Abstract
Background
Bacillus amyloliquefaciens B10-127 exhibited an excellent ability for industrial-scale microbial fermentation of 2,3-butanediol (2,3-BD) from biodiesel-derived glycerol. However, the accumulation of by-products (acetoin, acetoin, lactate and succinate) and the 2,3-BD yield remains prohibitively low for commercial production.
Results
Several strategies were developed to manipulate the carbon flux to 2,3-BD branch in a designed B. amyloliquefaciens. Firstly, extra copies of NADH/NAD+ regeneration system were introduced into B. amyloliquefaciens by co-overproduction of glycerol dehydrogenase and acetoin reductase, which resulting in improvement of 2,3-BD production and suppression of by-products accumulation. Subsequently, the transcriptional regulator ALsR under the control of a moderate promoter PbdhA was introduced into B. amyloliquefaciens, which increased carbon flux to 2,3-BD branch. Finally, a three-stage dissolved oxygen control strategy were proposed based on analysis of the characteristic of 2,3-BD fermentation, and a two-stage pH control strategy were proposed based on different pH preferences of ACR for reduction and oxidation. Following these strategies, a high titer (102.3 g/L), yield (0.44 g/g), and productivity (1.16 g/L/h) of 2,3-BD were achieved.
Conclusions
To our knowledge, this is the highest reported 2,3-BD production using biodiesel-derived glycerol as substrate, and this designed B. amyloliquefaciens should be an excellent candidate for producing 2,3-BD on an industrial scale.
Keywords: 2,3-butanediol; Glycerol; Cofactor regeneration; Manipulating carbon flux; Bacillus amyloliquefaciens
Background
2,3-Butanediol (2,3-BD) has potential applications in the manufacture of foods, pharmaceuticals fumigants, printing inks, moistening and softening agents, plasticizers [1]. Interest in microbial production of 2,3-BD has increased significantly because 2,3-BD has a wide range of industrial applications, and microbial production will alleviate the dependence on oil supply for the production of platform chemicals [2, 3].
To date, many studies on 2,3-BD production have focused on sugar fermentation. Despite the high-effective productivities that have been achieved via the conversion of glucose [4], the relatively high cost of conventional sugar substrates is still viewed as a major factor during 2,3-BD fermentation. Therefore, 2,3-BD production using cheaper alternative biomass-derived substrates under proper conditions is of high priority [2]. Biodiesel production from plant oils and animal fats reportedly generate large quantities of by-product waste glycerol [5]. For example, approximately 10 % of the biodiesel weight produced is waste glycerol [6]. In some European countries, the production of glycerol has increased significantly due to biodiesel uptake. However, those biodiesel companies have severe problems getting rid of excess glycerol and disposal is quite expensive. The collapse of glycerol prices causes major problems to these companies [7]. Since glycerol can be used as a carbon source in industrial microbiology, this by-product adds value to the productive chain of the biodiesel industry, contributing to their competitiveness [6]. Raw non-purified glycerol is an economical substitute for pure glycerol as a fermentation substrate. Therefore, converting the vast amounts of glycerol into cost-effective commercial products is an industrial priority. Some researchers have reported that Klebsiella strains can catalyze pure glycerol into 2,3-BD, but these reactions generate large quantities of 1,3-PD [8, 9]. Importantly, because Klebsiella pneumoniae is a pathogenic microorganism, it does not conform to the safety regulations of industrial-scale fermentation [3]. Metsoviti et al. [10] obtained a 2,3-BD concentration of 22 g/l and with a relatively high conversion yield on glycerol consumed of 0.40 g/g with a newly isolated Enterobacter aerogenes FMCC-10; however, the efficiency of the production was still much too low for an economic process.
Previously, we reported that B. amyloliquefaciens readily produces 2,3-BD from biodiesel-derived glycerol in the presence of beet molasses as a co-substrate [11]. In fed-batch fermentation, 2,3-BD production (83.3 g/L) from waste glycerol reached the highest level reported to date, but fermentation was accompanied by undesirably large production of acetoin, lactate, acetate, and succinate. However, both raw glycerol and molasses are easily available by-products from plant (mostly) biomass conversion, and they represent abundant renewable feed stocks and furthermore, they need no pretreatment before fermentation processes. So microbial production of 2,3-BD from these raw materials is potentially economically feasible if bacterial strain used for this purpose produces at least 100 g 2,3-BD per L (the recovery of 2,3-BD from culture broth is troublesome and usually energy-consuming distillation is necessary). However, neither process conditions have been optimized (it was just comparison of the effect of pure glucose or sucrose, or molasses on 2,3-BD biosynthesis yield) nor the strain was improved by genetic engineering methods.
The 2,3-BD pathway has been studied in various bacteria [12]. As shown in Scheme 1, assimilation of glycerol to produce 2,3-BD is also an oxido-reduction-associated process. In the oxidative pathway, the NAD+-dependent glycerol dehydrogenase (GDH) oxidizes glycerol to dihydroxyacetone, which is subsequently oxidized to pyruvate. Acetolactate synthase (ALS) catalyzes the in vivo coupling of two pyruvate molecules to form acetolactate, which is then decarboxylated to acetoin by acetolactate decarboxylase (ALDC). Finally, acetoin is reduced to 2,3-BD by an NADH-dependent acetoin reductase (ACR) [13]. Bacterial strains may accumulate acetoin for several reasons. One factor that limits acetoin degradation is low levels of ACR, assumed as the rate-limiting factor in the conversion of acetoin into 2,3-BD. Alternatively, low levels of NADH may limit the ACR reaction, since this coenzyme is preferentially used in 2,3-BD synthesis. However, some other by-products are also produced by B. amyloliquefaciens, such as succinate, lactate, and acetate, which negatively regulate the 2,3-BD yield and increase the costs of downstream separation and purification. Furthermore, large-scale microbial 2,3-BD production requires efficient and economical fermentation processes. Thus, in this work, we focused on improving strains to produce 2,3-BD with high yield, using low price substrates (raw glycerol and molasses) to lower the cost of feedstock, and optimizing the operation mode to make the process more efficient (Scheme 1).
Scheme 1.
