Abstract
Background
Although numerous efforts have been made, the pathogenesis underlying lung squamous-cell carcinoma (SCC) remains unclear. This study aimed to identify the CNV-driven genes by an integrated analysis of both the gene differential expression and copy number variation (CNV).
Results
A higher burden of the CNVs was found in 10–50 kb length. The 16 CNV-driven genes mainly located in chr 1 and chr 3 were enriched in immune response [e.g. complement factor H (CFH) and Fc fragment of IgG, low affinity IIIa, receptor (FCGR3A)], starch and sucrose metabolism [e.g. amylase alpha 2A (AMY2A)]. Furthermore, 38 TFs were screened for the 9 CNV-driven genes and then the regulatory network was constructed, in which the GATA-binding factor 1, 2, and 3 (GATA1, GATA2, GATA3) jointly regulated the expression of TP63.
Conclusions
The above CNV-driven genes might be potential contributors to the development of lung SCC.
Keywords: Lung squamous-cell carcinoma, Differentially expressed genes, Copy number variation, Copy number variation-driven genes
Background
Lung cancer is a leading cause of cancer mortalities worldwide [1] and the main type of lung cancer is non-small cell lung cancer (NSCLC), accounting for about 80 % of lung cancers [2]. NSCLC can be further divided into three histologic subtypes: squamous-cell carcinoma (SCC), adenocarcinoma (AC), and large-cell lung carcinoma (LCC) [3]. Of them, lung SCC represents about one third of the NSCLC burden [4]. Although numerous efforts have been made to elucidate the underlying pathogenesis and therapy of this disease over the past few decades, lung SCC is still incurable due to lack of effective therapeutic methods and late diagnosis [5, 6]. Therefore, further research in molecular pathology is still needed.
Copy number variants (CNVs) are DNA segments of ≥1 kb in length, which is present in the genome in a variable frequency [7]. Once, it was recognized that their frequency was low and associated with specific chromosome disorders directly. During the 1990s, copy number duplications and deletions were found to cause a quantity of single gene disorders [8]. Changes in DNA copy number, whether confined to specific genes or affecting whole chromosomes, have been identified as major causes of some developmental abnormalities and diseases [9]. Recently, studies related to CNVs and their roles in tumorigenesis have increased markedly [10]. For example, alteration in copy number of chromosomal regions such as 3q26.2-q29, 3p26.3-p11.1, 17p13.3-p11.2 and 9p13.3-p13.2 has been deemed as predictors of lung cancer [11]. Using molecular pathway analysis, copy number alterations in 11 genes are associated with the focal adhesion pathway in SCLC [12]. All these evidence has proven a vital role of CNV in lung cancer.
Expression profile analyses also have been performed to identify a mass of differentially expressed genes (DEGs) between normal and lung tumor samples. Wang et al. have identified 17 genes preferentially expressed in lung SCC, including four novel genes [13]. Takefumie et al. identified 40 DEGs that could separate cases with lymph-node metastasis from those without metastasis in NSCLC [14]. Furthermore, four mRNA expression subtypes including classical, basal, secretory and primitive are identified in lung LCC by gene expression profile analysis [15]. However, only few DEGs are commonly detected in different studies, which may be due to the differences in statistical methods or specimen characteristics. One possible way to increase homogeneity in these findings is an integrated analysis using both DEGs and CNVs [16, 17].
The integrated analysis of CNV and gene differential expression has previously been performed for lung AC [17], but not lung SCC. Thus, the goal of this study was to identify the CNV-driven genes in lung SCC by combining the transcriptional profile data contributed by Anders and Huber (GSE17710) [15] and CNVs data deposited in the Cancer Genome Atlas (TCGA). The identified CNV-driven genes may have a key role in elucidating the underlying mechanisms of lung SCC, thus providing a basis for developing new therapies against this disease.
Result
DEGs screening
A total of 428 DEGs were screened out with the cut-off value of |log2Ratio| > 1. Among them, 211 ones were up-regulated in lung SCC, while 217 ones were down-regulated. To explore their functions, they were subjected to GO— (p value <0.05) and KEGG (p value <0.05) pathway enrichment analyses. Noticeably, genes including matrix metallopeptidase 7 (MMP7), transferrin (TF) and serpin peptidase inhibitor clade A (SERPINA1) were enriched in several GO functional terms and pathways (Table 1).
Table 1.
