Abstract
Most aspects of RNA metabolism involve DEAD-box RNA helicases, enzymes that bind and remodel RNA and RNA-protein complexes in an ATP-dependent manner. Here we show that the DEAD-box helicase Ded1p oligomerizes in the cell and in vitro, and unwinds RNA as a trimer. Two protomers bind the single stranded region of RNA substrates and load a third protomer to the duplex, which then separates the strands. ATP utilization differs between the strand separating protomer and those bound to the single stranded region. Binding of the eukaryotic initiation factor 4G to Ded1p interferes with oligomerization and thereby modulates unwinding activity and RNA affinity of the helicase. Our data reveal a strict division of labor between the Ded1p protomers in the oligomer. This mode of oligomerization fundamentally differs from other helicases. Oligomerization represents a previously unappreciated level of regulation for DEAD-box helicase activities.
Introduction
DEAD-box helicases are the largest helicase family and play key roles in RNA metabolism (Linder and Jankowsky, 2011). These enzymes contain a structurally conserved helicase core with 12 characteristic sequence motifs, including the D-E-A-D signature (Linder et al., 1989; Putnam and Jankowsky, 2013b). Biochemical activities of DEAD-box helicases include RNA duplex unwinding, disruption of RNA-protein complexes, RNA-dependent ATP hydrolysis, and formation of stable, ATP-dependent complexes with RNA (Henn et al., 2012; Jarmoskaite and Russell, 2011; Linder and Jankowsky, 2011; Liu et al., 2014; Putnam and Jankowsky, 2013b). All of these activities are based on the ability of the enzymes to bind and remodel RNA and RNA-protein complexes in an ATP-dependent manner (Putnam and Jankowsky, 2013b).
DEAD-box helicases unwind RNA duplexes by local strand separation, which differs from the translocation-based strand separation seen for other helicases (Bizebard et al., 2004; Yang et al., 2007a; Yang and Jankowsky, 2006). DEAD-box helicases load directly to the duplex and pry apart a limited number of basepairs in an ATP-dependent fashion (Jarmoskaite et al., 2014; Mallam et al., 2012; Pan et al., 2014; Tijerina et al., 2006; Yang et al., 2007a). The remaining basepairs dissociate non-enzymatically (Chen et al., 2008; Rogers et al., 1999; Yang and Jankowsky, 2006). For several DEAD-box helicases strand opening has been shown to require only ATP binding (Chen et al., 2008; Liu et al., 2008). ATP hydrolysis promotes recycling of the enzymes (Liu et al., 2008). For many DEAD-box helicases, loading onto the duplex is facilitated by single stranded or structured RNA adjacent to the duplex (Tijerina et al., 2006; Yang and Jankowsky, 2006). The polarity of the single stranded regions is not critical for unwinding, but the loading region has to be proximal to the duplex (Yang et al., 2007a). Enzyme loading for several DEAD-box helicases is associated with protein domains outside the helicase core (Hardin et al., 2010; Mallam et al., 2011).
Many non-DEAD-box helicases work as oligomers (Enemark and Joshua-Tor, 2008). It is unknown whether oligomerization is also important for DEAD-box helicases. Potential of DEAD-box helicases to oligomerize has been suggested by two lines of evidence. First, for several DEAD-box helicases unwinding rate constants show a sigmoidal dependence on the enzyme concentration, an observation consistent with cooperative oligomerization (Halls et al., 2007; Yang et al., 2007a; Yang and Jankowsky, 2005, 2006). Second, multiple DEAD-box helicases appear to bind to themselves in the cell, also suggesting oligomerization (Ernoult-Lange et al., 2012; Minshall and Standart, 2004; Ogilvie et al., 2003; Rudolph et al., 2006). Whether and how oligomerization of DEAD-box helicases impacts their biochemical and biological functions is unknown.
Here, we mechanistically characterize oligomerization for the DEAD-box RNA helicase Ded1p from S. cerevisiae. Ded1p is involved in translation initiation and possibly other processes of RNA metabolism (Beckham et al., 2008; Burckin et al., 2005; Chuang et al., 1997; Hilliker et al., 2011; Jamieson et al., 1991). Ded1p orthologs are highly conserved in eukaryotes (Sharma and Jankowsky, 2014). The human ortholog DDX3X has been implicated in tumorigenesis and is targeted by several pathogens including HIV, HCV, and poxviridae (Sharma and Jankowsky, 2014).
We show that Ded1p oligomerizes in the cell and in vitro, and that it unwinds RNA as a trimer with a strict division of labor between the protomers. This mode of oligomerization fundamentally differs from previously characterized helicases. We further show that the eukaryotic initiation factor 4G interferes with oligomerization of Ded1p and thereby modulates its unwinding activity, indicating that oligomerization is an efficient mechanism for regulation of DEAD-box helicase activities.
RESULTS
Ded1p forms oligomers in the cell and in vitro
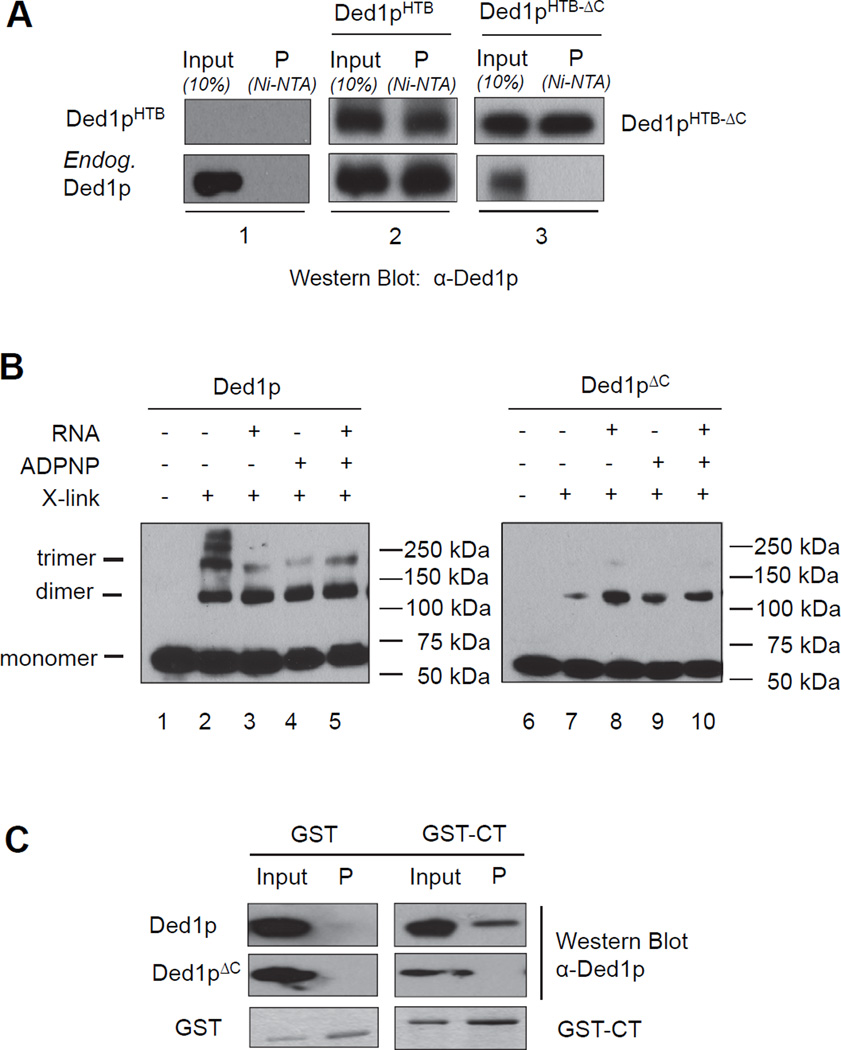
We first tested whether Ded1p oligomerizes in the cell. We transformed yeast cells with a plasmid encoding DED1 bearing a histidine-biotin (HTB) affinity tag. Adsorption of Ded1pHTB to a Ni-affinity column after RNase treatment showed both, Ded1pHTB and wtDed1p. No endogenous Ded1p was detected in the control without Ded1pHTB (Figure 1A). This observation indicates RNA-independent association of multiple Ded1p protomers in the cell.