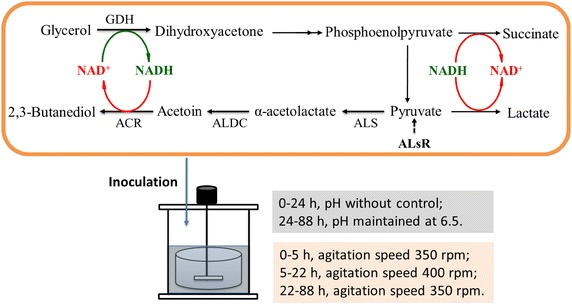
The strategy for designed B. amyloliquefaciens to manipulate the carbon flux to 2,3-BD branch. To make the 2,3-BD branch gains a competitive advantage over the end products of pyruvate-deriving pathways (such as acetic acid, lactic acid and succinic acid), several strategies were developed to manipulate the carbon flux to 2,3-BD branch in a designed B. amyloliquefaciens. Firstly, extra copies of NADH/NAD+ regeneration system were introduced into B. amyloliquefaciens by co-overproduction of glycerol dehydrogenase and acetoin reductase. Subsequently, the transcriptional regulator ALsR under the control of a moderate promoter PbdhA was introduced into B. amyloliquefaciens. Finally, a three-stage dissolved oxygen control strategy were proposed based on analysis of the characteristic of 2,3-BD fermentation, and a two-stage pH control strategy were proposed based on different pH preferences of ACR for reduction and oxidation
Results and discussion
Over-production of glycerol dehydrogenase and its effects on 2,3-BD production
Glycerol dehydrogenase (GDH) is an important polyol dehydrogenase for glycerol metabolism in diverse microorganisms, and for value-added utilization of glycerol in the industry that catalyzes the dehydrogenation of glycerol to dihydroxyacetone. This reaction is coupled to the reduction of oxidized NAD+ to NADH. So, over-production of glycerol dehydrogenase could increase not only the dehydrogenation of glycerol but also the level of available NADH.
In the genome sequence of K. pneumoniae ATCC 25955, there are two GDHs (DhaD and GldA) [14]. So, the two enzymes were separately introduced into B. amyloliquefaciens, and generated recombinant strains DH and GL, respectively. As shown in Table 1, the specific activities of GDH in strains DH and GL were separately 4.76 and 3.02 folds higher than in the parental strain. Also, the highest 2,3-BD concentration were increased by 10.7 % with strain DH and 6.35 % with strain GL, which suggested that overproduction of DhaD was more efficient for 2,3-BD production. Wang et al. [14] found that DhaD is highly induced by glycerol, and apart from catalyzing the dehydrogenation of glycerol to dihydroxyacetone, it also could catalyze the reduction of acetoin to 2,3-BD in the presence of NADH. In other words, DhaD plays a dual role in glycerol metabolism and 2,3-butanediol formation. Therefore, DhaD over-eproduction might enhance 2,3-BD production by increasing not only the level of available NADH but also catalytic activity of 2,3-BD formation.
Table 1.
Effect of over-production of GDH on 2,3-BD production in B. amyloliquefaciens
| Strains | CGly (g/L) | C Suc (g/L) | Specific activity of GDH (U/mg) | 2,3-BD (g/L) | Acetoin (g/L) | Lactate (g/L) | Acetate (g/L) | Succinate (g/L) | DCW (g/L) |
|---|---|---|---|---|---|---|---|---|---|
| B10-127 | 80 ± 2 | 10 ± 1 | 0.12 ± 0.01 | 31.3 ± 1.03 | 8.56 ± 0.36 | 2.12 ± 0.10 | 1.11 ± 0.06 | 3.65 ± 0.15 | 9.21 ± 0.26 |
| DH | 80 ± 2 | 10 ± 1 | 0.69 ± 0.06 | 34.6 ± 1.12 | 4.87 ± 0.21 | 2.46 ± 0.12 | 1.02 ± 0.05 | 3.96 ± 0.17 | 9.12 ± 0.21 |
| GL | 80 ± 2 | 10 ± 1 | 0.48 ± 0.04 | 33.3 ± 1.15 | 6.65 ± 0.29 | 2.38 ± 0.12 | 1.04 ± 0.06 | 3.81 ± 0.16 | 9.16 ± 0.22 |
C Gly glycerol consumption, C Suc sucrose consumption, GDH glycerol dehydrogenase
Batch fermentation was carried out at 37 °C in 250-mL flasks [95 g/l co-substrate; molasses to crude glycerol ratio 1.5:8 (w/w)] at 37 °C and shaking speed 180 rpm
Introduction of extra copies of DhaD/ACR enzymes into B. amyloliquefaciens and their effects on 2,3-BD production
The NAD+-dependent DhaD oxidizes glycerol to dihydroxyacetone, with concomitant reduction of NAD+ to NADH. In contrast, the NADH-dependent ACR reduces acetoin to 2,3-BD, with concomitant oxidation of NADH to NAD+. So, co-overproduction of DhaD and ACR may enhance 2,3-BD production [15]. Inspired by this idea, the dhaD gene harbored in pMA5-acr was overexpressed in the strain GA. The plasmid genetic rate remained about 95 %, indicating that the pMA5-acr-HapII-dhaD was stably expressed in the strain GA. The specific activities of GDH (0.67 ± 0.06 U/mg) and ACR (0.58 ± 0.05 U/mg) in the strain GA were 4.58-fold and 3.66-fold higher than in the strain B10-127, respectively.
The effects co-overexpression of dhaD and acr on cell growth and 2,3-BD production were also investigated. As shown in Fig. 1a, the strain GA grew at a slightly lower rate compared to that of the parent strain (B10-127), suggesting that cell growth was slightly inhibited by the over-production of DhaD/ACR. As shown in Fig. 1b, the strain GA consumes glycerol more slowly than the parental strain during the exponential growth phase. However, the fermentation rate of the engineered strain GA remarkably increased in the stationary phase, with consequent reduction in fermentation time. Furthermore, co-overproduction of DhaD and ACR in B. amyloliquefaciens increased the highest 2,3-BD titer by 13.6 %, while decreasing the acetoin concentration by 64.6 % (see Fig. 1c, d). In addition, other by-products production, such as acetate, lactate and succinate, were also suppressed. However, in both parental and engineered strains, the intracellular NADH and NAD+ concentration had no difference during the glycerol fermentation (data not shown), possibly because introducing extra copies of DhaD/ACR enzymes into B. amyloliquefaciens accelerated the NADH/NAD+ regeneration rate without influencing the NAD+/NADH levels. Two reasons may account for this phenomenon. First, over-production of NAD+-dependent DhaD increased the rate of glycerol consumption and NADH level. Second, in the glycerol metabolism, the 2,3-BD branch is primarily responsible for oxidizing NADH, and when NADH-dependent ACR is overproduced, the 2,3-BD branch gains a competitive advantage over the end products of pyruvate-deriving pathways (such as lactic acid and succinic acid), through the enhanced availability of NADH. So, when extra copies of DhaD/ACR enzymes were introduced into B. amyloliquefaciens, it was found that overexpressing the NADH/NAD+ regeneration system effectively improved 2,3-BD production and inhibited by-products accumulation.
