVBaV L RNA of 8905 nt in length (GenBank accession number KM819698). There is a single open reading frame (ORF) in the viral complementary strand (vc-strand) that encodes a deduced RdRp (L protein) of 2877 aa with a molecular mass of 331.6 kDa. The 5' untranslatable regions (UTRs) and 3'-UTRs of the L RNA were 241 and 30 nt in length, respectively. The 17-nt termini of the L RNA formed a panhandle structure with a mismatch at the 11th nucleotide.
The RdRp protein of MVBaV has a conserved region similar to those of other tospoviruses. The five conserved motifs, motif A (DXXKW), motif B (QGXXXXXSS), motif C (SDD), motif D (K), and motif E (EXXS), were all found within the RdRp of MVBaV. Furthermore, three motifs, Motif F1 (TDF), Motif F2 (KxQRTK) and Motif F3 (DREIY), which were found in the RdRp of CaCV, IYSV, TYRV, WBNV, GBNV, WSMoV, TZSV, and CCSV [21], were also present in the MVBaV RdRp (Fig 4).
Fig 4. The RdRp conserved motifs in viruses of the genus Tospovirus.

The consensus amino acid residues in each RdRp motif of the family Bunyaviridae are shown in bold and underlined. Identical amino acid (aa) residues are indicated with dots and deficient aa residues with hyphens. The positions of the conserved motifs in RdRp are indicated. Abbreviations and accession numbers of the analyzed sequences of tospoviruses are listed in Table 2.
M RNA
The complete nucleotide sequence of the MVBaV M RNA is 4731 nt in length and contains two ORFs encoding NSm and GP, in an ambisense coding strategy and separated by an IGR of 322 nts (GenBank accession number KM819699). The 5'- and 3'-UTRs of MVBaV M RNA contain 57 and 47 nts, respectively, forming a panhandle structure with 21 base pairs and 3 mismatches at the termini. The ORF on the viral strand codes for the NSm protein of 309 aa with a molecular mass of 34.3 kDa. The ORF on the vc-strand was 3,375 nt in length and codes for a 1,124 aa GP with a molecular mass of 128.1 kDa.
The NSm protein, probably involved in cell-to-cell movement, belongs to the 30 K movement protein superfamily [27, 28]. The conserved motifs of the 30 K movement protein superfamily, such as the D-motif and G-residue, were found in the NSm protein of MVBaV, while the P/D-L-X motif and the phospholipase A2 catalytic site (PLA2-motif) present in NSm proteins of TSWV, GRSV, CSNV, and TCSV [28, 29], was absent from the MVBaV (Fig 5).
Fig 5. The NSm conserved motifs in viruses of the genus Tospovirus.

The positions of the conserved motifs in NSm are indicated. Abbreviations and accession numbers of the analyzed sequences of tospoviruses are listed in Table 2.
In topology of MVBaV GP protein, four N-glycosylation sites (N294, N549, N930, and N1101) and five transmembrane domains (aa positions 4–26, 297–316, 321–343, 414–436, and 1051–1073) were predicted and two potential signal peptides with predicted cleavage sites at aa 24 (VYL-LN) and aa 436 (SIA-LQ) to yield the glycoproteins Gn (47.8 kDa) and Gc (77.7 kDa) were present, whereas Arg-Gly-Asp (RGD) motif was not found.
S RNA
The complete nucleotide sequence of the MVBaV S RNA is 3294 nt in length (GenBank accession code KM819701). The S RNA contains two ORFs in an ambisense coding strategy separated by an AU-rich IGR of 1008 nt in length spun from coordinates 1386–2393 in the v-strand. The 65- and 67-nt untranslated regions in the 5'- and 3'-UTRs can form a panhandle structure of 22 bp with 2 mismatches. The ORF on the v-strand codes for a 439 aa NSs protein with a molecular mass of 49.1 kDa and the ORF on the vc-strand encodes a 277 aa nucleoprotein (N protein) of 30.69 kDa. Three highly conserved motifs, Walker A (GxxxxGKT), Walker B (DEXX), and YL, were present in the NSs proteins of some tospoviruese [30, 31]. Walker A and YL were present (aa positions 175–182 and 416–417), while the Walker B motif was not found in the NSs protein of MVBaV (Fig 6).
Fig 6. The NSs conserved motifs in viruses of the genus Tospovirus.
The positions of the conserved motifs in NSs are indicated. Abbreviations and accession numbers of the analyzed sequences of tospoviruses are listed in Table 2.
Evolutionary relationship of the MVBaV to other tospoviruses
MVBaV RdRp shares the highest sequence homology to that of WSMoV (85.8%) and high sequence homology to GBNV, WBNV, and CaCV (83.9–84.9%). To the rest members of the WSMoV serogroup (MYSV, TZSV, CCSV), the homology were 76.1–77.8%. Intriguingly, the RdRp homology between MVBaV and topoviruses of different serotypes was all less than 70%, with members of TSWV and the distinct serotypes being the lowest (42.3–45.2%) (Table 3). An inspection of the proteins encoded by the M RNA and S RNA of different tospoviruses revealed that MVBaV was closest to WSMoV serogroup (M RNA, 64.6–83.2% homology; S RNA, 45.4–74.4% homology) and most distant from TSWV, GYSV, and Distinct (M RNA, 34.1–39.2% homology; S RNA, 15.9–29.9% homology), with IYSV serogroup in between (M RNA, 66.2–68.5% homology; S RNA, 41.9–49.4% homology).