Figure 1. Ded1p oligomerizes in the cell and in vitro.
(A) Affinity precipitation of endogenous Ded1p with HTB-tagged Ded1p or Ded1pΔC in the presence of RNAse. Inputs and affinity precipitations (P) were analyzed by western blot using a polyclonal Ded1p antibody (α-Ded1p).
(B) Formaldehyde crosslinking of purified recombinant wtDed1p or Ded1pΔC with and without RNA (16 bp, 25 nt unpaired region 3' to the duplex) and ADPNP, as indicated. Protein species were visualized by western blot (α-Ded1p). Mobilities for the Ded1p monomer, dimer, and trimer are indicated.
(C) Pull-down of Ded1p and Ded1pΔC by the GST-tagged Ded1p C-terminus. The left panel shows a control of GST only. See also Figure S1.
To probe whether Ded1p oligomerized in the absence of other proteins, we performed protein-protein crosslinking experiments with purified recombinant Ded1p in vitro (Figure 1B). Multiple protomers crosslinked without RNA and the non-hydrolyzable ATP analog ADPNP, indicating formation of complexes with up to five Ded1p protomers (Figure 1B, lane 2). Ded1p also oligomerized in the presence of RNA, and upon addition of ADPNP. The molecular weight of these oligomers was consistent with a dimer and, to a lesser extent, a trimer (Figure 1B, lanes 3–5). The data indicate that oligomerization of Ded1p does not depend on cellular co-factors, but represents an intrinsic ability of the enzyme.
Removal of the 69 C-terminal amino acids (Ded1pΔC, Figure S1), a region of low complexity (Sharma and Jankowsky, 2014), diminished oligomerization, both in the cell and in vitro (Figure 1A, panels 3, Fig. 1B, lanes 6–10). Removal of the C-terminus confers a temperature sensitive growth phenotype and alleviates growth inhibition caused by overexpression of full-length Ded1p (Hilliker et al., 2011). Ded1pΔC still formed dimers in vitro, albeit at lower efficiency than wtDed1p (Figure 1B, lanes 6–10). However, no complexes with more than two protomers were seen. The isolated C-terminus (CT) co-purified with full-length Ded1p, but not with Ded1pΔC (Figure 1C). These data suggest that the C-terminus, while not critical for dimerization of Ded1p, is part of the protein-protein interface in the trimer, and likely in the larger complexes seen in the absence of RNA and ATP analog.
Oligomerization is required for optimal helicase activity
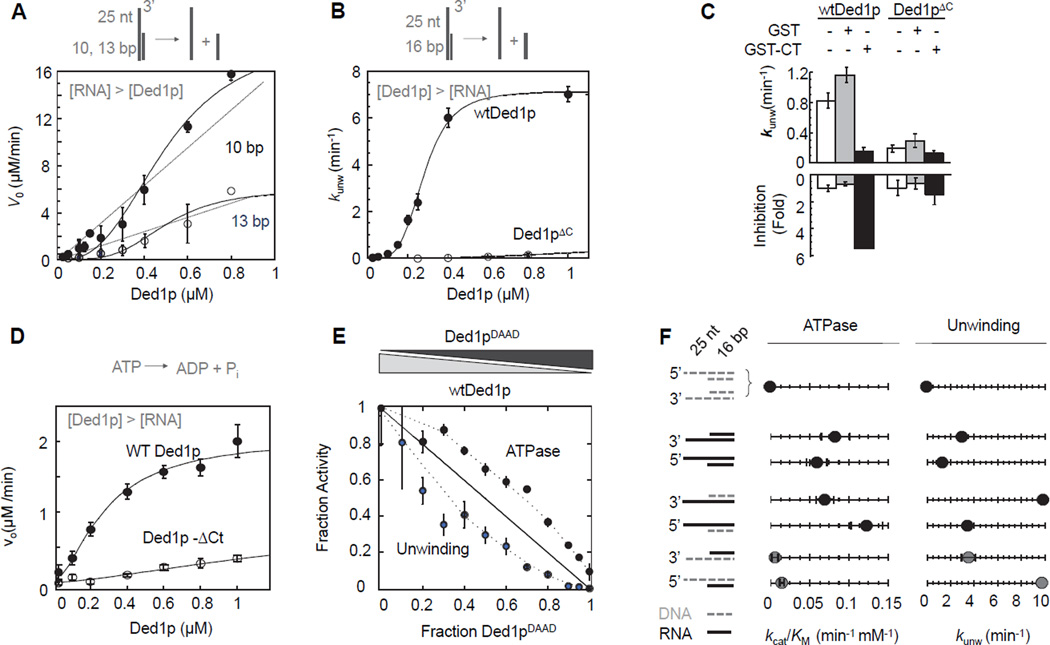
To test the impact of oligomerization on the catalytic activities of Ded1p, we measured ATP-dependent RNA unwinding with RNA excess, allowing for multiple RNA substrate turnovers per enzyme (Figures 2A, S2A–F). The initial unwinding rates (v0) for two duplexes with different lengths showed a sigmoidal dependence of v0 on the Ded1p concentration, indicating cooperative oligomerization (Figure 2A). For a monomeric enzyme, v0 would linearly depend on the Ded1p concentration.