Fig. 1.
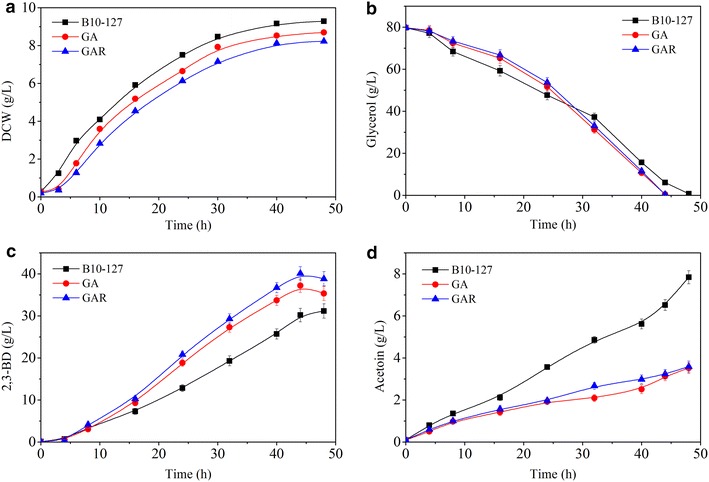
Time profiles of 2,3-BD fermentation (a cell growth; b glycerol consumption; c 2,3-BD production; d acetoin formation). Batch fermentation was carried out at 37 °C in a 5-L bioreactor containing 2.5-L initial medium [95 g/L co-substrate; molasses to crude glycerol ratio 1.5:8 (w/w)] at 37 °C, agitation speed 350 rpm, and airflow rate 0.66 vvm
Manipulating the carbon flux from pyruvic acid to 2,3-butanediol branch by moderate expression of the transcriptional regulator ALsR
There are three key enzymes involved in 2,3-BD branch, i.e. α-acetolactate synthase (ALS), α-acetolactate decarboxylase (ALDC), and acetoin reductase (ACR). To make the 2,3-BD branch gains a competitive advantage over the end products of pyruvate-deriving pathways (such as acetic acid, lactic acid and succinic acid), in our pre-test, we attempted to overexpress ALS and ALDC to enhance acetoin production. And we succeeded in increasing the activity of these enzymes by more than 50-fold, however, 2,3-BD production was not significantly enhanced and cell growth was markedly inhibited. ALS and ALDC are encoded by the alsSD operon in B. subtilis [16]. It has been reported that the transcriptional regulator ALsR is essential for the expression of alsSD [17] and that the disruption of alsR prevents the transcription of alsSD [16].
The recombinant plasmids pMA5-HpaII-alsR and pMA5-PbdhA-alsR constructed using the strong HpaII and moderate PbdhA promoters to express ALsR, respectively [18]. In the preliminary study, to improve the carbon flux to 2,3-BD branch, alsR under the control of two different promoters (HpaII and PbdhA) were cloned into B. amyloliquefaciens B10-127, respectively. 2,3-BD production was improved by regulating ALsR expression using either of the two promoters. However, although ALsR expression was higher under the control of the stronger promoter (HpaII) than the moderate promoter (PbdhA), moderate enhancement of ALsR expression was more benefic to improve 2,3-BD production. Compared with the parent strain, 2,3-BD concentration increased by 9.2 % and 15.4 % under the control of and PbdhA, respectively. Furthermore, the rates of cell growth and glucose consumption under the control of HpaII were lower than under the control of PbdhA. Zhang et al. [18] also reported that moderate enhancement of ALsR expression was more efficient for acetoin (precursor of 2,3-BD) production than strong over-expression of ALsR.
So, we selected the moderate PbdhA promoter to express ALsR in B. amyloliquefaciens GA. The plasmid genetic rate of the resulting GAR recombinant remained stable at about 95 %, indicating that the pMA5-dhaD-HapII-acr-PbdhA-alsR was stably expressed in the GAR strain. The specific activities of ALS (0.82 ± 0.07 U/mg) and ALDC (0.25 ± 0.03 U/mg) in the GAR strain were respectively 1.02-fold and 1.76-fold higher than in the GA strain.
As shown in Fig. 1a, the GA strain had a faster growth rate than the recombinant strains, suggesting that ALsR expression inhibited cell growth. All of the glycerol (80 g/L) was consumed within 44 h (Fig. 1b), and strain GAR produced approximately 38.8 g/L 2,3-BD (Fig. 1c), respectively. And main by-products production, such as acetoin, acetate, lactate and succinate, were further suppressed (Table 2). The main reason might be that the 2,3-BD branch gains a competitive advantage over the end products of pyruvate-deriving pathways. Thus, an excellent 2,3-BD producer from glycerol was redesigned through introducing extra copies of DhaD/ACR enzymes and improving catalytic activities of enzymes involved in 2,3-BD synthetic branch.
Table 2.
Metabolic flux distribution in B. amyloliquefaciens (unit: mol/mol substrate)
| Strains | Flux to | |||||
|---|---|---|---|---|---|---|
| 2,3-BD | Acetoin | Lactate | Succinate | Acetate | Biomass | |
| B10-127 | 0.695 | 0.174 | 0.027 | 0.036 | 0.022 | 0.048 |
| GAR | 0.862 | 0.061 | 0.012 | 0.015 | 0.013 | 0.043 |
Metabolic flux redistributions
In the aerobic glycerol metabolism of B. amyloliquefaciens, 2,3-BD plays a major role in oxidizing NADH. To secure NADH and C, it must compete with other end products of pyruvate-deriving pathways. Thus, this study characterized the metabolic flexibilities of B. amyloliquefaciens in response to over-expression of the dhaD, acr and alsR genes. For this purpose, the concentrations of major metabolites of both strains (B10-127, GAR) were determined (titers of 2,3-BD, acetoin, succinate, lactate and acetic acid). As shown in Table 2, compared with the parent strain, the molar yield of 2,3-BD was higher (24.1 %), and the molar yields of unwanted by-products were significantly lower in the mutant strain (64.9, 55.4, 58.3 and 36.9 % for acetoin, lactate, succinate and acetate, respectively). This observation suggests that moderate enhancement of ALsR and co-overproduction of DhaD, ACR in strain B10-127 make the 2,3-BD branch gain a competitive advantage over the end products of pyruvate-deriving pathways. And this result also indicates that glycerol fluxes are redistributed in B. amyloliquefaciens.