Table 3. Comparison of the RNAs and deduced proteins of MVBaV with those of other Tospoviruses.
| M RNA | Full length | 5'UTR | NSm | IGR | GPs | 3'UTR | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| nt | nt | nt | aa | Identity (%) | nt | nt | aa | Identity % | nt | |
| WSMoV serogroup | ||||||||||
| MVBaV | 8905 | 241 | 8634 | 2877 | 30 | |||||
| WSMoV | 8917 | 248 | 8637 | 2878 | 85.8 | 32 | ||||
| GBNV | 8911 | 245 | 8634 | 2877 | 84.9 | 32 | ||||
| WBNV | 8916 | 247 | 8637 | 2878 | 84.6 | 32 | ||||
| CaCV-AIT | 8912 | 247 | 8634 | 2877 | 84.5 | 31 | ||||
| CCSV | 8911 | 230 | 8652 | 2883 | 77.8 | 29 | ||||
| TZSV | 8919 | 233 | 8658 | 2885 | 76.9 | 28 | ||||
| MYSV | 8918 | 273 | 8613 | 2870 | 76.1 | 32 | ||||
| IYSV serogroup | ||||||||||
| HCRV | 8908 | 250 | 8622 | 2873 | 69.0 | 33 | ||||
| PolRSV | 8893 | 230 | 8631 | 2876 | 68.8 | 32 | ||||
| TYRV | 8877 | 223 | 8622 | 2873 | 68.5 | 32 | ||||
| IYSV | 8880 | 225 | 8622 | 2873 | 67.8 | 33 | ||||
| TSWV serogroup | ||||||||||
| CSNV | 8955 | 397 | 8625 | 2874 | 45.2 | 33 | ||||
| GRSV-US | 8876 | 217 | 8625 | 2874 | 45.2 | 34 | ||||
| TCSV | 8868 | 215 | 8622 | 2873 | 44.8 | 31 | ||||
| TSWV | 8897 | 236 | 8628 | 2875 | 43.1 | 33 | ||||
| Other serotype | ||||||||||
| INSV | 8780 | 147 | 8598 | 2865 | 44.9 | 35 | ||||
| SVNaV | 9010 | 184 | 8796 | 2931 | 42.5 | 30 | ||||
| BeNMV | 9040 | 220 | 8799 | 2932 | 42.3 | 21 | ||||
| M RNA | Full length | 5'UTR | NSm | IGR | GPs | 3'UTR | ||||
| nt | nt | nt | aa | Identity (%) | nt | nt | aa | Identity (%) | nt | |
| WSMoV serogroup | ||||||||||
| MVBaV | 4731 | 57 | 930 | 309 | — | 322 | 3375 | 1124 | — | 47 |
| CaCV-AIT | 4823 | 56 | 927 | 308 | 82.6 | 427 | 3366 | 1121 | 80.9 | 47 |
| CaCV-HT-1 | 4780 | 29 | 927 | 308 | 82.3 | 436 | 3369 | 1122 | 78.9 | 19 |
| GBNV | 4801 | 56 | 924 | 307 | 81.9 | 408 | 3366 | 1121 | 84.3 | 47 |
| WBNV | 4794 | 55 | 924 | 307 | 79.9 | 402 | 3366 | 1121 | 81.6 | 47 |
| TZSV | 4945 | 54 | 930 | 309 | 77.3 | 546 | 3369 | 1122 | 73.2 | 46 |
| WSMoV | 4877 | 55 | 939 | 312 | 75.2 | 470 | 3366 | 1121 | 80.7 | 47 |
| CCSV | 4704 | 54 | 930 | 309 | 75.1 | 303 | 3372 | 1123 | 73.6 | 45 |
| TNRV | 4716 | 59 | 933 | 310 | 73.1 | 307 | 3369 | 1122 | 65.3 | 48 |
| MYSV | 4815 | 58 | 927 | 308 | 64.6 | 398 | 3384 | 1127 | 63.4 | 48 |
| IYSV serogroup | ||||||||||
| TYRV | 4786 | 62 | 927 | 308 | 68.5 | 354 | 3393 | 1130 | 62.9 | 50 |
| IYSV | 4821 | 63 | 936 | 311 | 68.1 | 394 | 3411 | 1136 | 60.3 | 17 |
| PolRSV | 4710 | 62 | 927 | 308 | 66.2 | 263 | 3408 | 1135 | 60.0 | 50 |
| HCRV | 4741 | 47 | 927 | 308 | 65.9 | 330 | 3390 | 1129 | 61.9 | 47 |
| TSWV serogroup | ||||||||||
| GRSV-SA | — | — | 912 | 303 | 39.2 | — | 3402 | 1133 | 33.4 | — |
| GRSV-US | 4848 | 99 | 912 | 303 | 38.6 | 348 | 3405 | 1334 | 34.1 | 84 |
| CSNV | 4830 | 101 | 912 | 303 | 38.6 | 326 | 3408 | 1135 | 34.5 | 83 |
| TCSV | — | — | 912 | 303 | 38.6 | — | 3405 | 1134 | 34.1 | — |
| TSWV | 4767 | 100 | 909 | 302 | 37.5 | 266 | 3408 | 1135 | 34.1 | 84 |
| ZLCV | — | — | 912 | 303 | 34.3 | — | 3408 | 1135 | 34.5 | — |