Figure 2. Connection between oligomerization, ATPase and helicase activity.
(A) Unwinding of two RNA substrates (2 µM) with differing duplex length under steady-state conditions with increasing concentrations of Ded1p. Data points here and in all subsequent figures are averages from multiple independent measurements (N ≥ 3) and error bars represent one standard deviation, unless indicated otherwise. Dashed lines indicate the dependence of the unwinding rate expected for a monomeric enzyme. Solid lines mark a trend.
(B) Unwinding of a 16 bp RNA substrate (0.5 nM) under pre-steady state conditions with increasing concentrations of Ded1p and Ded1pΔC. Solid lines are the best fits to the Hill equation (for wt Ded1p:kunwmax = 7.4 ± 0.2 min−1,K1/2 = 320 ± 40 nM, H = 3.2 ± 0.1; for Ded1pΔC: no meaningful datafit)
(C) Inhibition of unwinding by Ded1p, but not Ded1pΔC with the C-terminal domain of Ded1p. Measurements were performed with 0.5 nM RNA (13bp, 25 nt 3’ ssRNA), 300 nM Ded1p,900 nM Ded1pΔC, 0.4 mM ATP, 1.0 µM GST-Ded1p-Ct or GST.
(D) RNA-stimulated ATPase activity with 50 nM RNA (16 bp, 25 nt 3’ ssRNA) at indicated concentrations of Ded1p and Ded1pΔC. Solid lines are the best fits to the Hill equation (for wt Ded1p:Vmax = 2.1 ± 0.2 µM·min−1, K1/2 = 220 ± 60 nM, H = 1.6 ± 0.2; for Ded1pΔC: no meaningful datafit)
(E) Inhibition of helicase and RNA-stimulated ATPase activities of Ded1p with Ded1pDAAD, measured at 0.5 nM (unwinding) or 50 nM (ATPase) RNA (16 bp duplex with 3' 25 ntssRNA), 1 µM total protein concentration ([Ded1p]+ [Ded1pDAAD]), and 2 mM ATP. Activities are plotted as a fraction of the activity of 1 µM wtDed1p. The solid line indicates the expected result for a monomeric enzyme. Dashed mark a trend.
(F) Comparison of unwinding and ATPase activity of wtDed1p for a series of RNA-DNA hybrid substrates (cartoons on the left - grey broken line: DNA; black solid line: RNA). Values for kcat/Km for ATP hydrolysis were obtained with 2 µM nucleic acid substrate, 0.4 µM Ded1p, as in Fig. 3A, values for kunwmax for unwinding with 0.1 nM substrate, 2 mM ATP/Mg2+, as in Fig. 2B. See also Figure S2.
RNA unwinding under pre-steady state conditions, with enzyme excess resulted in a sigmoidal functional binding isotherm for wtDed1p (Figure 2B), as previously reported (Halls et al., 2007; Yang et al., 2007a; Yang and Jankowsky, 2005, 2006). The Hill coefficient of H = 3.2 ± 0.2 (Figure 2B) indicates cooperation between at least three Ded1p protomers. Similar Hill coefficients were measured for a range of different RNA substrates (Figure S2G). Ded1pΔC showed significantly lower unwinding activity than wtDed1p at comparable concentrations (Figure 2B). The isolated Ded1p C-terminus (CT) inhibited the unwinding activity of wtDed1p, but not of Ded1pΔC (Figure 2C). These data provide further direct evidence that oligomerization of Ded1p is critical for optimal helicase activity, and that the C-terminus is important for the unwinding oligomer.
ATPase and helicase activities are differently affected in the Ded1p oligomer
We then measured functional binding isotherms for RNA-dependent ATPase activity with enzyme excess. Functional affinity of wtDed1p for RNA was similar to that for RNA unwinding, but the Hill coefficient was only H = 1.6 ± 0.2 (Figure 2D). Similar Hill coefficients were measured for a range of different RNA substrates (Figure S2K). The data suggest that optimal RNA-stimulated ATPase activity requires cooperation between fewer Ded1p protomers than unwinding activity.
Deletion of the C-terminus decreased the RNA-stimulated ATPase activity by a factor of approximately 4 (protein concentration: 1 µM), but helicase activity by a factor of roughly 80, compared to wtDed1p (Figure 2B,D). Discrepancies between ATPase and helicase activities were anticipated, given that unwinding by Ded1p requires only ATP binding, but not turnover (Chen et al., 2008; Liu et al., 2008). However, a simple lack of ATP hydrolysis during unwinding would not cause differences in Hill coefficients in unwinding and ATPase reactions or such large differences between wtDed1p and Ded1pΔC in ATPase vs. helicase activities. Our data thus suggested a more intricate link between ATPase and helicase activities in the Ded1p oligomer.
To probe the connection between unwinding and ATPase activities, we mixed wtDed1p with mutant Ded1pDAAD. The mutation renders the protein deficient for both, ATPase and helicase activities (Iost et al., 1999). We varied the ratio between wtDed1p and Ded1pDAAD, while maintaining constant overall protein concentration (Figure 2E). Additive inhibition would result in a linear decrease of activity with increasing concentration of inactive protomer (Moreau et al., 2007). The inhibitory effect of Ded1pDAAD on the RNA-stimulated ATPase activity was smaller than expected from simple additive inhibition. In contrast, the effect on unwinding was larger than expected from additive inhibition and larger than the effect on the ATPase activity (Figure 2E). These data suggest that ATP-binding deficiency in at least one protomer is beneficial for RNA-stimulated ATPase activity, but detrimental to unwinding. This result raised the possibility that individual protomers participate differently in ATPase and helicase activity.
To confirm and extend these observations, we compared unwinding and ATPase activity for a series of RNA-DNA hybrid substrates (Figure 2F). Previous data had shown that unpaired DNA facilitated unwinding, even though DNA does not stimulate ATPase activity of Ded1p (Iost et al., 1999; Yang and Jankowsky, 2006). However, the connection between unwinding and ATPase activities for these substrates remained unclear. Here, we observed similar unwinding rate constants for comparable substrates with unpaired RNA or DNA tails (Figure 2F). ATP hydrolysis activity was considerably weaker for substrates with DNA tails, indicating that ATPase activity is mainly associated with unpaired RNA (Fig. 2F). Given that Ded1p protomers bind DNA tails (Figure S2J), our finding supports the notion that individual Ded1p protomers participate differently in ATPase and helicase activities.