Manipulating the carbon flux from acetoin to 2,3-butanediol by using a three-stage oxygen control strategy
Oxygen supply is one of the most important variables in the 2,3-BD fermentation [2, 3]. Jansen et al. [19] found that high oxygen supply favored cell mass formation at the expense of 2,3-BD production. Decreasing the oxygen supply would increase 2,3-BD yield, but it would decrease the overall conversion rate due to lower cell concentrations [20, 21]. Therefore, it is necessary to establish a proper oxygen supply control strategy to ensure efficient 2,3-BD production.
As it is easier to control dissolved oxygen concentration by changing agitation speed than by varying aeration rate, in this study, we investigated the characteristics of 2,3-BD fermentation by B. amyloliquefaciens GAR under different oxygen supply methods by changing agitation speeds. Based on the analysis of two kinetic parameters including specific cell growth rate (μx) specific glycerol consumption rate (μs) and specific 2,3-BD formation rate (μp) (Fig. 2), a three-stage agitation speed control strategy, aimed at achieving high concentration and high yield 2,3-BD, was proposed. At the first 5 h, agitation speed was controlled at 350 rpm, subsequently agitation speed was raised to 400 rpm until 22 h, and then, agitation speed was reduced to 350 rpm (Fig. 3). Finally, the maximum concentration of 2,3-BD reached 42.6 g/L, which were 9.85 % over the best results controlled by constant agitation speeds. What’s more, titer of acetoin was reduced by 61.5 %. The proposed three-stage agitation speed control strategy was therefore proved to be successful to enhance 2,3-BD production. The idea developed in this paper could be applied to the other industrial biotechnological process to achieve high product concentration and high yield simultaneously.
Fig. 2.
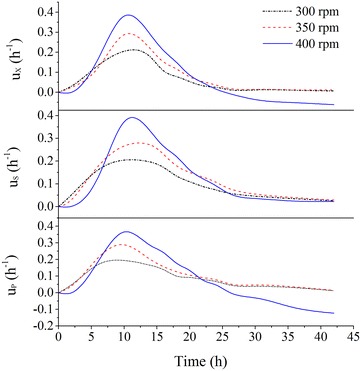
Comparison of kinetic parameters in 2,3-BD fermentation at different agitation speeds. (Batch fermentation was carried out at 37 °C in a 5-L bioreactor containing 2.5-L initial medium (95 g/L co-substrate; molasses to crude glycerol ratio 1.5:8 (w/w)) at 37 °C, and airflow rate 0.66 vvm.)
Fig. 3.
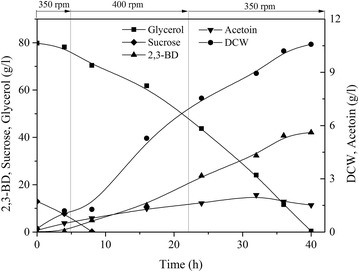
Time course of 2,3-BD fermentation by B. amyloliquefaciens GAR using three-stage agitation speed control strategy. (Batch fermentation was carried out at 37 °C in a 5-L bioreactor containing 2.5-L initial medium (95 g/L co-substrate; molasses to crude glycerol ratio 1.5:8 (w/w)) at 37 °C, airflow rate 0.66 vvm, and agitation speed: 0–5 h, 350 rpm; 5–22 h, 400 rpm; 22–88 h, 350 rpm.)
Some other parameter OTR [22], kLa [23], OUR [24] and RQ [25, 26] guided oxygen supply control strategies were successfully applied in 2,3-BD fermentation and proved to be effective. However, the parameters of OTR, kLa, OUR and RQ are not easy to control, thus restricting the application of those strategies [20]. In this study, a simple oxygen supply method based on agitation speed control was set up to realize efficient 2,3-BD fermentation.
Manipulating the carbon flux from acetoin to 2,3-butanediol by using a two-stage pH control strategy in fed-batch fermentation
Firstly, fed-batch fermentation was performed under the combined feeding strategy (initial addition of beet molasses and later co-feeding with glycerol and molasses) [11]. The representative time courses of fed-batch fermentation by B. amyloliquefaciens GAR are presented in Fig. 4a. The 2,3-BD reached 89.5 g/L within 90 h, however, it was accompanied by undesirably large production of acetoin (~20 g/L). However, as shown in Fig. 4a, since glycerol is rapidly synthesized within 80 h of fermentation, it was also found that time profiles of 2,3-BD production could be divided into two stages. At the first 24 h, pH values were below 7.0, and 2,3-BD was quickly accumulated, while acetoin was produced very slowly. After this point, the pH value gradually rise to about 8.0, which suppressed 2,3-BD formation, while enhanced acetoin accumulation. Since 2,3-BD is produced from pyruvate in a mixed acid fermentation process, the first pH decline may be related to rapid secretion of organic acids (such as lactate and acetate). Subsequent 2,3-BD synthesis reverses the intracellular acidification and raises the pH [27].
Fig. 4.
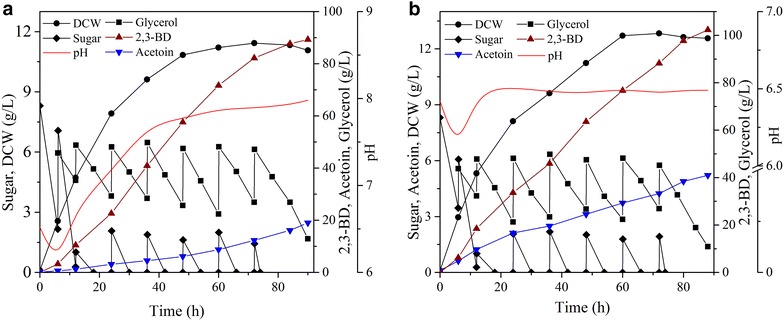
Effect of pH control strategy on 2,3-BD production by B. amyloliquefaciens GAR in fed-batch fermentation. a Without pH control; b At the first 16 h, the pH value was not under controlled, and after that point, pH was adjusted to 6.5.)