GO and KEGG pathway enrichment anal ysis of DEGs in lung SCC
| Gene expression | Category | Term | Count | P value |
|---|---|---|---|---|
| Up-regulated | GOTERM_BP_FAT | GO:0006955, immune response | 58 | 3.95E−33 |
| GOTERM_BP_FAT | GO:0006952, defense response | 42 | 5.74E−20 | |
| GOTERM_BP_FAT | GO:0009611, response to wounding | 36 | 6.34E−17 | |
| GOTERM_BP_FAT | GO:0006954, inflammatory response | 28 | 1.41E−15 | |
| GOTERM_BP_FAT | GO:0019882, antigen processing and presentation | 13 | 2.68E−10 | |
| GOTERM_CC_FAT | GO:0005576, extracellular region | 70 | 8.37E−15 | |
| GOTERM_CC_FAT | GO:0044421, extracellular region part | 44 | 1.06E−12 | |
| GOTERM_CC_FAT | GO:0005615, extracellular space | 34 | 1.17E−10 | |
| GOTERM_CC_FAT | GO:0042611, MHC protein complex | 11 | 3.75E−09 | |
| GOTERM_CC_FAT | GO:0042613, MHC class II protein complex | 7 | 2.00E−06 | |
| GOTERM_MF_FAT | GO:0008009, chemokine activity | 10 | 2.21E−09 | |
| GOTERM_MF_FAT | GO:0042379, chemokine receptor binding | 10 | 3.99E−09 | |
| GOTERM_MF_FAT | GO:0005125, cytokine activity | 16 | 7.17E−09 | |
| GOTERM_MF_FAT | GO:0032395, MHC class II receptor activity | 6 | 1.97E−06 | |
| GOTERM_MF_FAT | GO:0019865, immunoglobulin binding | 4 | 6.21E−04 | |
| KEGG_PATHWAY | hsa05330: allograft rejection | 12 | 1.53E−11 | |
| KEGG_PATHWAY | hsa05332: graft-versus-host disease | 12 | 4.09E−11 | |
| KEGG_PATHWAY | hsa04940: type I diabetes mellitus | 12 | 1.00E−10 | |
| KEGG_PATHWAY | hsa05320: autoimmune thyroid disease | 12 | 9.74E−10 | |
| KEGG_PATHWAY | hsa04514: cell adhesion molecules (CAMs) | 17 | 1.01E−09 | |
| Down-regulated | GOTERM_BP_FAT | GO:0055088, lipid homeostasis | 10 | 1.22E−08 |
| GOTERM_BP_FAT | GO:0042632, cholesterol homeostasis | 9 | 1.87E−08 | |
| GOTERM_BP_FAT | GO:0055092, sterol homeostasis | 9 | 1.87E−08 | |
| GOTERM_BP_FAT | GO:0065005, protein-lipid complex assembly | 6 | 2.18E−07 | |
| GOTERM_BP_FAT | GO:0034377, plasma lipoprotein particle assembly | 6 | 2.18E−07 | |
| GOTERM_CC_FAT | GO:0005615, extracellular space | 35 | 4.43E−12 | |
| GOTERM_CC_FAT | GO:0044421, extracellular region part | 39 | 1.70E−10 | |
| GOTERM_CC_FAT | GO:0005576, extracellular region | 52 | 3.66E−07 | |
| GOTERM_CC_FAT | GO:0034385, triglyceride-rich lipoprotein particle | 6 | 4.09E−06 | |
| GOTERM_CC_FAT | GO:0034361, very-low-density lipoprotein particle | 6 | 4.09E−06 | |
| GOTERM_MF_FAT | GO:0008289, lipid binding | 19 | 6.35E−06 | |
| GOTERM_MF_FAT | GO:0004866, endopeptidase inhibitor activity | 11 | 8.62E−06 | |
| GOTERM_MF_FAT | GO:0005496, steroid binding | 8 | 1.02E−05 | |
| GOTERM_MF_FAT | GO:0004867, serine-type endopeptidase inhibitor activity | 9 | 1.31E−05 | |
| GOTERM_MF_FAT | GO:0030414, peptidase inhibitor activity | 11 | 1.38E−05 | |
| KEGG_PATHWAY | hsa04610: complement and coagulation cascades | 7 | 3.32E−04 | |
| KEGG_PATHWAY | hsa00830: retinol metabolism | 6 | 8.02E−04 | |
| KEGG_PATHWAY | hsa00860: porphyrin and chlorophyll metabolism | 5 | 9.97E−04 | |
| KEGG_PATHWAY | hsa00053: ascorbate and aldarate metabolism | 4 | 0.0015 | |
| KEGG_PATHWAY | hsa00040: pentose and glucuronate interconversions | 4 | 0.0018 |
GO gene ontology, KEGG kyoto encyclopedia of genes and genomes, DEGs differentially expressed genes, SCC squamous-cell carcinoma, BP biological process, CC cellular component, MF molecular function
Identification of CNV-driven DEGs
It is found that regions of 1–10 kb long had the most copy number deletions (Table 2), while regions of >50 kb long had the most copy number duplications (Table 3). Overall, the ratio of copy number deletions in tumors to those in controls was larger in regions of 10–50 kb long, mostly larger than 6, and the ratio of copy number duplications in tumors to those in controls was lowest in regions of >50 kb long (Fig. 1).
Table 2.