Unwinding involves distinct loading and strand separating protomers
To better understand functional differences between individual protomers, we examined whether protomers interacting with the unpaired tail function distinctly from those interacting with the duplex region. We functionally separated a protomer interacting with the unpaired tail from an unwinding protomer by "clamping" Ded1p to the single stranded region and testing whether clamped protomers facilitate or prevent unwinding. To clamp protomers to the unpaired RNA we utilized the ability of Ded1p to form complexes with single stranded RNA and the non-hydrolyzable ATP analog ADPNP that have half lives of many hours (Liu et al., 2014).
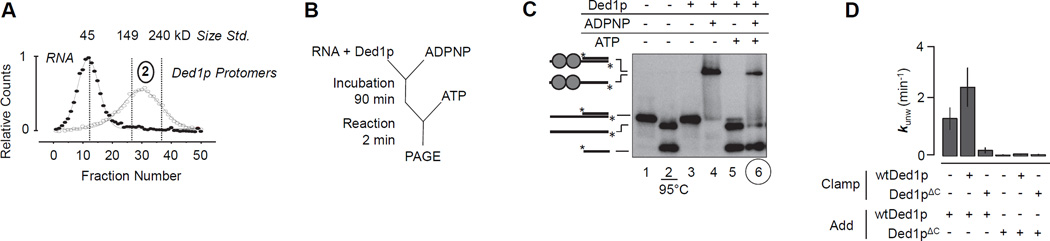
We clamped Ded1p to the 25 nt unpaired tail of a substrate with a 16 bp duplex (Figure 3). Ded1p unwinds substrates with shorter tails less efficiently than substrates with 25 nt, while substrates with longer tails do not further enhance unwinding activity (Yang et al., 2007a). The 25 nt tail binds two Ded1p protomers (Figure 3A). Shorter tails bind fewer Ded1p protomers, longer tails more protomers (Figure S3A,B). Collectively, these observations suggest that binding of two Ded1p protomers to an unpaired RNA tail is optimal for unwinding.
Figure 3. Unwinding involves distinct loading and unwinding protomers.
(A) Fractionation of of RNA (16 bp, filled circles) and RNA with Ded1p and ADPNP (open circles) on a sucrose gradient (6–40%). Curves represent best fits to a Gaussian distribution. Dashed lines indicate peak fraction numbers of protein markers sedimented under identical conditions.
(B) Reaction scheme for testing RNA unwinding with pre-bound (clamped) Ded1p protomers.
(C) Representative non-denaturing PAGE for RNA unwinding with pre-bound (clamped) Ded1p protomers, and associated controls (0.6 µM Ded1p; 0.5 nM RNA, both strands radiolabeled, 50 µM ADPNP/Mg2+, where applicable; and 2 mM ATP/Mg2+, where applicable). To samples in lanes 4 – 6, 1 µM 73 ntRNA scavenger was added after the reaction to remove loosely associated Ded1p from the labeled RNA.
(D) Impact of pre-bound (clamped) Ded1p or Ded1pΔC on the unwinding rate constant kunwmax. See also Figure S3.
After clamping two Ded1p protomers to the single stranded tail we added ATP and measured strand separation by the remaining free protein (Figure 3B). Reaction and associated controls were monitored by non-denaturing PAGE to visualize paired RNA, separated strands, and RNA clamped to Ded1p (Figure 3C). Upon addition of ATP to clamped Ded1p, strand separation occurred while Ded1p remained stably bound to the bottom strand (Figure 3C, lane 6). As expected, addition of ADPNP only induced a stable complex, but no strand separation (lane 4). Addition of ATP led to strand separation, but not to stable complex formation (lane 5). No significant Ded1p-RNA binding was induced by ADPNP during the unwinding reaction (Figure S3C). These data directly show that clamped Ded1p protomers participate in the unwinding reaction.
To further examine cooperation between clamped and added Ded1p protomers, we compared unwinding rate constants with and without clamped Ded1p (Figure 3D). Clamped protomers increased the unwinding rate constant, compared to the reaction without clamp (Figure 3D). In contrast, clamped Ded1pΔC did not facilitate loading of full-length Ded1p to the duplex, and clamped full-length Ded1p did not facilitate loading of Ded1pΔC (Figure 3D). Collectively, our observations suggest that clamped protomers aid the loading of at least one additional Ded1p protomer to the duplex. This protomer separates the duplex in an ATP dependent-fashion, and appears to be functionally distinct from the protomers bound to the unpaired tail, which can work with ADPNP. Loading of the third protomer involves the C-terminus of Ded1p on the unwinding protomer and at least on one protomer bound to the unpaired substrate tail. For simplicity, we will refer to the protomers bound to the unpaired tail as loading protomers, and to the protomer(s) separating the duplex as unwinding protomer(s). We first focused on characterizing the loading protomers.
Two loading protomers favor binding of a single ATP
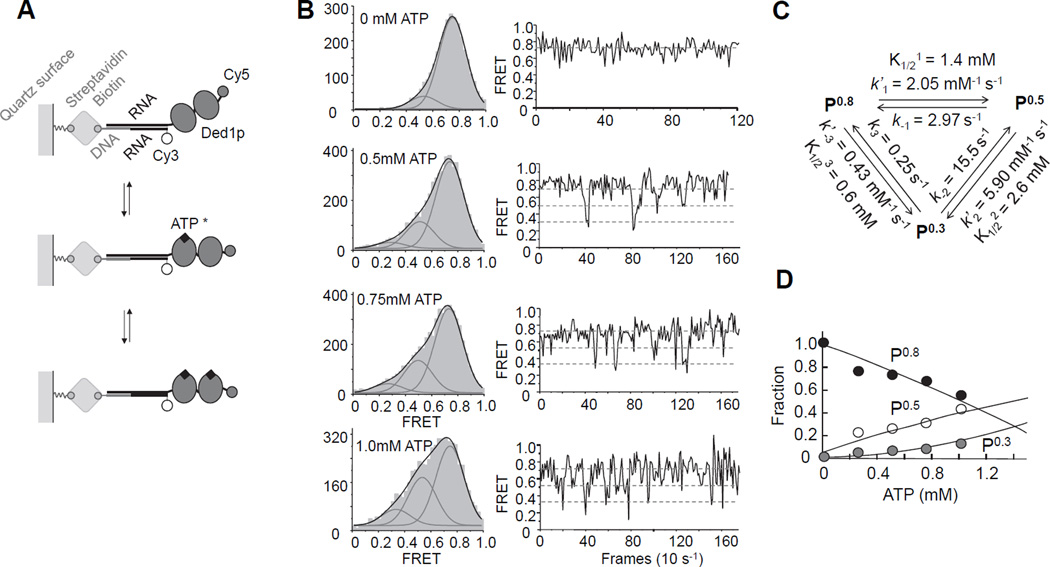
The data above had shown that unwinding readily occurred without ATP exchange in the loading protomers. However, when ATP exchange is not prevented, ATP hydrolysis is mainly associated with the loading protomers (Figure 2F). To resolve this apparent contradiction, it was critical to clarify how ATP binding and hydrolysis impacts the interaction of Ded1p with the unpaired RNA. To accomplish this, we used single molecule FRET (smFRET) and monitored the ATP-dependence of the interaction of Ded1p with an unpaired RNA attached to a long duplex region (Figure 4A). This duplex is not appreciably unwound during the observation time (Liu et al., 2014). We used saturating concentrations of Ded1p (1 µM), and monitored binding reactions at increasing ATP concentrations (Figure 4B).