Another major influence on 2,3-BD production is the pH [3]. Nakashimada et al. [27] reported that 2,3-BD synthesis is induced under acid supplementation, which may suggest that 2,3-BD, as a neutral metabolite, counteracts too high acidification. According to Garg and Jain [12] alkaline conditions favour formation of organic acids, with a simultaneous decrease in the 2,3-BD yield. In contrary, organic acid synthesis is reduced (over tenfold) and diol synthesis is increased (3–7-fold) under acidic conditions. Biebl et al. [8] observed that in Klebsiella sp., at neutral pH, it synthesizes acetic acid and ethanol, but below pH 6, 2,3-BD and ethanol are produced. However, the optimum pH for 2,3-BD production strongly depends on the microorganism and substrate used. Voloch et al. [28] found the pH range from 5 to 6 was more beneficial to 2,3-BD production by K. oxytoca. For E. aerogenes, Converti et al. [21] and Perego et al. [29] all experimentally determined a pH value of 6 as the optimum for the production of 2,3-BD. Stormer [30] found that in K. pneumoniae a pH above 6 causes a sharp decrease in the activity of α-acetolactate synthase (one of the key enzymes in the 2,3-BD pathway). Previously, the results also clearly showed that pH-dependent 2,3-BD production from glucose of B. amyloliquefaciens with the maximum production was at initial pH 6.5, and in the initial stage of fermentation, it was good for 2,3-BD synthesis without external pH control [31]. Industrial-scale fermentation requires obeying safety regulations, therefore, an urgent need for class 1 microorganisms (safe) is pronounced. The Gram-positive bacterium B. amyloliquefaciens has been classified as GRAS (generally regarded as safe) by the US Food and Drug Administration [32]. Therefore, increasing 2,3-BD production by B. amyloliquefaciens is an economically valuable goal.
During the production of 2,3-BD from glycerol, acetoin is the precursor of 2,3-BD. So, it is very important that maintain the suitable conditions to steadily biosynthesize 2,3-BD from acetoin. Previously, it was found that the ACR, which catalyzes the interconversion between acetoin and 2,3-butanediol [31, 33]. We further determined the effects of pH on ACR activity. As shown in Fig. 5, ACR showed very different pH preferences of pH 6.5 for reduction and pH 8.5 for oxidation. In other words, this enzyme is critical for 2,3-BD biosynthesis, exhibits the highest activity at pH 6.5, whereas at pH 7.0, about 20 % of its activity is lost, and at pH 8.0, about half of its activity is lost. This pH property of ACR from other bacterium was also reported [34–36].
Fig. 5.
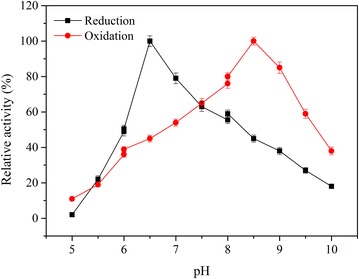
Effect of pH on acetoin reductase activity
Basing on the above pH preference, a two-stage control strategy was proposed to improve 2,3-BD formation (Fig. 4b). At the first stage, the pH value was not under controlled for induction of 2,3-BD formation, and then when the pH value reached about 6.5, the pH control device began to work to maintained pH value at 6.5. As expected, through the strategy, the 2,3-BD production was up to 102.3 g/L (achieved at 88 h) with corresponding productivity of 1.16 g/L h and a yield of 0.44 g/g substrate, which were 14.8, 18.4 and 15.5 % higher than that under without pH control. Thus, the two-stage pH control strategy proposed in this work is conducive to 2,3-BD formation. To our knowledge, these are the highest reported levels of 2,3-BD fermentation from biodiesel-derived glycerol.
Conclusion
Overexpressing the NADH/NAD+ regeneration system effectively improved 2,3-BD production and inhibited by-products accumulation. Medorate expression of ALsR made the 2,3-BD branch gain a competitive advantage over the end products of pyruvate-deriving pathways. The carbon flux from acetoin to 2,3-BD was manipulated by using two-stage controlled pH and dissolved oxygen strategies. Finally, a high titer (102.3 g/L), yield (0.44 g/g), and productivity (1.16 g/L/h) of 2,3-BD were achieved. To our knowledge, this is the highest reported 2,3-BD production using biodiesel-derived glycerol as a substrate, and this designed B. amyloliquefaciens should be an excellent candidate for producing 2,3-BD on an industrial scale.
Methods
Strains and plasmids
Strains, plasmids and primers used in this study are listed in Table 3. The parent strain was B. amyloliquefaciens B10-127, which has been deposited in the China Center for Type Culture Collection (CCTCC) under collection number CCTCC M 2012349. K. pneumoniae ATCC 25955 was obtained from American Type Culture Collection. The recombinant derivatives of Escherichia coli/B. subtilis shuttle plasmid vector pMA5-HapII were hosted in E.coli JM109. This shuttle vector introduced the expression cassette for hyper-expression of dhaD, gldA, acr and alsR into B. amyloliquefaciens B10-127.
Table 3.