| Other serotype | ||||||||||
| BeNMV | 4886 | 64 | 954 | 317 | 37.6 | 299 | 3486 | 1161 | 33.1 | 83 |
| INSV | 4962 | 85 | 912 | 303 | 36.9 | 465 | 3333 | 1110 | 32.9 | 86 |
| SVNaV | 4955 | 57 | 951 | 316 | 34.1 | 267 | 3588 | 1195 | 30.1 | 92 |
| S RNA | Full length | 5'UTR | NSs | IGR | N | 3'UTR | ||||
| nt | nt | nt | aa | Identity (%) | nt | nt | aa | Identity (%) | nt | |
| WSMoV serogroup | ||||||||||
| MVBaV | 3294 | 65 | 1320 | 439 | 1008 | 834 | 277 | 67 | ||
| CaCV-AIT | 3477 | 66 | 1320 | 439 | 71.3 | 1199 | 828 | 275 | 74.4 | 67 |
| CaCV-HT-1 | — | — | 1320 | 439 | 69.5 | — | 834 | 277 | 72.9 | — |
| TNeV | — | — | — | — | — | — | 828 | 275 | 74.0 | — |
| WSMoV | 3558 | 67 | 1320 | 439 | 70.8 | 1278 | 828 | 275 | 70.0 | 65 |
| GBNV | 3057 | 66 | 1320 | 439 | 70.6 | 773 | 831 | 276 | 71.5 | 67 |
| WBNV | 3401 | 66 | 1320 | 439 | 70.4 | 1120 | 828 | 275 | 71.8 | 67 |
| TNSV | 3012 | 66 | 1380 | 459 | 61.7 | 664 | 834 | 277 | 61.5 | 68 |
| TZSV | 3279 | 64 | 1380 | 459 | 60.6 | 934 | 837 | 278 | 59.4 | 64 |
| CCSV | 3172 | 66 | 1383 | 460 | 59.6 | 825 | 834 | 277 | 60.4 | 64 |
| TNRV | 3023 | 65 | 1356 | 451 | 47.3 | 690 | 846 | 281 | 56.2 | 66 |
| MYSV | 3232 | 68 | 1410 | 469 | 45.4 | 847 | 840 | 279 | 58.4 | 67 |
| PhySMV | 3257 | 68 | 1410 | 469 | 45.0 | 872 | 840 | 279 | 58.4 | 67 |
| IYSV serogroup | ||||||||||
| IYSV | 3105 | 70 | 1332 | 443 | 49.4 | 811 | 822 | 273 | 44.8 | 70 |
| TYRV | 3061 | 71 | 1332 | 443 | 49.4 | 762 | 825 | 274 | 44.1 | 71 |
| HCRV | 2744 | 73 | 1338 | 445 | 48.1 | 437 | 825 | 274 | 42.7 | 71 |
| PolRSV | 2485 | 73 | 1332 | 443 | 46.7 | 183 | 825 | 274 | 41.9 | 72 |
| TSWV serogroup | ||||||||||
| MeSMV | 3283 | 80 | 1368 | 455 | 20.2 | 887 | 789 | 262 | 28.5 | 159 |
| TSWV | 2955 | 88 | 1404 | 467 | 17.7 | 535 | 777 | 258 | 29.9 | 151 |
| GRSV-SA | — | — | 1404 | 467 | 17.5 | — | — | 235 | 28.5 | — |
| GRSV-US | 3049 | 87 | 1404 | 467 | 17.7 | 630 | 777 | 258 | 29.2 | 151 |
| CSNV | 2947 | 79 | 1404 | 467 | 17.3 | 529 | 783 | 260 | 27.0 | 152 |
| PNSV | 2949 | 87 | 1404 | 467 | 17.0 | 529 | 777 | 258 | 28.5 | 153 |
| ZLCV | — | — | — | — | — | — | 783 | 260 | 28.5 | — |
| TCSV | — | — | — | — | — | — | 777 | 258 | 28.1 | — |
| ANSV | — | — | — | — | — | — | 777 | 258 | 28.1 | — |
| GYSV serotype | ||||||||||
| GCFSV | 2833 | 67 | 1419 | 472 | 16.3 | 455 | 813 | 270 | 20.8 | 79 |
| GYSV | 2970 | 57 | 1443 | 480 | 15.9 | 653 | 741 | 246 | 22.9 | 76 |
| Other serotype | ||||||||||
| INSV | 2992 | 62 | 1350 | 449 | 19.9 | 642 | 789 | 262 | 24.3 | 149 |
| BeNMV | 2584 | 60 | 1320 | 439 | 19.1 | 315 | 813 | 270 | 27.3 | 76 |
| SVNaV | 2603 | 58 | 1323 | 440 | 18.8 | 318 | 834 | 277 | 29.8 | 70 |
| LNRV | 2768 | 85 | 1326 | 441 | 17.7 | 385 | 807 | 268 | 19.9 | 159 |
Of particular interest is the envelop glycoproteins (GPs). Although the aa homology between MVBaV and most members of WSMoV serogroup is greater than 70%, two members, TNRV and MYSV, share only 63.4% and 65.3% homology, compatible to the 62.9% of TYRV, a member belonged to IYSV serogroup.