Distribution of copy number deleti ons on chromosomes
| Chr | Deletions only | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Observed CNV in cases and controls | Ratio of case/control | p values | Observed CNV in cases and controls | Ratio of case/control | p values | Observed CNV in cases and controls | Ratio of case/control | p values | |
| 1–10 kb | 10–50 kb | >50 kb | |||||||
| 1 | 1709 | 2.49539 | 0.8626 | 384 | 6.08127 | 0.3246 | 170 | 3.72008 | 0.2772 |
| 2 | 1642 | 3.13118 | 1.00E−04 | 509 | 6.81826 | 0.0332 | 237 | 1.80857 | 0.0413 |
| 3 | 1611 | 3.01258 | 0.7392 | 500 | 7.48133 | 0.9189 | 266 | 2.15987 | 0.8298 |
| 4 | 1904 | 2.77845 | 0.8719 | 520 | 7.42289 | 0.9481 | 251 | 1.86097 | 0.3325 |
| 5 | 1483 | 2.52167 | 0.0617 | 543 | 5.93514 | 0.0453 | 249 | 2.01002 | 0.6479 |
| 6 | 1440 | 2.82868 | 0.2306 | 362 | 6.5556 | 0.0581 | 118 | 3.96903 | 0.5206 |
| 7 | 1248 | 3.57254 | 0.7114 | 368 | 8.09861 | 0.6195 | 113 | 2.40595 | 0.8872 |
| 8 | 1388 | 2.79406 | 0.0007 | 371 | 7.95181 | 0.882 | 306 | 2.41128 | 0.4959 |
| 9 | 1195 | 2.66351 | 1.00E−04 | 391 | 7.41424 | 0.0011 | 495 | 1.69698 | 0.7884 |
| 10 | 1107 | 2.5371 | 0.2834 | 269 | 4.27999 | 0.5673 | 208 | 2.3065 | 0.0204 |
| 11 | 1183 | 2.28072 | 0.0046 | 282 | 6.60225 | 0.0169 | 138 | 2.7495 | 0.0148 |
| 12 | 893 | 2.49211 | 0.0323 | 152 | 6.80094 | 0.0364 | 35 | 2.42966 | 0.0054 |
| 13 | 932 | 2.23366 | 0.3331 | 272 | 4.2262 | 0.0271 | 94 | 1.51505 | 0.5151 |
| 14 | 741 | 2.25316 | 0.0106 | 221 | 8.66705 | 0.2036 | 118 | 2.3046 | 0.6436 |
| 15 | 623 | 2.4265 | 0.06 | 157 | 6.87918 | 0.0042 | 139 | 6.42084 | 0.0004 |
| 16 | 647 | 2.54643 | 0.5412 | 216 | 6.04562 | 0.0075 | 122 | 3.28046 | 0.1362 |
| 17 | 594 | 2.33232 | 0.2435 | 121 | 3.90574 | 0.3034 | 72 | 3.95823 | 0.0278 |
| 18 | 698 | 2.30858 | 0.9919 | 135 | 10.792 | 0.7888 | 74 | 1.79206 | 0.2417 |
| 19 | 405 | 2.39362 | 0.9983 | 121 | 3.42131 | 0.9996 | 44 | 2.54418 | 0.7119 |
| 20 | 427 | 1.88614 | 0.0405 | 78 | 5.83578 | 0.4637 | 24 | 3.52891 | 0.1368 |
| 21 | 345 | 1.94557 | 0.4154 | 85 | 4.8969 | 0.0999 | 82 | 1.77643 | 0.025 |
| 22 | 319 | 2.5579 | 0.0145 | 81 | 6.01787 | 0.0148 | 44 | 4.84881 | 0.0547 |
Table 3.
Distribution of copy number duplications in chromosomes
| Chr | Duplications only | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Observed CNV in cases and controls | RATIO of Case/Control | p values | Observed CNV in cases and controls | Ratio of Case/Control | p values | Observed CNV in cases and controls | Ratio of Case/Control | p values | |
| 1 ~ 10 kb | 10 ~ 50 kb | >50 kb | |||||||
| 1 | 1924 | 4.56541 | 1.00E−04 | 2621 | 4.97426 | 1.00E−04 | 11,911 | 1.98385 | 0.0022 |
| 2 | 1912 | 3.84581 | 0.0086 | 2627 | 3.91763 | 1.00E−04 | 13,136 | 1.71774 | 1.00E−04 |
| 3 | 2070 | 3.17944 | 0.0012 | 2669 | 3.17624 | 1.00E−04 | 13,120 | 1.56874 | 1.00E−04 |
| 4 | 1724 | 4.56274 | 0.8782 | 2410 | 4.06176 | 0.2453 | 11,164 | 1.80262 | 0.999 |
| 5 | 1272 | 3.06097 | 1 | 1866 | 4.18235 | 0.9991 | 9063 | 1.87245 | 0.9003 |
| 6 | 1560 | 3.12497 | 0.0007 | 2049 | 3.86111 | 1.00E−04 | 9809 | 1.71417 | 0.0002 |