Strains, plasmids and primers used in this study
| Bacterial strain, plasmid or primer names | Relevant characteristic or sequence | Source or enzyme site |
|---|---|---|
| Strains | ||
| B. amyloliquefaciens | ||
| B10-127 | Host strain | Lab stock |
| DH | B10-127 with pMA5 -dhaD | This study |
| GL | B10-127 with pMA5-gldA | This study |
| GA | B10-127 with pMA5-acr-HapII-dhaD | This study |
| GAR | B10-127 with pMA5-acr-HapII-dhaD-PbdhA-alsR | This study |
| K. pneumoniae ATCC 25955 | Source of dhaD gene | Lab stock |
| E. coli JM109 | Lab stock | |
| Plasmids | ||
| pMA5-HapII | Expression vector (in E. coli, Apr; in B. amyloliquefaciens, Kanr) | Lab stock |
| pMA5-acr | pMA5-HapII with acr (in E. coli, Apr; in B. amyloliquefaciens, Kanr) | Lab stock |
| pMA5-PbahA-alsR | pMA5 containing PbahA-alsR (in E. coli, Apr; in B. amyloliquefaciens, Kanr) | Lab stock |
| pMA5-HpaII-alsR | pMA5 containing HpaII-alsR (in E. coli, Apr; in B. amyloliquefaciens, Kanr) | Lab stock |
| pMA5-gldA | pMA5-HapII with gldA (in E. coli, Apr; in B. amyloliquefaciens, Kanr) | This study |
| pMA5-dhaD | pMA5-HapII with dhaD (in E. coli, Apr; in B. amyloliquefaciens, Kanr) | This study |
| pMA5-acr-HapII-dhaD | Apr, Kanr; pMA5-HapII with acr and HapII-dhaD (in E. coli, Apr; in B. amyloliquefaciens, Kanr) | This study |
| pMA5-acr-HapII-dhaD-PbahA-alsR | Apr, Kanr; pMA5-acr-HapII-dhaD with PbahA-alsR (in E. coli, Apr; in B. amyloliquefaciens, Kanr) | This study |
| Primers | ||
| P1 | 5′-CGGGATCCATGAAGCCTGAAGATATCG-3′ | BamHI |
| P2 | 5′-CGACGCGTCTATCCTGTCTTTTGCGC-3′ | MluI |
| P3 | 5′-CGACGCGTTTTTGAGTGATCTTCTC-3′ | MluI |
| P4 | 5′-CGGGATCCATGGATCGCATTATTCAATC-3′ | BamHI |
| P5 | 5′-CGACGCGTTTATTCCCATTCCTGCAGG-3′ | MluI |
Underlined nucleotides are the restriction enzyme sites
Culture conditions
B. amyloliquefaciens and E. coli were cultured in Luria-Bertain (LB) medium. When necessary, ampicillin or kanamycin was added into the culture medium. For 2,3-BD production, B. amyloliquefaciens was inoculated into 10 mL LB medium added with 40 g/L glycerol and cultivated overnight with agitation (180 rpm, rotary shaker) at 37 °C. After 12 h, 2.5 mL of the seed culture (OD600 = 5.0–6.0) was inoculated into fermentation medium (crude glycerol (80 g/L), beet molasses (15 g/L) (Addition of beet molasses could enhance glycerol assimilation [11] ), corn steep liquor (30 g/L), soybean meal (20 g/L), ammonium citrate (5 g/L), K2HPO4 (3 g/L), MgSO4·7H2O (0.3 g/L), FeSO4·7H2O (0.05 g/L), pH6.5). Waste glycerol comprised of 88 % (w/w) glycerol, 6–9 % (w/w) water, 4–6 % (w/w) ash, 4 % (w/w) chlorides, and 0.2 % (w/w) methanol. The composition of beet molasses was 56.6 % (w/w) sucrose, 0.08 % (w/w) glucose, 23.8 % (w/w) water, 10 % (w/w) sulfated ash, 7.3 % (w/w) colloidal substances, and 2.1 % (w/w) nitrogen.
Batch fermentation was carried out at 37 °C in 250-mL flasks or a 5-L bioreactor (BIOTECH-2002, Baoxing Biological Equipment Co., Shanghai, China) containing 2.5-L initial medium (95 g/l co-substrate; molasses to crude glycerol ratio 1.5:8 (w/w)) at 37 °C and airflow rate 0.66 vvm. The fed-batch (inoculated with 4 % v/v seed culture) was cultivated in a 5-L stirring bioreactor with a working volume of 2.5 l (15 g/l molasses). Glycerol at 80 % (w/v), or a solution of 80 % glycerol, 15 % molasses, and 5 % H2O, was fed into the bioreactor to maintain the glycerol concentration between 20 and 50 g/l from 5 to 88 h. Supplementation was ceased after 88 h to minimize the glycerol residue in the final broth [11].
Plasmids construction
The gene dhaD and gldA (encoding glycerol dehydrogenases) from K. pneumoniae ATCC 25955 were amplified by PCR technique using primers P1/P2 and P4/P5, respectively. The purified PCR products were separately double-digested by BamH I and Mlu I, and then ligated to the corresponding sites of the pMA5 plasmid. The recombinant plasmid pMA5-dhaD and pMA5-gldA were generated.
The dhaD gene, containing the HapII promoter from the pMA5-dhaD plasmid, was then PCR-amplified using primers P2 and P3. The amplified HapII-dhaD gene was inserted into the Mlu I site of the previously constructed plasmid pMA5-acr [15] to create the pMA5-acr-HapII-dhaD plasmid. The PbdhA-alsR gene cut from the previously constructed plasmid pMA5-PbdhA-alsR [18] was inserted into the EcoR V and Hind III sites of the pMA5-acr-HapII-dhaD plasmid to create the pMA5-acr-HapII-dhaD-PbdhA-alsR plasmid. These constructed plasmids were isolated from E. coli JM109 and subsequently transformed into B. amyloliquefaciens according to published method [37]. Furthermore, the stability of plasmids in B. amyloliquefaciens was also tested as described in our previous study [15].
Enzyme assays
The cell pellets collected by centrifugation were suspended and washed with 0.1 M potassium phosphate buffer (pH7.0) at least for three times. For determining DhaD or GldA activity [14], the cell pellets were resuspended in binding buffer (20 mM potassium phosphate, 500 mM NaCl, 20 mM imidazole, pH 7.4). For determining acetoin reductase (ACR) activity [38], the cell pellets were suspended in 0.1 M potassium phosphate buffer (pH 6.5) containing 0.1 mM β-mercaptoethanol and 2 μg/mL phenylmethylsulfonyl fluoride. For acetolactate synthase (ALS) and acetolactate decarboxylase (ALDC) enzymes, the cell pellets were washed three times with wash buffer (0.2 mM NaH2PO4, 2.2 mM Na2HPO4, and 8.5 mM NaCl; pH 7.4) and then suspended in this buffer. Cells were disrupted using a sonicator (SONICS, Newtown, CT) for 20 min with chilling. The lysed cells were centrifuged at 12,000 rpm for 25 min at 4 °C, and the supernatant was used for enzyme assays. The ALS and ALDC assays were performed according to published procedures [39].
Analytical methods
The cell mass concentration was determined from the OD600 in a UV–visible spectroscopy system (UV-2000, UNICO, America). The dry cell weight (DCW) was calculated from the optical density using a calibration curve for the strain. The composition of the fermentation broth (glycerol, 2,3-BD, acetoin, acetate, lactate and succinate) was determined by high-performance liquid chromatography (HPLC) [31]. The intracellular NAD+ and NADH concentrations were measured by procedures described previously [40]. All assays were performed by triplicate cultures.