The phylogenetic trees constructed with RdRps, NSm, GnG, NSs, and N all clearly showed that MVBaV was a distict member of WSMoV serogroup, more closely related to a subgroup composed of CaCV, WBNV, WSMoV, and GBNV (Fig 7).
Fig 7. Phylogenetic trees of tospoviruses.
Amino acid sequences of the RNA-dependent RNA Polymerase (RdRp), GP protein (GP),NSm protein (NSm),NSs protein (NSs), and N protein (N) were used to constructed the trees. The dendrograms were produced using the Neighbour—Joining algorithm with 1000 bootstrap replicates. The tospovirus species belonging to WSMoV serogroup are circled. The scale bar represents a relative genetic distance. Numbers above critical branches are significant bootstrap values (>70%). Abbreviations and accession numbers of the analyzed sequences of tospoviruses are listed in Table 2.
S RNA sequence diversity
To assess the genome sequence diversity of MVBaV, the S RNA of nine additional MVBaV isolates were cloned, sequenced, and compared with that of the type isolate XCSY-3. As shown in Table 3, the lengths of the S RNA of these isolates range from 3273 to 3324 nt with six length patterns and the IGRs from 987 to1038 nt with 7 patterns among a total of 10 isolates. However, the 5'-UTR and 3'-UTR were completely conserved at length of 65 and 67 nt, respectively. The N and NSs proteins were highly conserved with only one aa substitution in the 277-aa N protein in two isolates and 2–6 aa substitutions in the 439-aa NSs proteins in eight isolates (Table 4). Therefore, the rich sequence diversity of the S RNA was reflected mainly in the length of the RNA fragment and the IGS.
Table 4. Comparison of the S RNA from isolate XCSY-3 to those of nine additional MVBaV isolates.
| Isolates (Accession number) | Full length | 5'UTR | NSs | IGR | N | 3'UTR | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| nt | nt | Nt | aa | Identity (%) | nt | nt | aa | Identity (%) | nt | |
| XCSY-3 (KM819701) | 3294 | 65 | 1320 | 439 | 1008 | 834 | 277 | - | 67 | |
| XCBY-1 (KM819709) | 3273 | 65 | 1320 | 439 | 99.5 | 987 | 834 | 277 | 100 | 67 |
| XZDL-1 (KM819704) | 3293 | 65 | 1320 | 439 | 99.3 | 1007 | 834 | 277 | 99.6 | 67 |
| SL-3 (KM819700) | 3324 | 65 | 1320 | 439 | 99.3 | 1038 | 834 | 277 | 100 | 67 |
| HX-2 (KM819702) | 3294 | 65 | 1320 | 439 | 100 | 1008 | 834 | 277 | 99.6 | 67 |
| NN-5 (KM819705) | 3288 | 65 | 1320 | 439 | 99.1 | 1002 | 834 | 277 | 100 | 67 |
| NN-10 (KM819706) | 3288 | 65 | 1320 | 439 | 98.4 | 1002 | 834 | 277 | 100 | 67 |
| NN-16 (KM819708) | 3276 | 65 | 1320 | 439 | 99.3 | 990 | 834 | 277 | 100 | 67 |
| YZ-3 (KM819703) | 3293 | 65 | 1320 | 439 | 98.6 | 1007 | 834 | 277 | 100 | 67 |
| YZ- 4(KM819707) | 3286 | 65 | 1320 | 439 | 99.8 | 1000 | 834 | 277 | 100 | 67 |
Discussion
The N protein sequence, in combination with biological characters such as the thrips vector species and host range, represents the main classification criteria for the establishment of a new tospovirus species. In term of molecular differentiation, N protein sequence identity of 90% has been set as the threshold to establish a new species of tospovirus. As of to date, nine definitive species and fourteen tentative species of tospoviruses have been recognized by the International Committee on Taxonomy of Viruses [7, 32]. Among the tentative species, Iris yellow spot virus and Watermelon bud necrosis virus are pending for final approval as definitive species [32]. The recently reported vain-banding syndrome on mulberry (Fig 1) added a new plant to the host list of tospovirus [3]. Data of genome analysis presented in this study clearly demonstrates that MVBaV is indeed a new member of Tospovirus.