Figure 4. Analysis of ATP binding to the loading protomers by single molecule FRET.
(A) Schematics for the single molecule FRET measurements.
(B) Left column: smFRET Histograms for Ded1p binding to the RNA with increasing concentrations of ATP/Mg2+, as indicated. Grey curves represent best fits to individual Gaussian distributions, black curves the sum of the individual distributions. Right column: representative time traces for reactions in the presence of increasing concentrations of ATP/Mg2+, as indicated in the histograms. Dashed lines mark the three FRET states.
(C) Association and dissociation rate constants for ATP/Mg2+, and associated equilibrium dissociation constants.
(D) Fraction of each FRET state calculated from smFRET histograms as function of the ATP/Mg2+ concentration. Lines are the best fit of the experimental data to the model shown in panel (C). See also Figure S4.
Without ATP, a major smFRET peak at E ~ 0.8 (P0.8) was seen, consistent with observations from a previous study (Liu et al., 2014). This state represents both free RNA and RNA bound to Ded1p without ATP, a complex that is highly dynamic (Liu et al., 2008; Liu et al., 2014; Putnam and Jankowsky, 2013a). At the saturating Ded1p concentrations used here (Figure S4A–F), the free RNA state is not appreciably populated, and P0.8 corresponds to Ded1p bound to RNA without ATP. With increasing ATP concentrations, peaks emerged at E ~ 0.5 (P0.5) and E ~ 0.3 (P0.3) (Figure 4B). P0.3 corresponds to the smFRET state upon complete binding of the RNA by Ded1p in the presence of non-hydrolyzable ATP analogs (Liu et al., 2014). Given that the 25 nt tail binds two Ded1p protomers (Figure 3A), P0.3 represents two Ded1p protomers bound to RNA and ATP. P0.5 also increased with the ATP concentration (Fig. 4B), suggesting that this state corresponds to one ATP bound to either one or two Ded1p protomers. The ratio between P0.3 and P0.5 state increased only slightly with the Ded1p concentration, but P0.3 remained always significantly smaller than P0.5 (Figure 4D). This observation implies a mechanism that disfavors simultaneous binding of two ATP to two Ded1p protomers.
To illuminate this mechanism, we determined the kinetic parameters for the smFRET transitions from smFRET time traces (Figure 4B,C). The rate constants showed that binding of Ded1p to the RNA is fast, compared to ATP binding to Ded1p at the reaction conditions (konDed1p > 40µM−1s−1 vs. konATP = 0.002µM−1s−1, Figure S4F). The kinetic parameters faithfully recapitulate the dependence of the fractions of the three smFRET states on the ATP concentration (Figure 4D), thus reflecting association and dissociation of ATP to RNA-bound Ded1p. Binding of the second ATP is modestly anti-cooperative with the first bound ATP and dissociation of the second ATP is markedly faster than dissociation of the first ATP (Fig. 4C). This arrangement counteracts a saturation of two RNA-bound Ded1p protomers with ATP, favoring the state with only one bound ATP (Figure 4D).
Notably, dissociation of the second ATP (k−2 = 15.5 s−1) was more than one order of magnitude faster than the rate constant for ATP turnover (kcatATP= 0.23 s−1, Figure S4G,H), and therefore unlikely to reflect ATP turnover. The rate constant for the transition P0.3→P0.8 (k3 = 0.25 s−1) closely matches the ATP turnover rate constant and might represent ATP turnover. The notion that the P0.3→P0.8 transition corresponds to ATP turnover was supported by a marked decline in the frequency of P0.3→P0.8 transitions upon mutation of the arginine finger in motif VI (Figure S4I–L). This mutation disrupts the transition state for ATP hydrolysis, thereby preventing ATP hydrolysis while allowing ATP binding (Liu et al., 2008). Together, the data suggest that rapid dissociation of the second ATP curtails the frequency of ATP turnover. Collectively, the smFRET approach revealed that two ATP can be bound by the loading protomers. However, the state with a single bound ATP is favored over two bound ATP even at increasing ATP concentrations (Figure 4D).
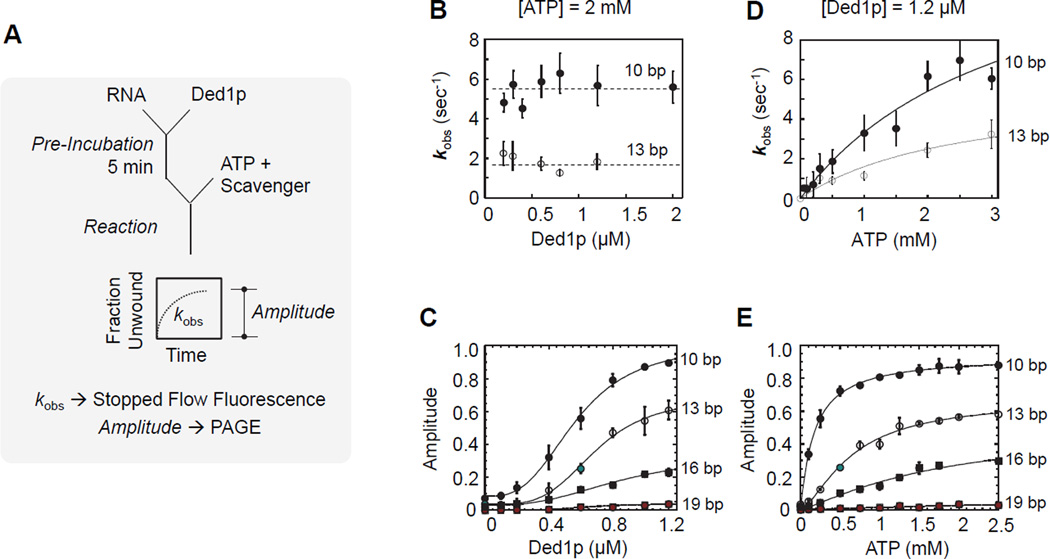
The unwinding protomer displays weak affinity for ATP
We next examined the unwinding protomer. To distinguish between strand separation step and loading process, we pre-incubated Ded1p with the RNA substrate, and started the reaction with ATP and RNA scavenger that precludes re-association of Ded1p with the RNA substrate during the reaction (Figures 5A; S5A,B). Because only a single RNA-protein binding cycle is monitored, this reaction regime is referred to as single cycle (Jankowsky et al., 2000; Liu et al., 2008). Reaction amplitudes and strand separation rate constants under these conditions reflect characteristics of the unwinding protomer.