Kinetic parameters calculation
The specific cell growth rate (μX, h−1), specific substrate consumption rate (μS, h−1) and specific 2,3-BD formation rate (μP, h−1) were estimated from experimental or fitted data of cell growth (X, g/L), residual substrate concentration (S, g/l), and 2,3-BD production (P, g/L) by Eqs. (1)–(3), respectively. The fitted data were obtained by interposing between experimental data of cell growth, residual substrate concentration or 2,3-BD production at definite time (dt = 0.1 h) with the approximation method of cubic spline interpolation in Origin software (Version 8.0, OriginLab Corp., Northampton, MA, USA) [20].
| 1 |
| 2 |
| 3 |
Authors’ contributions
TY conceived of the study, executed the experimental work and data analysis, and coordinated the manuscript draft and revision. ZR, ZX helped to revise and proofread the manuscript. XZ, MX helped with data analysis. SY coordinated the manuscript draft and revision. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the High-tech Research and Development Programs of China (2015AA021004), the National Basic Research Program of China (973 Program) (2012CB725202), the National Natural Science Foundation of China (31400082, 21276110), the Research Project of Chinese Ministry of Education (113033A), Fundamental Research Funds for the Central Universities (JUSRP11544), the National Training Programs of Innovation and Entrepreneurship for Undergraduates (201510295071), the Priority Academic Program Development of Jiangsu Higher Education Institutions,the 111 Project (111-2-06),and the Jiangsu province “Collaborative Innovation Center for Advanced Industrial Fermentation” industry development program.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests
Contributor Information
Taowei Yang, Email: ytw1228@163.com.
Zhiming Rao, Phone: +86-0510-85910886, Email: raozhm@jiangnan.edu.cn.
Xian Zhang, Email: zxshengwu@126.com.
Meijuan Xu, Email: xumeijuan@jiangnan.edu.cn.
Zhenghong Xu, Email: zhenghxu@163.com.
Shang-Tian Yang, Email: yang.15@osu.edu.
References
- 1.Syu MJ. Biological production of 2,3-butanediol. Appl Microbiol Biotechnol. 2001;55(1):10–18. doi: 10.1007/s002530000486. [DOI] [PubMed] [Google Scholar]
- 2.Ji XJ, Huang H, Ouyang PK. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol Adv. 2011;29(3):351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Celinska E, Grajek W. Biotechnological production of 2,3-butanediol–current state and prospects. Biotechnol Adv. 2009;27(6):715–725. doi: 10.1016/j.biotechadv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Ma C, Wang A, Qin J, Li L, Ai X, Jiang T, et al. Enhanced 2,3-butanediol production by Klebsiella pneumoniae SDM. Appl Microbiol Biotechnol. 2009;82(1):49–57. doi: 10.1007/s00253-008-1732-7. [DOI] [PubMed] [Google Scholar]
- 5.Papanikolaou S, Fakas S, Fick M, Chevalot I, Galiotou-Panayotou M, Komaitis M, et al. Biotechnological valorisation of raw glycerol discharged after bio-diesel (fatty acid methyl esters) manufacturing process: production of 1,3-propanediol, citric acid and single cell oil. Biomass Bioenerg. 2008;32(1):60–71. doi: 10.1016/j.biombioe.2007.06.007. [DOI] [Google Scholar]
- 6.Da Silva GP, Mack M, Contiero J. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv. 2009;27(1):30–39. doi: 10.1016/j.biotechadv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Willke T, Vorlop KD. Industrial bioconversion of renewable resources as an alternative to conventional chemistry. Appl Microbiol Biotechnol. 2004;66(2):131–142. doi: 10.1007/s00253-004-1733-0. [DOI] [PubMed] [Google Scholar]
- 8.Biebl H, Zeng AP, Menzel K, Deckwer WD. Fermentation of glycerol to 1,3-propanediol and 2,3-butanediol by Klebsiella pneumoniae. Appl Microbiol Biotechnol. 1998;50(1):24–29. doi: 10.1007/s002530051251. [DOI] [PubMed] [Google Scholar]
- 9.Petrov K, Petrova P. High production of 2,3-butanediol from glycerol by Klebsiella pneumoniae G31. Appl Microbiol Biotechnol. 2009;84(4):659–665. doi: 10.1007/s00253-009-2004-x. [DOI] [PubMed] [Google Scholar]
- 10.Metsoviti M, Paramithiotis S, Drosinos EH, Galiotou-Panayotou M, Nychas G-JE, Zeng A-P, et al. Screening of bacterial strains capable of converting biodiesel-derived raw glycerol into 1,3-propanediol, 2,3-butanediol and ethanol. Eng Life Sci. 2012;12(1):57–68. doi: 10.1002/elsc.201100058. [DOI] [Google Scholar]
- 11.Yang T-W, Rao Z-M, Zhang X, Xu M-J, Xu Z-H, Yang S-T. Fermentation of biodiesel-derived glycerol by Bacillus amyloliquefaciens: effects of co-substrates on 2,3-butanediol production. Appl Microbiol Biotechnol. 2013;97(17):7651–7658. doi: 10.1007/s00253-013-5048-x. [DOI] [PubMed] [Google Scholar]
- 12.Garg SK, Jain A. Fermentative production of 2,3-butanediol: a review. Bioresour Technol. 1995;51:103–109. doi: 10.1016/0960-8524(94)00136-O. [DOI] [Google Scholar]
- 13.Xiao Z, Xu P. Acetoin metabolism in bacteria. Crit Rev Microbiol. 2007;33(2):127–140. doi: 10.1080/10408410701364604. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Tao F, Xu P. Glycerol dehydrogenase plays a dual role in glycerol metabolism and 2,3-butanediol formation in Klebsiella pneumoniae. J Biol Chem. 2014;289(9):6080–6090. doi: 10.1074/jbc.M113.525535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang T-W, Rao Z-M, Zhang X, Xu M-J, Xu Z-H, Yang S-T. Improved production of 2,3-butanediol in Bacillus amyloliquefaciens by over-expression of glyceraldehyde-3-phosphate dehydrogenase and 2,3-butanediol dehydrogenase. PLoS ONE. 2013;8(10):e76149. doi: 10.1371/journal.pone.0076149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renna MC, Najimudin N, Winik LR, Zahler SA. Regulation of the Bacillus subtilisalsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175(12):3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fradrich C, March A, Fiege K, Hartmann A, Jahn D, Hartig E. The transcription factor AlsR binds and regulates the promoter of the alsSD operon responsible for acetoin formation in Bacillus subtilis. J Bacteriol. 2012;194(5):1100–1112. doi: 10.1128/JB.06425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Zhang R, Bao T, Yang T, Xu M, Li H, et al. Moderate expression of the transcriptional regulator ALsR enhances acetoin production by Bacillus subtilis. J Ind Microbiol Biotechnol. 2013;40(9):1067–1076. doi: 10.1007/s10295-013-1303-5. [DOI] [PubMed] [Google Scholar]