The genome of MVBaV is composed of L, M, and S RNAs. The genome organization strategy and the size as well as the proteins encoded are typical of the genus Tospovirus, particularly the WSMoV serogroup (Table 3). Phylogenetic analyses with five functional proteins encoded in L, M, and S RNAs all show that MVBaV is a distinct member of the WSMoV serogroup (Fig 7). This conclusion was also supported by the serological investigation. RdRp is responsible for the genome replication and may associates with host factors to fulfill its function [4]. Alignment of RdRp aa sequences revealed that all six conserved motifs for tosopoviruses were present in the RdRp of MVBaV (Fig 4). The highest homology of viral proteins between MVBaV and other topoviruses was found in RdRp, 83.9–85.8% to that of WSMoV, GBNV, WBNV, and CaCV. These five species form a subgroup within the WSMoV serogroup, while the rest members in the same serogroup share homology of 76.1–77.8% with MVBaV, compared with 67.8–69% for viruses from the IYSV serogroup and 42.5–45.2% for viruses from TSWV and distinct serotypes (Table 3). High homology in RdRp may suggest a conserved functional mechanism for this protein.
The NSm protein encoded by M RNA is thought to be probably involved in cell-to-cell movement and belongs to the 30 K movement protein superfamily [27, 28]. Two of the four feature motifs, P/D-L-X motif and PLA2-motif, characteristic in TSWV serotype viruses did not found their counterparts in viruses of other seropgroups. For P/D-L-X motif, rather than DLF, all other tospovurses except BeNMV and SVNaV, have a motif of DSL; for PLA2-motif, instead of CCQLHLMC, most WSMoV serogroup viruses contain CMQLNLTS, following the motif pattern CXXLNLTS. Of particular note is that MYSV contains a sequence of CTQLIFTS in this motif positions, very different from all other members in the WSMoV serogroup (Fig 5).
Tospoviruses are transmitted by insects belonging to the order Thysanoptera [33]. Glycoproteins encoded by the M RNA had been shown to be involved in vector transmission in TSWV virus [34, 35]. The glycoproteins (GPs) of MVBaV are 84.3% homologous to GBMV and 78.7–81.6% to CaCV, WSMoV, and WBNV (Table 3). Similarity in glycoproteins may imply a similar insect vector. Members of WSMoV serogroup all have apparent origin in Asia and are predominantly transmitted by only two species of the genus Thrips that have been proposed as WSMoV-Thrips-Asian type[4]. Thrips palmi, the major vector of the several tospoviruses in the WSMoV serogroup, was found abundantly in agriculture fields in China [36, 37]. Since Pseudodendrothrips mori is widely distributed in the mulberry field in China [38], it may well be the vector for MVBaV. Confirmation of the disease transmissibility by this thrips is currently under investigation.
N proteins encoded by the S RNA contain antigenic determinants that form the bases for the serotype differentiation in tospoviruses [18, 19]. The N protein of MVBaV shares the highest homology of 74.4% with that of CaCV, 70.0–71.8% with those of WSMoV, GBNV, and WBNV, and 56.2–61.5% to the rest of members in the WSMoV serotype (Table 3). It was proposed that tospovirus species sharing N homology identity above 51.8% were serologically related [39].
The NSs protein encoded in S RNA has been shown to participate in viral movement, replication, and suppression of the host defense mechanism in TSWV and GBNV [30, 40, 41]. The NSs protein of MVBaV shares 70.2–71.3% homology with CaCV, WSMoV, GBNV, and WBNV, 54.4–61.7% with the rest members of the WSMoV serogroup. This protein also shares homology of 46.7–49.6% with members of the IYSV serogroup, but only 17.7% homology with TSWV (Table 3). Alignment of NSs proteins showed that WSMoV has the typical Walker A and YL motifs, but not the Walker B motif (Fig 6).
Phylogenetic analysis and homology calculation of the viral proteins RdRp, NSm, GP, NSs, and N of tospoviruses places MVBaV between the CaCV subgroup composed of CaCV, GBNV, WSMoV, and the CCSV subgroup composed of CCSV, TZSV, TRNV, and MYSV within the WSMoV serogroup of Tospovirus (Fig 7, Table 3). Thus MVBaV may represent a species that bridges the lineage within the WSMoV serogroup of Tospovirus.
Genetic diversity of MVBaV was investigated by sequencing the S RNA of 10 MVBaV isolates collected from mulberry orchards in different locations in this study. As shown in Table 4, the sizes of the RNA range from 3273 to 3324 nt, showing a disperse pattern of 3288 (2 isolates), 3293 (2 isolates), 3294 (2 isolates), 3273 (1 isolate), 3276 (1 isolate), 3286 (1 isolate), and 3324 nt (1 isolate). However, the 5’ UTR (65 nt), 3’ UTR (67 nt), the NSs (1320 nt, 439 aa), and N (834 nt, 277 aa) are completely conserved in length among all isolates. Minor substitutions in NSs (2–6 in 439 aa, 9 isolates) and N (1 in 277 aa, 2 isolates) were observed. Variations in size of the S RNA were caused by the varied length in the IGR, which ranges from 987 to 1038 nt. Intriguingly, the IGR length pattern was perfectly matched with that of S RNA size pattern, i.e., S RNA size of 3288 nt with IGR of 1002 nt (isolates NN-5 and NN-10); S RNA size of 3293 nt with IGR of 1007 nt (isolates YZ-3 and XZDL-1); S RNA size of 3294 nt with IGR of 1008 nt (isolates XCSY-3 and HX-2). Thus, it is suggested that IGR of S RNA can be used as a genetic marker for MVBaV population analysis.