| 7 | 1782 | 3.99707 | 0.9365 | 2523 | 4.25108 | 0.1637 | 9972 | 1.86011 | 0.9153 |
| 8 | 1667 | 4.06267 | 1.00E−04 | 2150 | 3.30777 | 1.00E−04 | 10,659 | 1.61986 | 1.00E−04 |
| 9 | 1178 | 4.59415 | 0.0016 | 1552 | 4.38777 | 0.1466 | 7928 | 1.83387 | 0.0005 |
| 10 | 986 | 5.63493 | 0.0195 | 1243 | 4.28617 | 0.0254 | 7138 | 1.89335 | 1.00E−04 |
| 11 | 1167 | 3.06759 | 1.00E−04 | 1598 | 3.44469 | 1.00E−04 | 8478 | 1.52969 | 1.00E−04 |
| 12 | 1254 | 3.18495 | 0.3166 | 1432 | 3.62956 | 0.0677 | 7431 | 1.70661 | 0.0044 |
| 13 | 700 | 3.91295 | 0.1717 | 1011 | 3.52657 | 0.0039 | 5535 | 1.60301 | 0.0112 |
| 14 | 655 | 3.43045 | 0.342 | 1070 | 4.14655 | 0.5122 | 5294 | 1.66644 | 0.0106 |
| 15 | 574 | 4.72049 | 0.3773 | 930 | 4.28337 | 1.00E−04 | 4371 | 2.0948 | 1.00E−04 |
| 16 | 726 | 5.96031 | 0.0747 | 1094 | 6.71092 | 0.1114 | 5387 | 2.4445 | 1.00E−04 |
| 17 | 597 | 5.26255 | 0.0105 | 995 | 4.69484 | 1.00E−04 | 5477 | 1.89667 | 0.0003 |
| 18 | 719 | 3.61849 | 1 | 849 | 4.09121 | 1 | 4894 | 1.54215 | 1 |
| 19 | 520 | 4.72055 | 0.8769 | 943 | 4.15877 | 0.9467 | 5166 | 1.67776 | 1 |
| 20 | 594 | 4.01529 | 0.0188 | 728 | 4.42725 | 0.0907 | 3890 | 1.58039 | 0.0172 |
| 21 | 340 | 5.28483 | 0.9071 | 558 | 3.61798 | 0.8819 | 2724 | 1.8263 | 0.2397 |
| 22 | 526 | 6.95908 | 0.0003 | 689 | 9.38731 | 0.0032 | 2976 | 2.84862 | 1.00E−04 |
Fig. 1.
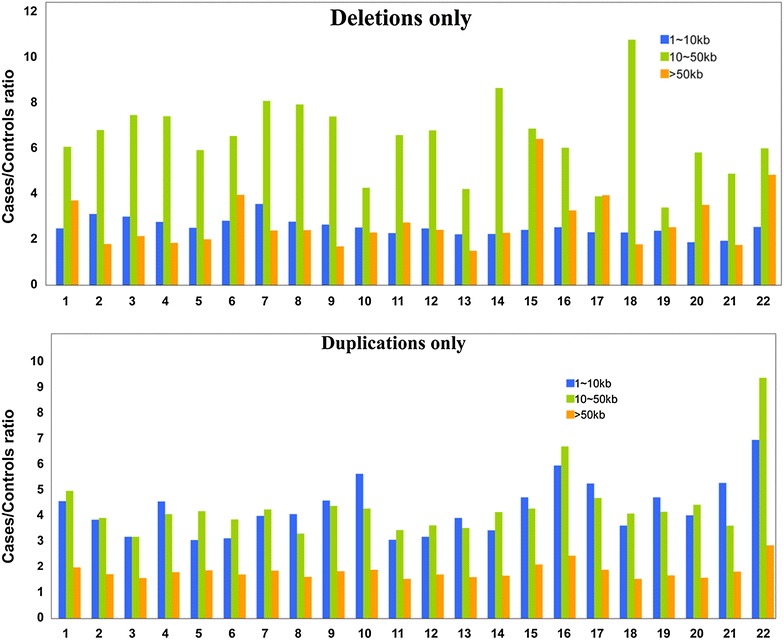
The ratios of the number of copy number deletions/duplications in cases to that in controls at different lengths
A total of 313 CNV genes related to lung SCC were detected, which were only found in more than 80 % cases, while not in the controls. Then these CNVs were checked for overlap with the DEGs. A total of 163 overlapping CNV genes that were also detected by microarrays were obtained, of which 24 genes displayed significantly different expression. Furthermore, 16 CNV-driven genes were identified with the same expressional tendencies, namely copy number increasing with the expression level (Table 4). Among them, seven (FCGR3A, FCGR2B, AMY2A, AMY2B, CFH, LCE1D, and CFHR3) were located on chr 1, three on chr 3 (TP63, MUC4, and MUC20), and one on chr 4, 5, 6, 7, 16 and 19, respectively (Fig. 2).
Table 4.
CNV-driven genes in lung SCC
| Chr | Gene | log2(copy number) | log2(FC) | Karyotype |
|---|---|---|---|---|
| 1 | FCGR3A | 1.13334 | 3.91384 | q23.3 |
| 1 | FCGR2B | 1.14834 | 2.52075 | q23.3 |
| 1 | AMY2A | 1.06865 | 2.50174 | p21.1 |
| 1 | AMY2B | 1.58959 | 1.94116 | p21.1 |
| 1 | CFH | 1.06624 | 1.47934 | q31.3 |
| 1 | LCE1D | 1.23411 | 1.41679 | q21.3 |
| 1 | CFHR3 | 1.20817 | 1.35526 | q31.3 |
| 3 | TP63 | 1.04434 | 3.18336 | q28 |
| 3 | MUC4 | 1.35013 | 2.38177 | q29 |
| 3 | MUC20 | 1.63525 | 2.25619 | q29 |
| 4 | TMPRSS11E | 1.11569 | 1.78099 | p16.3 |
| 5 | ZDHHC11 | 1.07998 | 1.03782 | q14.1 |
| 6 | HLA-DQA1 | 1.12618 | 1.73272 | p21.32 |
| 7 | ARHGEF5 | 1.07093 | 1.46641 | q35 |
| 16 | CES1 | 1.68967 | 2.06824 | q12.2 |
| 19 | LILRB5 | 1.08841 | 1.39769 | q13.42 |
CNV copy number variation, FC fold change
Fig. 2.