Figure 5. Characterization of the unwinding protomer under single cycle conditions.
(A) Reaction scheme for a single cycle unwinding reaction.
(B) Observed unwinding rate constants (kobs) under single cycle conditions for two substrates containing a 25 nt unpaired region 3' to duplex with 10, and 13 bp, respectively, as function of increasing Ded1p concentrations. Unwinding reactions were measured by stopped-flow fluorescence spectroscopy.
(C) Unwinding amplitudes for a series of substrates (duplex lengths as indicated, identical 25 nt unpaired regions 3' to the duplex) as function of increasing Ded1p concentrations. Amplitudes were measured by PAGE (Figure S5).
(D) Observed unwinding rate constants (kobs) under single cycle conditions for the substrate used in panel (C), as function of increasing ATP concentration at 1.2 µM Ded1p. The black solid line marks a binding isotherm.
(E) Unwinding amplitudes for the substrates in panel (C) as function of increasing ATP concentrations at 1.2 µM Ded1p. Deconvolution of unwinding and dissociation rate constants, and resulting amplitudes as function of increasing ATP concentrations are shown in Figure S5.
We measured apparent strand separation rate constants (kobs) under single cycle conditions by stopped flow fluorescence (Figure S5B), because reactions were too fast for manual measurements. However, reaction amplitudes measured by stopped flow fluorescence were indistinguishable from manually obtained amplitudes (Figure S5D). At constant ATP (2 mM), kobs showed no significant dependence on the Ded1p concentration (Figure 5B), as expected for single cycle conditions. To assess the impact of duplex length on strand separation, we measured unwinding amplitudes for a set of substrates with duplexes from 10 to 19 bp. Amplitudes increased with Ded1p concentration, reflecting the concentration-dependent binding of Ded1p to the substrates (Figure 5C). K1/2 values and Hill coefficients (H ~ 3) were similar for all duplexes (Figure 5C). This observation indicates that duplex length does not significantly impact loading of the unwinding protomer and confirms participation of at least three Ded1p protomers in the unwinding reaction. The apparent affinity (K1/2 = 0.54 µM) is the affinity of Ded1p for RNA without ATP, which is approximately twofold larger than the affinity with ATP (K1/2 = 0.32 µM, Figure 2B), consistent with previous observations for different RNA substrates (Putnam and Jankowsky, 2013a). Amplitudes at saturating concentrations of Ded1p decreased with increasing duplex length (Figure 5C), because the strand separation rate constant decreases with duplex length (Chen et al., 2008; Yang et al., 2007a).
We next probed the ATP affinity of the unwinding protomer. The apparent strand separation constant (kobs) increased almost linearly with the ATP concentration over the experimentally accessible range ([ATP] < 3 mM) (Figure 5D). This observation indicates a significantly weaker ATP affinity for the unwinding protomer than for the loading protomers (Figure 4C). Amplitudes for duplexes of different length showed a scaling with the ATP concentration (Figure 5E), which indicated that duplex length did not significantly alter the weak ATP affinity of the unwinding protomer (Figure S5F). The data reveal that physiological ATP concentrations are only sub-saturating for the unwinding protomer.
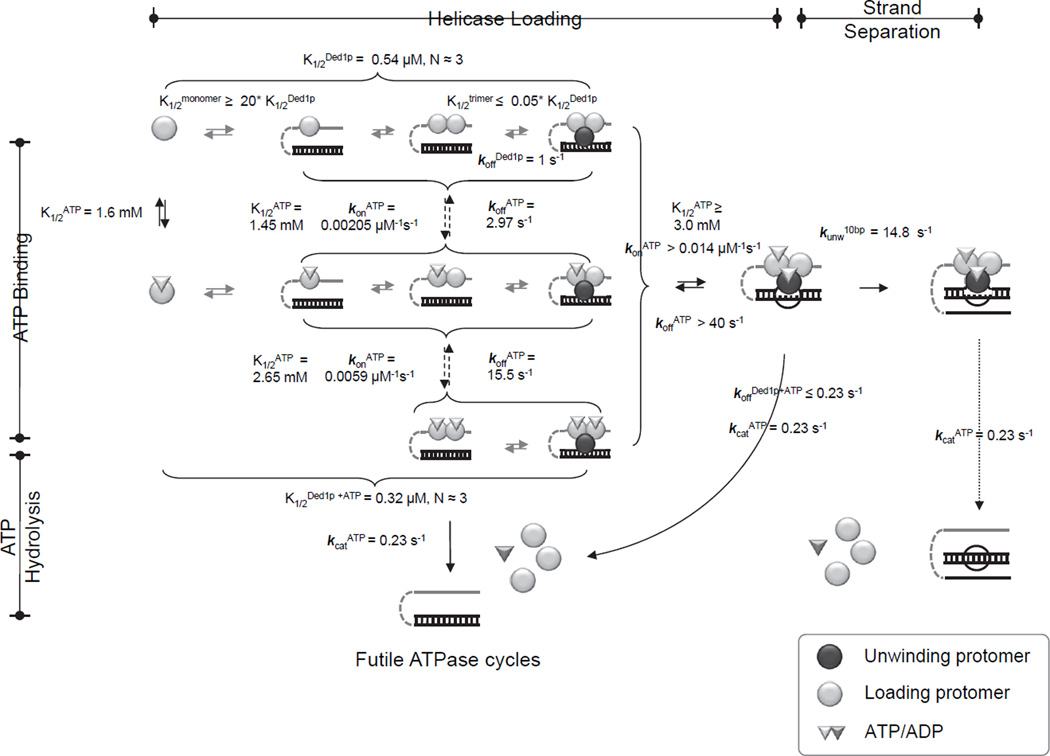
Coordination between loading and unwinding protomers
To quantitatively describe coordination between unwinding and loading protomers, we combined the smFRET data with multiple cycle and single cycle unwinding datasets in a unified kinetic and thermodynamic framework (Figure 6). Model building, fitting strategy and assessment of model quality are described in Figure S6, and in the Supplementary Methods. The framework shows that the Ded1p oligomer can readily assemble on the RNA under physiological ATP and Ded1p concentrations (Firczuk et al., 2013). The differing ATP affinities of loading and the unwinding protomers render the unwinding activity more sensitive to inhibition than the ATPase activity, explaining previous observations showing that AMP inhibits unwinding stronger than ATPase activity (Putnam and Jankowsky, 2013a).