Genome reassortment and recombination play a role in the evolution of RNA virus. In this regard, genome reassortment and recombination have been observed to take place in several species of Tospovirus [42–43]. It was noted that the proteins GPs and NSm encoded by the M RNA of GRSV-US, a strain from the south Florida of the United State [43], do not group with the South African isolate (GRSV-SA), but with those of TCSV (Fig 7), demonstrating a genome fragment reassortment. Indeed, a large survey of the GRSV in vegetables in Florida and the Southeastern United States showed that all GRSV contained a reassorted genome with M RNA from TCSV [44]. Although no genome reassortment event has been observed for any of the ten Tospovirus members reported in mainland China [6, 12, 13, 45, 46], detailed survey by screening the genome components is required to evaluate the epidemic potential of these viruses, as genome reassortment may change the virus-vector or virus—host specificity.
We did not succeed in transmitting MVBaV by sap inoculation to any plants including mulberry. However, the virus could be transmitted by grafting (data not shown), establishing a positive correlation of MVBaV to the disease. Of important note, 32 out of 48 (66.7%) mulberry plants with viral disease-like symptoms were found to be infected with MVBaV in our preliminary survey, demonstrating the dominant nature of MVBaV in the field [3].The high incidence of MVBaV in the mulberry orchards and the vast importance of tospovirus to crops in general imply that this virus may represent a substantial threat to the silkworm industry in China [3]. Future studies should focus on the biology of MVBaV, including the plant-virus interaction and vector-virus interaction, disease epidemiology, and practical measures for the disease control. In these regards, elucidation of the genome organization of MVBaV lays a solid foundation for future studies.
Supporting Information
(DOC)
Acknowledgments
This work was supported in part by grants from the Guangxi Natural Science Foundation (2011GXNSFD018019, 2014GXNSFAA118085), and the Key Laboratory of Ministry of Education of China for Microbial and Plant Genetic Engineering (JB1301).
Data Availability
All relevant data are within the paper and all sequence data are available from the GenBank database with accession numbers KM819698-KM819709.
Funding Statement
This work was partially supported by grants from the Natural Science Foundation of Guangxi Province (2011GXNSFD018019, 2014GXNSFAA118085), and the Key Laboratory of Ministry of Education of China for Microbial and Plant Genetic Engineering (JB1301). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tsuchizaki T, Saito Y. Mulberry latent virus isolated from mulberry (Morus alba L.). Annu Phytopath Soc Japan 1976; 42: 304–309. [Google Scholar]
- 2. Tsuchizarci T, Hlbino H, Saito Y. Mulberry ringspot virus isolated from mulberry showing ringspot symptom. Annu Phytopath Soc Japan 1971; 37: 266–271. [Google Scholar]
- 3. Meng JR, Liu PP, Zou CW, Wang ZQ, Liao YM, Cai JH, et al. First report of a Tospovirus in mulberry. Plant Dis. 2013; 97: 1001. [DOI] [PubMed] [Google Scholar]
- 4. Pappu HR, Jones RAC, Jain RK. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 2009; 141: 219–236. 10.1016/j.virusres.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 5. Quito-Avila DF, Peralta EL, Martin RR, Ibarra MA, Alvarez RA, Mendoza A, et al. Detection and occurrence of melon yellow spot virus in Ecuador: an emerging threat to cucurbit production in the region. Eur J Plant Pathol. 2014; 140:193–197. [Google Scholar]
- 6. Dong JH, Cheng XF, Yin YY, Fang O, Ding M, Li TT, et al. Characterization of tomato zonate spot virus, a new tospovirus in China. Arch Virol. 2008; 153: 855–864. 10.1007/s00705-008-0054-5 [DOI] [PubMed] [Google Scholar]
- 7. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Amsterdam: Elsevier Academic Press; San Diego: Elsevier, 2011; pp:725–741. [Google Scholar]
- 8. Chen TC, Li JT, Lin YP, Yeh YC, Kang YC, Huang LH, et al. Genomic characterization of Calla lily chlorotic spot virus and design of broad-spectrum primers for detection of tospoviruses. Plant Pathol. 2012; 61: 183–194. [Google Scholar]