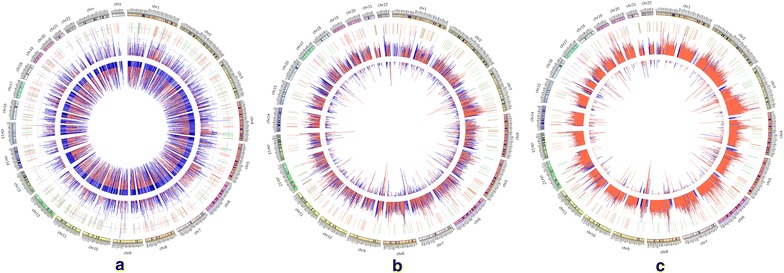
Genomic distributions of differentially expressed genes (DEGs) and copy number variations (CNV) related to lung squamous-cell carcinoma using Circos-plots. a, b and c Represent the genomic distribution of CNV regions of 1–10 kb, 10–50 kb and >50 kb, respectively. The outermost bars in a circle labeled with numbers represent chromosomes; the second outermost circle represents DEGs (red and green indicating up-regulated and down-regulated DEGs, respectively); the first innermost circle (inward) represent copy number deletions, and the second innermost circle (outward) representing copy number duplications (a red line indicates a CNV occurring in controls, a blue line indicates a CNV occurring in cases, with the length of the line determined by the copy number)
GO and pathway analysis of CNV-driven genes
To functionally understand the CNV-driven genes, GO (p value <0.05) and KEGG pathway (p value <0.05) enrichment analyses were also performed. According to GO annotation, the CNV-driven genes were mainly functionally related to the biological process term immune response (such as CFH, FCGR3A), and cellular component term amylase activity (AMY2A and AMY2B), as well as molecular function terms amylase activity and IgG binding. Meanwhile, the CNV-driven genes were significantly enriched in pathways of systemic lupus erythematosus, starch and sucrose metabolism pathway (amylase alpha 2A, AMY2A) (Table 5).
Table 5.
GO and KEGG pathway enrichment analysis of CNV-driven genes
| Category | Term | P value | Genes |
|---|---|---|---|
| GOTERM_BP_FAT | GO:0006955, immune response | 0.002390231 | FCGR2B, LILRB5, CFH, FCGR3A, HLA-DQA1 |
| GOTERM_CC_FAT | GO:0005576, extracellular region | 0.001678775 | CFHR3, TMPRSS11E, MUC20 |
| CFH, AMY2B, FCGR3A, AMY2A, MUC4 | |||
| GOTERM_MF_FAT | GO:0016160, amylase activity | 0.002309112 | AMY2B, AMY2A |
| GOTERM_MF_FAT | GO:0004556, alpha-amylase activity | 0.002309112 | AMY2B, AMY2A |
| GOTERM_MF_FAT | GO:0019864, IgG binding | 0.006146971 | FCGR2B, FCGR3A |
| KEGG_PATHWAY | hsa05322: systemic lupus erythematosus | 0.00534888 | FCGR2B, FCGR3A, HLA-DQA1 |
| KEGG_PATHWAY | hsa00500: starch and sucrose metabolism | 0.048568802 | AMY2B, AMY2A |
Discussion
In the present study, using both transcriptional profile data and CNV data, we identified genes with differential expression that may be caused by CNV. DEGs such as MMP7, TF and SERPINA1 might be associated with lung SCC. The up-regulated MMP7 was enriched in several significant GO functional terms such as negative regulation of cellular protein metabolic process, male gonad development and sterol homeostasis in our study. MMP7 belongs to metalloproteinase (MMP) family and plays a role in the breakdown of extracellular matrix [18]. It is shown that the polymorphism in MMP7 promoter increases susceptibility to esophageal SCC [19]. Moreover, down-regulated TF enriched in significant GO functional terms including extracellular space and extracellular region part. It functioned as an iron transporter [20]. The receptor of TF contributes to NSCLCs and it may be an indicator of poorer prognosis in certain groups of patients [21]. Up-regulated SERPINA1 was enriched in several significant GO function terms such as extracellular region and extracellular region part as well. It is a serine protease inhibitor and contributes to chronic obstructive pulmonary disease [22]. There is evidence that SERPINA1 is a biomarker for progression of cutaneous SCC [23]. All these evidences suggest that MMP7, TF and SERPINA1 may play pivotal roles in lung SCC.
CNV analysis presents a higher burden of the CNVs in length of 10–50 kb in lung SCC. A significant increase in CNV burden was observed in most of the individual chromosomes. It is reported that the increased burden of structural variation is a genetic risk factor for cancer [24]. Hence, no wonder the structural variation in length is correlated with lung SCC. Our study provides a hint for the etiology of lung SCC, which is the fragile or disordered genomes, specifically due to the structural variations of copy number in length of 10–50 kb.