Figure 6. Coordination between loading and unwinding protomers.
Kinetic and thermodynamic framework for oligomerization and duplex unwinding by Ded1p. Parameters were calculated from experimental data as described in Supplemental Methods, using Kintek Global Kinetic Explorer (Kintek Co., Texas). See also Figure S6.
The framework delineates the complex link between strand separation and ATP binding and hydrolysis. ATP binding by one of the loading protomers is favorable for optimal unwinding. However, unwinding can also occur without ATP bound by the loading protomers. ATP binding is required for the unwinding protomer, but hydrolysis is not necessary for strand separation (Chen et al., 2008; Liu et al., 2008). Accordingly, the rate constant for the strand separation step for the 10 bp duplex (kunw at Ded1p and ATP saturation) exceeds the rate constant for ATP turnover (kcat) by a factor of approximately 60. ATP-bound Ded1p dissociates from the RNA with a rate constant similar to that for ATP turnover, supporting the notion that ATP hydrolysis promotes Ded1p dissociation and recycling for subsequent unwinding cycles (Liu et al., 2008).
The framework rationalizes the cooperativity of Ded1p oligomerization. A Ded1p trimer binds RNA tighter than monomeric or dimeric Ded1p. Dissociation of the Ded1p oligomer without ATP is ~5 fold faster than ATP turnover, consistent with weaker affinity of Ded1p for RNA without ATP. Rapid dissociation of Ded1p without ATP explains the inability to detect stable Ded1p-RNA complexes by smFRET.
The rate constants for dissociation of Ded1p from RNA with and without ATP provide insight into the scaling of futile binding cycles with duplexes of different length. For a 10 bp duplex, 95% of binding events produce strand separation, consistent with previous data comparing ATP turnover and unwinding under steady state conditions (Chen et al., 2008). For a 16 bp duplex only ~ 30% of binding events are productive. For the 19 bp substrate, less than one percent of binding events result in strand separation.
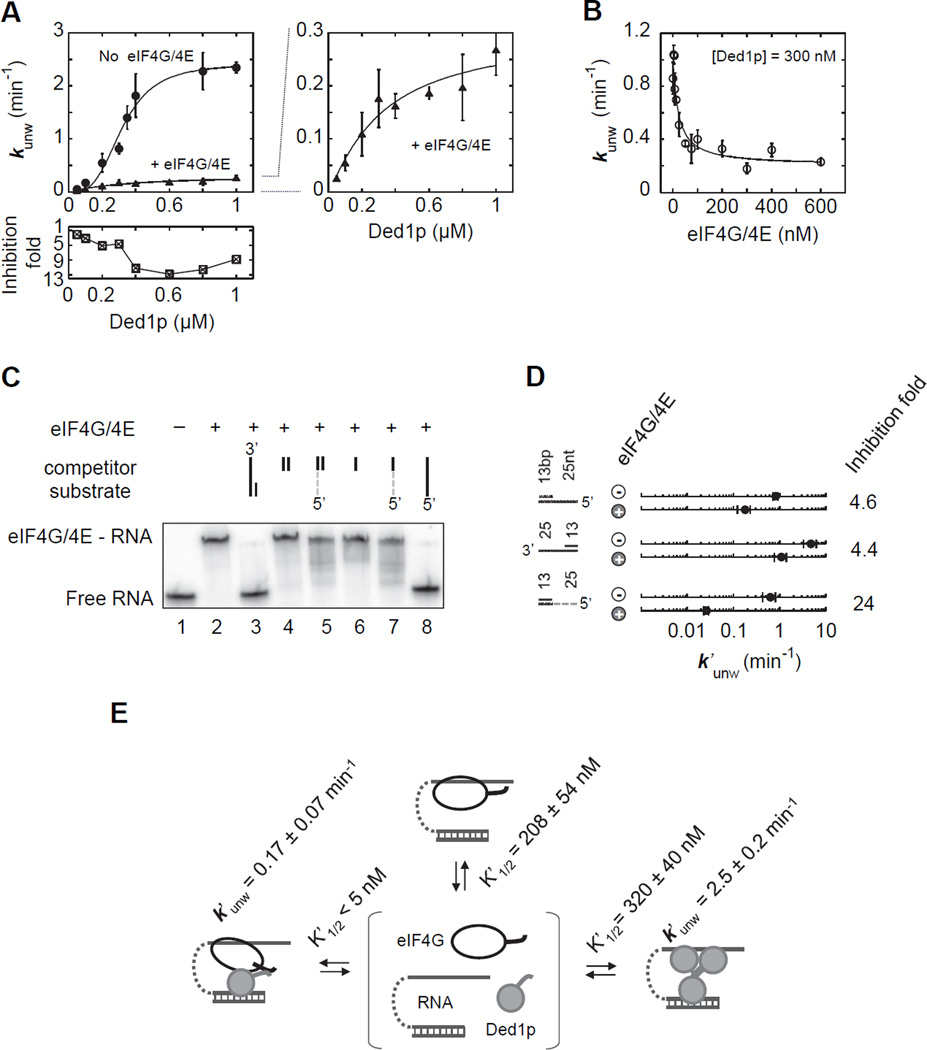
eIF4G interferes with oligomerization of Ded1p
The significance of the Ded1p C-terminus for the formation of the unwinding oligomer raised the question whether and how the Ded1p co-factor eIF4G, which binds Ded1p at its C-terminus (Hilliker et al., 2011), affected oligomerization and thereby the Ded1p activity. To address this question, we examined the effect of eIF4G on unwinding reactions by Ded1p (Figure 7A). eIF4G was purified and tested in complex with eIF4E, because eIF4G alone is unstable (Hilliker et al., 2011). Unwinding rate constants were lowered by eIF4G/E, and inhibition increased with the Ded1p concentration (Figures 7A, S7A,B). With eIF4G/E, the unwinding rate constant did not depend on the Ded1p concentration in the characteristic sigmoidal fashion seen for Ded1p alone. This observation suggests that eIF4G/E, which can bind RNA (Figure S7D), does not inhibit unwinding by Ded1p through competition for the RNA substrate, since competition would not change the sigmoidal shape of curve. Instead, the data suggest formation of a eIF4G-Ded1p complex that can unwind the duplex, but does not involve multiple Ded1p.
Figure 7. eIF4G interferes with oligomerization of Ded1p.