- 9. Hedil M, Hassani-Mehraban A, Lohuis D, Kormelink R. Analysis of the A-U rich hairpin from the Intergenic Region of Tospovirus S RNA as Target and Inducer of RNA Silencing. PloS One 2014; 9: e106027 10.1371/journal.pone.0106027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Torres R, Larenas J, Fribourg C, Romero J. Pepper necrotic spot virus, a new tospovirus infecting solanaceous crops in Peru. Arch Virol. 2012; 157:609–615. 10.1007/s00705-011-1217-3 [DOI] [PubMed] [Google Scholar]
- 11. Zhou J, Kantartzi SK, Wen RH, Newman M, Hajimorad R, Rupe JC, et al. Molecular characterization of a new tospovirus infecting soybean. Virus Genes 2011; 43: 289–295. 10.1007/s11262-011-0621-9 [DOI] [PubMed] [Google Scholar]
- 12. Xu Y, Lou SG, Li XL, Zheng YX, Wang WC, Liu YT. The complete S RNA and M RNA nucleotide sequences of a Hippeastrum chlorotic ringspot virus (HCRV) isolate from Hymenocallis littoralis (Jacq.) Salisb in China. Arch Virol. 2013; 158: 2597–2601. 10.1007/s00705-013-1756-x [DOI] [PubMed] [Google Scholar]
- 13. Xu Y, Lou SG, Li QF, Hu Q, Sun LP, Li ZY, et al. Molecular characterization of the hippeastrum chlorotic ringspot virus L segment and its protein. Arch Virol. 2014; 159: 2805–2807. 10.1007/s00705-014-2100-9 [DOI] [PubMed] [Google Scholar]
- 14. Dong JH, Yin YY, Fang Q, McBeath JH, Zhang ZK. A new tospovirus causing chlorotic ringspot on Hippeastrum sp. in China. Virus Genes 2013; 46: 567–570. 10.1007/s11262-012-0873-z [DOI] [PubMed] [Google Scholar]
- 15. de Oliveira AS, Bertran AG, Inoue-Nagata AK, Nagata T, Kitajima EW, Oliveira RR. An RNAdependent RNA polymerase gene of a distinct Brazilian tospovirus. Virus Genes 2011; 43: 385–389. 10.1007/s11262-011-0639-z [DOI] [PubMed] [Google Scholar]
- 16. Yin Y, Zheng K, Dong J, Fang Q, Wu S, Wang L, et al. Identification of a new tospovirus causing necrotic ringspot on tomato in China. Virology J. 2014; 11: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shimomoto Y, Kobayashi K, Okuda M. Identification and characterization of Lisianthus necrotic ringspot virus, a novel distinct Tospovirus species causing necrotic disease of lisianthus (Eustoma grandiflorum). J Gen Plant Pathol. 2014; 80: 169–175. [Google Scholar]
- 18. Chen TC, Lu YY, Cheng YH, Li JT, Yeh YC, Knag YC, et al. Serological relationship between melon yellow spot virus and Watermelon silver mottle virus and differential detection of the two viruses in cucurbits. Arch Virol. 2010; 155: 1085–1095. 10.1007/s00705-010-0688-y [DOI] [PubMed] [Google Scholar]
- 19. Kang YC, Yeh SD, Liao CH, Chou WC, Liu FL, Dong JH, et al. Verification of serological relationship between two phylogenetically related peanut-infecting Tospovirus species. Eur J Plant Pathol. 2014; 140: 815–828. [Google Scholar]
- 20. Khatabi B, Wen RH, Hershman DE, Kennedy BS, Newman MA, Hajimorad MR. Generation of polyclonal antibodies and serological analyses of nucleocapsid protein of Soybean vein necrosis-associated virus: A distinct soybean infecting tospovirus serotype. Eur J Plant Pathol. 2012; 133: 783–790. [Google Scholar]
- 21. Chen TC, Li JT, Fan YS, Yeh YC, Yeh SD, Kormelink R. Molecular characterization of the full-length L and M RNAs of Tomato yellow ring virus, a member of the genus Tospovirus . Virus Genes 2013; 46: 487–95. 10.1007/s11262-013-0880-8 [DOI] [PubMed] [Google Scholar]
- 22. de Oliveira AS, Melo FL, Inoue-Nagata AK, Tatsuya Nagata T, Kitajima EW, Resende RO.Characterization of Bean necrotic mosaic virus: A member of a novel evolutionary lineage within the genus Tospovirus. PloS One 2012; 7: e38634 10.1371/journal.pone.0038634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Margaria P, Miozzi L, Ciuffo M, Pappu H, Turina M. The complete genome sequence of polygonum ringspot virus. Arch Virol. 2014; 159: 3149–3152. 10.1007/s00705-014-2166-4 [DOI] [PubMed] [Google Scholar]
- 24. Dullemans AM, Verhoeven JT, Kormelink R, van der Vlugt RA. The complete nucleotide sequence of chrysanthemum stem necrosis virus. Arch Virol. 2015; 160: 605–608. 10.1007/s00705-014-2282-1 [DOI] [PubMed] [Google Scholar]
- 25. Kikkert M, van Poelwijk F, Storms M, Kassies W, Bloksma H, van Lent J, et al. A protoplast system for studying tomato spotted wilt virus infection. J Gen Virol. 1997; 78: 1755–1763. [DOI] [PubMed] [Google Scholar]
- 26. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis Version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh P, Indi SS, Savithri HS. Groundnut Bud Necrosis Virus encoded NSm associates with membranes via its C-terminal domain. PLoS One 2014; 9: e99370 10.1371/journal.pone.0099370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li W, Lewandowski DJ, Hilf ME, Adkins S. Identification of domains of the Tomato spotted wilt virus NSm protein involved in tubule formation, movement and symptomatology. Virology 2009; 390: 110–121. 10.1016/j.virol.2009.04.027 [DOI] [PubMed] [Google Scholar]