- 19.Jansen NB, Flickinger MC, Tsao GT. Application of bioenergetics to modelling the microbial conversion of D-xylose to 2,3-butanediol. Biotechnol Bioeng. 1984;26(6):573–582. doi: 10.1002/bit.260260603. [DOI] [PubMed] [Google Scholar]
- 20.Ji XJ, Huang H, Du J, Zhu JG, Ren LJ, Hu N, et al. Enhanced 2,3-butanediol production by Klebsiella oxytoca using a two-stage agitation speed control strategy. Bioresour Technol. 2009;100(13):3410–3414. doi: 10.1016/j.biortech.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 21.Converti A, Perego P, Del Borghi M. Effect of specific oxygen uptake rate on Enterobacter aerogenes energetics: carbon and reduction degree balances in batch cultivations. Biotechnol Bioeng. 2003;82:370–377. doi: 10.1002/bit.10570. [DOI] [PubMed] [Google Scholar]
- 22.Beronio PB, Tsao GT. Optimization of 2,3-butanediol production by Klebsiella oxytoca through oxygen transfer rate control. Biotechnol Bioeng. 1993;42(11):1263–1269. doi: 10.1002/bit.260421102. [DOI] [PubMed] [Google Scholar]
- 23.Fages J, Mulard D, Rouquel JJ, Wilhelm JL. 2,3-Butanediol production from Jerusalem artichoke, Heliunthus tuberosus, by Bucillus polymyxa ATCC 12321. Optimization of KLa profile. Appl Microbiol Biotechnol. 1986;25:197–202. doi: 10.1007/BF00253648. [DOI] [Google Scholar]
- 24.Zeng A, Biebl H, Deckwer W. 2,3-Butanediol production by Enterobacter aerogenesin continuous culture: role of oxygen supply. Appl Microbiol Biotechnol. 1990;33:264–268. doi: 10.1007/BF00164519. [DOI] [Google Scholar]
- 25.Zeng AP, Byun TG, Posten C, Deckwer WD. Use of respiratory quotient as a control parameter for optimum oxygen supply and scale-up of 2,3-butanediol production under microaerobic conditions. Biotechnol Bioeng. 1994;44(9):1107–1114. doi: 10.1002/bit.260440912. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Yang Y, Sun J, Shen Y, Wei D, Zhu J, et al. Microbial production of 2,3-butanediol by a mutagenized strain of Serratia marcescens H30. Bioresour Technol. 2010;101(6):1961–1967. doi: 10.1016/j.biortech.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 27.Nakashimada Y, Marwoto B, Kashiwamura T, Kakizono T, Nishio N. Enhanced 2,3-butanediol production by addition of acetic acid in Paenibacillus polymyxa. J Biosci Bioeng. 2000;90(6):661–664. doi: 10.1016/S1389-1723(00)90013-6. [DOI] [PubMed] [Google Scholar]
- 28.Voloch M, Jansen NB, Ladish MR, Tsao GT, Narayan R, Rodwell VW. 2,3-Butanediol. In: Blanch HW, Drew S, Wang DC, editors. Comprehensive biotechnology; the principles, applications and regulations of biotechnology in industry, agriculture and medicine. Oxford: Pergamon/Elsevier; 1985. pp. 933–944. [Google Scholar]
- 29.Perego P, Converti A, Borghi AD, Canepa P. 2,3-Butanediol production by Enterobacter aerogenes: selection of the optimal conditions and application to food industry residues. Bioprocess Eng. 2000;23:613–620. doi: 10.1007/s004490000210. [DOI] [Google Scholar]
- 30.Stormer FC. Evidence for induction of the 2,3-butanediol-forming enzymes in Aerobacter aerogenes. FEBS Lett. 1968;2(1):36–38. doi: 10.1016/0014-5793(68)80094-8. [DOI] [PubMed] [Google Scholar]
- 31.Yang T, Rao Z, Zhang X, Lin Q, Xia H, Xu Z, et al. Production of 2,3-butanediol from glucose by GRAS microorganism Bacillus amyloliquefaciens. J Basic Microbiol. 2011;51(6):650–658. doi: 10.1002/jobm.201100033. [DOI] [PubMed] [Google Scholar]
- 32.de Boer AS, Diderichsen B. On the safety of Bacillus subtilis and B. amyloliquefaciens: a review. Appl Microbiol Biotechnol. 1991;36(1):1–4. doi: 10.1007/BF00164689. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Yang T, Lin Q, Xu M, Xia H, Xu Z, et al. Isolation and identification of an acetoin high production bacterium that can reverse transform 2,3-butanediol to acetoin at the decline phase of fermentation. World J Microbiol Biotechnol. 2011;27(12):2785–2790. doi: 10.1007/s11274-011-0754-y. [DOI] [Google Scholar]
- 34.Zhang L, Xu Q, Zhan S, Li Y, Lin H, Sun S, et al. A new NAD(H)-dependent meso-2,3-butanediol dehydrogenase from an industrially potential strain Serratia marcescens H30. Appl Microbiol Biotechnol. 2013 doi: 10.1007/s00253-013-4959-x. [DOI] [PubMed] [Google Scholar]
- 35.Gao J, Yang HH, Feng XH, Li S, Xu H. A 2,3-butanediol dehydrogenase from Paenibacillus polymyxa ZJ-9 for mainly producing R, R -2,3-butanediol: purification, characterization and cloning. J Basic Microbiol. 2012 doi: 10.1002/jobm.201200152. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez E, Fernandez MR, Larroy C, Pares X, Biosca JA. Characterization and functional role of Saccharomyces cerevisiae 2,3-butanediol dehydrogenase. Chem Biol Interact. 2001;130–132(1–3):425–434. doi: 10.1016/S0009-2797(00)00282-9. [DOI] [PubMed] [Google Scholar]
- 37.Vojcic L, Despotovic D, Martinez R, Maurer K-H, Schwaneberg U. An efficient transformation method for Bacillus subtilis DB104. Appl Microbiol Biotechnol. 2012;94(2):487–493. doi: 10.1007/s00253-012-3987-2. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson WL. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl Environ Microbiol. 2008;74(22):6832–6838. doi: 10.1128/AEM.00881-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atsumi S, Li Z, Liao JC. Acetolactate synthase from Bacillus subtilis serves as a 2-ketoisovalerate decarboxylase for isobutanol biosynthesis in Escherichia coli. Appl Environ Microbiol. 2009;75(19):6306–6311. doi: 10.1128/AEM.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snoep J, Joost M, de Mattos T, Neijssel O. Effect of the energy source on the NADH/NAD ratio and on pyruvate catabolism in anaerobic chemostat cultures of Enterococcus faecalis NCTC 775. FEMS Microbiol Lett. 1991;81:63–66. doi: 10.1111/j.1574-6968.1991.tb04713.x. [DOI] [Google Scholar]