Among the 16 overlapping genes, 7 were located on chr 1, and 3 on chr 3, indicating CNV related to lung SCC may mainly occur on chromsomes 1 and 3. According to GO functional analysis and KEGG pathway annotation, these 16 genes may be mainly involved in lung SCC by influencing starch and sucrose metabolism (e.g. AMY2A, AMY2B), and the immune response (e.g. FCGR3A, FCGR2B, CFH, HLA-DQA1). AMY2A and AMY2B encoding amylases that hydrolyze 1,4-α-glucoside bonds in oligosaccharides and polysaccharides, thus is necessary for the digestion of dietary starch and glycogen [25]. Kang et al. have reported that AMY2A is a possible tumor-suppressor gene of 1p21.1 in gastric carcinoma [26]. Furthermore, Xi et al. have detected CNV of AMY2A using the Bayesian information criterion [27]. Thus, it may be inferred alteration in AMY2B and AMY2A copy numbers may have a role in lung SCC occurrence via the starch and sucrose metabolism pathway. FCGR3A encoding a cell surface molecule CD16a, a member CD16 gene family, which is much similar to FCGR3B on chromosome 1. Zhou et al. have reported that it is CNV of FCGR3A other than FCGR3B and FCGR2B that is involved in anti-GBM disease [28]. Meanwhile, a recent study also discovered a role of CD16 signaling receptor in antibody-dependent cancer cell killing [29]. Thus, it may be speculated that alterations in the copy number of these two genes might influence the immune processes, further contributing to lung SCC. However, CNV of the two genes have never been reported in lung SCC. More significantly, CFH at chr 1 is a member of the regulator of complement activation gene cluster. It was found that CFH sensitizes NSCLC cells to complement attack and inhibit the growth of tumor cells [30]. Therefore, the presence of CNV located in chr 1 and chr 3 might be potential contributory factors to the development of lung SCC.
TP63 (transformation-related protein 63) encoded by TP63 gene, along with p53 and p73, constitutes the p53 gene family of transcription factors [31]. Noticeably, TAp63 may functionally similar to p53 as it is reported to transactivate multiple p53 downstream targets. However, p53 has only one promoter with three conserved domains, whereas either p63 or p73 has two promoters, thus each having two isotypes: one containing TA domain (TAp63, TAp73), and the other containing no TA domain (ΔNp63 and ΔNp73). Wu et al. investigated the cell functions by introducing TAp63α and ΔNp63α into Saos2 cells using adenovirus expression vectors and subsequently detecting the gene profiles using DNA microarrays, and they have found that p63 can regulate a wide range of various cellular functions, such as cell cycle control, stress, and signal transduction, which are critical events in cancer and development [32]. It thus may be inferred that CNV of TP63 might have a role in lung SCC by altering the expression of its downstream genes, although no copy number variation has been reported in p63 gene so far.
Conclusions
In this study we conducted an integrated analysis of transcriptional profile and CNV of lung SCC, and finally screened 16 CNV-driven genes. The variation in these gene copy number is speculated to have a role in lung SCC occurrence. For example, FCGR3A, FCGR2B and HLA-DQA1 may function via the systemic lupus erythematosus pathway, and AMY2B and AMY2A may participate in lung SCC via starch and sucrose metabolism pathway. Our work provides new insights into the mechanisms underlying lung SCC, and also suggest some new targets for therapy of lung SCC. However, their roles in lung SCC require further experimental validation. The genes AMY2A, CFH, and TP63 However, more in-depth studies are needed in order to verify our findings.
Methods
As the paper did not involve any human or animal study, ethical approval was not required.
Source of gene expression data and CNV data
Transcriptional profile of GSE17710 [33] was downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), which was annotated using the platform of GPL9053 (Agilent-UNC-custom-4X44k). This dataset was collected from both the tumor tissues and adjacent normal tissues of 56 lung SCC patients. CNV data were downloaded from the TCGA database (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm?mode=ApplyFilter&showMatrix=true&diseaseType=LUSC&tumorNormal=TN&tumorNormal=T&tumorNormal=NT&platformType=1&platformType=4&platformType=40) in Dec., 2013. Only data of level 3 were accessible and downloaded, which included CNV information (segment mean value) in both the tumor issues and matched adjacent normal tissues (control) from 513 patients with lung SCC using Affymetrix Genome-Wide Human SNP 6.0 array containing 1.8 million SNP and CNV probes. A segment mean value is log2 transformed ratio of the detected copy number in either the tumor or normal tissues to the copy number 2 that is detected in a normal human using hg19 as reference genome. A value larger than one indicates a copy number increase, and a value smaller than one means a copy number depletion.
Preprocessing of transcriptional profile data
The probe-level data of transcriptional profile were first converted into expression measures. DEGs were identified between the tumor tissues and adjacent normal tissues with the cut-off criteria of |log2FC (Fold change)| > 1.
Preprocessing of CNV microarray data
First, the distribution of CNVs (copy number deletion or increase) on the 22 chromosomes was investigated in both the cases and controls, and the number of CNV regions of 1–10 kb, 10–50 kb and >50 kb in length on each chromosome was calculated, respectively. Permutation test was performed to calculate the P value of CNVs in either the cases or controls in each chromosome based on 1000 replicates. Next, Circos software was used to display the distribution of on each chromosome in both tumors and controls.