- 29. Silva MS, Martins CR, Bezerra IC, Nagata T, de Avila AC, Resende RO. Sequence diversity of NS(M) movement protein of tospoviruses. Arch Virol. 2001; 146: 1267–1281. [DOI] [PubMed] [Google Scholar]
- 30. Lokesh B, Rashmi PR, Amruta BS, Srisathiyanarayanan D, Murthy MRN, Savithri HS. NSs encoded by Groundnut bud necrosis virus is a bifunctional enzyme. PLoS One 2010; 5: e9757 10.1371/journal.pone.0009757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhai Y, Bag S, Mitter N, Turina M, Pappu HR. Mutational analysis of two highly conserved motifs in the silencing suppressor encoded by tomato spotted wilt virus (genus Tospovirus, family Bunyaviridae). Arch Virol. 2014; 159: 1499–1504. 10.1007/s00705-013-1928-8 [DOI] [PubMed] [Google Scholar]
- 32.International Committee on Taxonomy of Viruses. Available: http://www.ictvonline.org/virusTaxonomy.asp
- 33. Whitfield AE, Ullman DE, German TL. Tospovirus thrips interactions. Annu Rev Phytopathol. 2005; 43: 459–489. [DOI] [PubMed] [Google Scholar]
- 34. Montero-Astúa M, Rotenberg D, Leach-Kieffaber A, Schneweis BA, Park S, Park JK, et al. Disruption of vector transmission by a plant-expressed viral glycoprotein. Mol Plant Microbe Interact. 2014; 27: 296–304. 10.1094/MPMI-09-13-0287-FI [DOI] [PubMed] [Google Scholar]
- 35. Whitfield AE, Ullman DE, German TL. Expression, purification, and characterization of a soluble form of Tomato spotted wilt virus glycoprotein GN. J Virol. 2004; 78: 13197–13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones DR. Plant viruses transmitted by thrips. Eur J Plant Pathol. 2005; 113: 119–157. [Google Scholar]
- 37. Reitz SR, Gao YL, Lei ZR. Thrips: pests of concern to China and the United States. Agr Sci China 2011; 10: 867–892. [Google Scholar]
- 38. Etebari K, Matindoost L, Singh RN. Decision Tools for mulberry thrips Pseudodendrothrips mori (Niwa, 1908) management in sericultural regions; an overview. Insect Science 2004; 11: 243–255. [Google Scholar]
- 39. Seepiban C, Gajanandana O, Attathom T, Attathom S. Tomato necrotic ringspot virus, a new tospovirus isolated in Thailand. Arch Virol. 2011; 156: 263–274. 10.1007/s00705-010-0856-0 [DOI] [PubMed] [Google Scholar]
- 40. de Ronde D, Butterbach P, Lohuis D, Hedil M, van Lent JW, Kormelink R. Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the tomato spotted wilt virus . Mol Plant Pathol. 2013; 14: 405–415. 10.1111/mpp.12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Ronde D, Pasquier A, Ying S, Butterbach P, Lohuis D, Kormelink R. Analysis of Tomato spotted wilt virus NSs protein indicates the importance of the N-terminal domain for avirulence and RNA silencing suppression. Mol Plant Pathol. 2014; 15: 185–195. 10.1111/mpp.12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lian S, Lee JS, Cho WK, Yu J, Kim MK, Choi HS, et al. Phylogenetic and recombination analysis of tomato spotted wilt virus . PLoS One 2013; 8:e63380 10.1371/journal.pone.0063380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Webster CG, Reitz SR, Perry KL, Adkins S. A natural M RNA reassortant arising from two species of plant- and insect-infecting bunyaviruses and comparison of its sequence and biological properties to parental species. Virology 2011; 413:216–225. 10.1016/j.virol.2011.02.011 [DOI] [PubMed] [Google Scholar]
- 44. Webster CG, Frantz G, Reitz SR, Funderburk JE, Mellinger HC, McAvoy E, et al. Emergence of Groundnut ringspot virus and Tomato chlorotic spot virus in vegetables in Florida and the southeastern United States. Phytopathology 2015; 105:388–398. 10.1094/PHYTO-06-14-0172-R [DOI] [PubMed] [Google Scholar]
- 45. Rao X, Wu Z, Li Y. Complete genome sequence of a Watermelon silver mottle virus isolate from China. Virus Genes 2013; 46:576–580. 10.1007/s11262-013-0885-3 [DOI] [PubMed] [Google Scholar]
- 46. Hu ZZ, Feng ZK, Zhang ZJ, Liu YB, Tao XR. Complete genome sequence of a tomato spotted wilt virus isolate from China and comparison to other TSWV isolates of different geographic origin. Arch Virol. 2011; 156:1905–1908. 10.1007/s00705-011-1078-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and all sequence data are available from the GenBank database with accession numbers KM819698-KM819709.