Identification of lung SCC-related CNVs
First, genes located within CNV regions were identified according to the human hg19 reference genome, and its copy vari in an identified gene was also calculated. Next, a gene that is related to lung SCC with CNV was retained only when its CNV was not detected in controls but detected in more than 80 % ceases. The segment mean value and the copy number were denoted as 0 and 1, respectively when missing in a sample.
Identification of CNV-driven genes
The association between the copy number difference data and differential expression data was analyzed to screen CNV-driven genes. A CNV-driven gene was defined only when the differential expression trend was consistent with the copy number change, namely an up-regulated gene should also have an increased copy number; vice versa.
Functional annotation and pathway enrichment analysis of CNVs
The functional enrichment analysis of the DEGs and CNV-driven genes was carried out using Database for Annotation, Visualization and Integrated Discovery (DAVID) software based on the gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) pathway databases [34]. P < 0.05 was set as the cut-off.
Authors’ contributions
ZY and BZ participated in the design of this study, and they both performed the statistical analysis. YY and SMJ carried out the study and collected important background information. TW drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81170049, 81470252, 81160011) and Natural Science Foundation of Ningxia (NZ11151).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Contributor Information
Zhao Yang, Email: zhaoyyd@163.com.
Bing Zhuan, Email: bingzhuanan@126.com.
Ying Yan, Email: yingyanand@yeah.net.
Simin Jiang, Email: siminjjj@126.com.
Tao Wang, Phone: +86-027-85488882, Email: taowangtts@163.com.
References
- 1.Bray F, Jemal A, Grey N, Ferlay J, Forman D. Global cancer transitions according to the Human Development Index (2008–2030): a population-based study. Lancet Oncol. 2012;13(8):790–801. doi: 10.1016/S1470-2045(12)70211-5. [DOI] [PubMed] [Google Scholar]
- 2.Peters S, Adjei A, Gridelli C, Reck M, Kerr K, Felip E. Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(suppl 7):vii56–64. [DOI] [PubMed]
- 3.Reinmuth N, Brandt B, Semik M, Kunze W-P, Achatzy R, Scheld HH, et al. Prognostic impact of Cyfra21-1 and other serum markers in completely resected non-small cell lung cancer. Lung Cancer. 2002;36(3):265–270. doi: 10.1016/S0169-5002(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 4.Cumberbatch M, Tang X, Beran G, Eckersley S, Wang X, Ellston RP, et al. Identification of a subset of human non-small cell lung cancer patients with high PI3Kβ and low PTEN expression, more prevalent in squamous cell carcinoma. Clin Cancer Res. 2014;20(3):595–603. doi: 10.1158/1078-0432.CCR-13-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Moreno P, Brambilla E, Thomas R, Soria J-C. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res. 2012;18(9):2443–2451. doi: 10.1158/1078-0432.CCR-11-2370. [DOI] [PubMed] [Google Scholar]
- 6.Drilon A, Rekhtman N, Ladanyi M, Paik P. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol. 2012;13(10):e418–e426. doi: 10.1016/S1470-2045(12)70291-7. [DOI] [PubMed] [Google Scholar]
- 7.Kirov G, Rees E, Walters JT, Escott-Price V, Georgieva L, Richards AL, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry. 2014;75(5):378–385. doi: 10.1016/j.biopsych.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riccardi VM, Lupski JR. Duplications, deletions, and single-nucleotide variations: the complexity of genetic arithmetic. Genet Med. 2012;15(3):172–173. doi: 10.1038/gim.2012.124. [DOI] [PubMed] [Google Scholar]
- 9.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43(9):838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenman CD, Bignell G, Butler A, Edkins S, Hinton J, Beare D, et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics. 2010;11(1):164–175. doi: 10.1093/biostatistics/kxp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Boerdonk RA, Sutedja TG, Snijders PJ, Reinen E, Wilting SM, van de Wiel MA, et al. DNA copy number alterations in endobronchial squamous metaplastic lesions predict lung cancer. Am J Respir Crit Care Med. 2011;184(8):948–956. doi: 10.1164/rccm.201102-0218OC. [DOI] [PubMed] [Google Scholar]
- 12.Ocak S, Yamashita H, Udyavar A, Miller A, Gonzalez A, Zou Y, et al. DNA copy number aberrations in small-cell lung cancer reveal activation of the focal adhesion pathway. Oncogene. 2010;29(48):6331–6342. doi: 10.1038/onc.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Hopkins D, Schmidt C, Silva S, Houghton R, Takita H, et al. Identification of genes differentially over-expressed in lung squamous cell carcinoma using combination of cDNA subtraction and microarray analysis. Oncogene. 2000;19(12):1519–1528. doi: 10.1038/sj.onc.1203457. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi T, Daigo Y, Katagiri T, Tsunoda T, Okada K, Kakiuchi S, et al. Expression profiles of non-small cell lung cancers on cDNA microarrays: identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene. 2003;22(14):2192–2205. doi: 10.1038/sj.onc.1206288. [DOI] [PubMed] [Google Scholar]
- 15.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan B, Dachrut S, Coral H, Yuen ST, Chu KM, Law S, et al. Integration of DNA copy number alterations and transcriptional expression analysis in human gastric cancer. PLoS One. 2012;7(4):e29824. doi: 10.1371/journal.pone.0029824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu T-P, Lai L-C, Tsai M-H, Chen P-C, Hsu C-P, Lee J-M, et al. Integrated analyses of copy number variations and gene expression in lung adenocarcinoma. PLoS One. 2011;6(9):e24829. doi: 10.1371/journal.pone.0024829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quan T, Little E, Quan H, Voorhees JJ, Fisher GJ. Elevated matrix metalloproteinases and collagen fragmentation in photodamaged human skin: impact of altered extracellular matrix microenvironment on dermal fibroblast function. J Invest Dermatol. 2013;133(5):1362. doi: 10.1038/jid.2012.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Jin X, Fang S, Wang R, Li Y, Wang N, et al. The functional polymorphism in the matrix metalloproteinase-7 promoter increases susceptibility to esophageal squamous cell carcinoma, gastric cardiac adenocarcinoma and non-small cell lung carcinoma. Carcinogenesis. 2005;26(10):1748–1753. doi: 10.1093/carcin/bgi144. [DOI] [PubMed] [Google Scholar]
- 20.Canonne-Hergaux F, Gros P. Expression of the iron transporter DMT1 in kidney from normal and anemic mk mice. Kidney Int. 2002;62(1):147–156. doi: 10.1046/j.1523-1755.2002.00405.x. [DOI] [PubMed] [Google Scholar]
- 21.Whitney JF, Clark JM, Griffin TW, Gautam S, Leslie KO. Transferrin receptor expression in nonsmall cell lung cancer. Histopathologic and clinical correlates. Cancer. 1995;76(1):20–25. doi: 10.1002/1097-0142(19950701)76:1<20::AID-CNCR2820760104>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Chappell S, Daly L, Morgan K, Guetta Baranes T, Roca J, Rabinovich R, et al. Cryptic haplotypes of SERPINA1 confer susceptibility to chronic obstructive pulmonary disease. Hum Mutat. 2006;27(1):103–109. doi: 10.1002/humu.20275. [DOI] [PubMed] [Google Scholar]
- 23.Farshchian M, Kivisaari A, Ala-aho R, Riihilä P, Kallajoki M, Grénman R, et al. Serpin peptidase inhibitor clade A member 1 (SerpinA1) is a novel biomarker for progression of cutaneous squamous cell carcinoma. Am J Pathol. 2011;179(3):1110–1119. doi: 10.1016/j.ajpath.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang R, Chen B, Pfütze K, Buch S, Steinke V, Holinski-Feder E, et al. Genome-wide analysis associates familial colorectal cancer with increases in copy number variations and a rare structural variation at 12p12. 3. Carcinogenesis. 2014;35(2):315–323. doi: 10.1093/carcin/bgt344. [DOI] [PubMed] [Google Scholar]
- 25.Sawai T, Hehre EJ. A novel amylase (Candida transglucosyl-amylase) that catalyzes glucosyl transfer from starch and dextrins. J Biol Chem. 1962;237(7):2047–2052. [PubMed] [Google Scholar]
- 26.Kang JU, Koo SH, Kwon KC, Park JW. AMY2A: a possible tumor-suppressor gene of 1p21. 1 loss in gastric carcinoma. Int J Oncol. 2010;36(6):1429. doi: 10.3892/ijo_00000637. [DOI] [PubMed] [Google Scholar]
- 27.Xi R, Hadjipanayis AG, Luquette LJ, Kim T-M, Lee E, Zhang J, et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci. 2011;108(46):E1128–E1136. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.X-j Zhou, Lv J-c Bu, D-f YuL, Y-r Yang, Zhao J, et al. Copy number variation of FCGR3A rather than FCGR3B and FCGR2B is associated with susceptibility to anti-GBM disease. Int Immunol. 2010;22(1):45–51. doi: 10.1093/intimm/dxp113. [DOI] [PubMed] [Google Scholar]
- 29.Kudo K, Imai C, Lorenzini P, Kamiya T, Kono K, Davidoff AM, et al. T lymphocytes expressing a CD16 signaling receptor exert antibody-dependent cancer cell killing. Cancer Res. 2014;74(1):93–103. doi: 10.1158/0008-5472.CAN-13-1365. [DOI] [PubMed] [Google Scholar]
- 30.Ajona D, Hsu Y-F, Corrales L, Montuenga LM, Pio R. Down-regulation of human complement factor H sensitizes non-small cell lung cancer cells to complement attack and reduces in vivo tumor growth. J Immunol. 2007;178(9):5991–5998. doi: 10.4049/jimmunol.178.9.5991. [DOI] [PubMed] [Google Scholar]
- 31.Murray-Zmijewski F, Lane D, Bourdon J. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13(6):962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 32.Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee C-CR, et al. ΔNp63α and TAp63α regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003;63(10):2351–2357. [PubMed] [Google Scholar]
- 33.Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16(19):4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]