(A) Impact of eIF4G/E (300 nM) on unwinding of an RNA substrate (13 bp, 25 nt overhang 3') under pre-steady-state conditions with increasing concentrations of Ded1p (0.4 mM ATP). Solid lines mark the trend. The lower panel shows the inhibition by eIF4G/E at the corresponding Ded1p concentrations. The right plot zooms into the plot with eIF4G/E.
(B) Unwinding of the RNA substrate used in panel (A) at a constant concentration of Ded1p (0.4 mM ATP) and increasing concentrations of eIF4G/E, as indicated. The line is a fit to a binding isotherm (K1/2 = 21 ± 5 nM, kunwmin = 0.26 ± 0.13 min−1).
(C) Binding of eIF4G/E (200 nM) to RNA (71 nt unpaired RNA, 1 nM, lane 2) and competition of RNA and RNA-DNA chimeric oligonucleotides (4 µM, lanes 2–8). DNA regions are marked by the grey broken line, RNA regions by a solid black line.
(D) Impact of eIF4G/E (300 nM) on the unwinding activity of Ded1p (300 nM, 0.4 mM ATP) with RNA substrates and a RNA-DNA chimeric substrate (DNA marked by broken grey line). Inhibition is noted on the right.
(E) Basic scheme for the reaction with Ded1p and eIF4G/E. Apparent affinities (K1/2) were calculated by including eIF4G, eIF4G/E-RNA and Ded1p-eIF4G/E-RNA in the model shown in Fig. 6, at the ATP concentration (0.4 mM) used in panels (A) and (B). K1/2 for RNA binding by the Ded1p-eIF4G/E complex represents binding of eIF4G/E to RNA-bound Ded1p. See also Figure S7.
This notion was supported by experiments where the concentration of eIF4G/E was varied at a constant Ded1p concentration (Figure 7B). Inhibition did not reach completion, as would have been expected from competition by the RNA binding protein eIF4G for the RNA, and as seen for the DEAD-box helicase Mss116p, which does not interact with eIF4G (Figure S7C). Instead, the inhibition of Ded1p at higher eIF4G/E concentrations leveled at a constant value (Figures 7B, S7A). These data were consistent with a Ded1p-eIF4G/E complex that unwinds the duplex, but with a lower rate constant than the Ded1p trimer. Notably, this Ded1p-eIF4G/E complex showed higher apparent affinity for the substrate than the Ded1p trimer (K1/2'[Ded1p-eIF4G/E]= 21 ± 5 nM, Figure 7B, vs. K1/2'Ded1p = 322 ± 18 nM, Figure 7A). Upon removal of the C-terminus of Ded1p (Ded1pΔC), the major binding site for eIF4G (Hilliker et al., 2011), eIF4G affected the activity of Ded1pΔC predominantly through competition for the RNA, as seen with the unrelated Mss116p (Figure S7C). This result indicates that deletion of the C-terminus of Ded1p diminishes formation of the Ded1p-eIF4G/E complex.
To further characterize the Ded1p-eIF4G/E complex, we examined unwinding of a chimeric substrate containing an RNA duplex with DNA overhang (Figure 7C,D). Ded1p unwound this substrate with an observed rate constant similar to that seen for the corresponding RNA substrate (Figure 7D), as previously observed (Yang et al., 2007a; Yang and Jankowsky, 2006). Although eIF4G did not bind the DNA-RNA chimeras (Figure 7C), eIF4G inhibited unwinding by Ded1p markedly stronger than with RNA substrates (Figure 7C). Since the inability of eIF4G to bind this substrate rules out competition with Ded1p, we conclude that the eIF4G-Ded1p complex forms, but that eIF4G in the complex is unable to bind the DNA region. eIF4G thus replaces the loading protomers in the Ded1p trimer. This notion was supported by crosslinking of Ded1p with and without eIF4G, which showed interference of eIF4G with oligomerization of Ded1p (Figure S7E).
Collectively, our observations reveal an unwinding-competent Ded1p-eIF4G/E complex. eIF4G/E helps to load a Ded1p protomer to the duplex (Figure 7E). Both, Ded1p oligomerization and formation of the eIF4G-Ded1p complex involve the C-terminus of Ded1p in a mutually exclusive fashion. Although the eIF4G-Ded1 complex unwinds RNA with an apparent rate constant considerably lower than the Ded1p trimer, the apparent affinity of the eIF4G-Ded1 complex for the RNA is higher than that of the Ded1p oligomer (Figure 7E). These data mark oligomerization as powerful mechanism to regulate DEAD-box helicase activity.
DISCUSSION
While DEAD-box, and perhaps other eukaryotic RNA helicases are not usually viewed as oligomers, our data emphasize the significance of oligomerization for activity and regulation for at least a subset of these enzymes. For Ded1p, the K1/2 for oligomerization in vitro (on RNA: K1/2 = 0.3 µM with ATP, Fig. 6) are below the physiological Ded1p concentration of roughly 1µM (Firczuk et al., 2013; Sharma and Jankowsky, 2014), albeit in a similar range. Ded1p oligomers should thus readily form in the cell, consistent with our findings. However, Ded1p would not need to be exclusively present in oligomers, and thus available for interactions with other proteins.
The mode of oligomerization for Ded1p is, to our knowledge, unprecedented for other helicases. While other helicases form hexameric rings or dimers (Enemark and Joshua-Tor, 2008; Lohman et al., 2008), Ded1p forms a trimer. In other oligomeric helicases, each protomer usually cycles through the various stages of ATP and nucleic acid binding and during the course of an unwinding reaction, each protomer performs all functions (Enemark and Joshua-Tor, 2008). In the Ded1p oligomer, the protomers do not alternate their roles. Two Ded1p protomers bind single stranded RNA and thereby facilitate the loading of the third protomer to the duplex region. The third protomer separates the duplex. This strict division of labor between protomers is a distinct feature of the Ded1p oligomer.
In other helicases, ATP binding and hydrolysis are closely coordinated between protomers (Enemark and Joshua-Tor, 2008). In Ded1p, no comparable coordination is seen. In fact, the exact state of the ATPase cycle in the loading protomers appears not to be critical for the unwinding reaction. The loading protomers only have to remain bound to the unpaired region. Unwinding is therefore most efficient when the two loading protomers are clamped to the RNA. Long residence times of the loading protomer on the single stranded RNA increase the probability for loading of the third protomer to the duplex. We speculate that the anti-cooperativity in ATP binding by the two loading protomers serves the same purpose. Simultaneous ATP binding to both loading protomers is minimized, because ATP hydrolysis from this state causes disassembly of the oligomer, an obvious disadvantage for the unwinding reaction. In contrast, hydrolysis of only one bound ATP by both loading protomers appears to not completely disassemble the oligomer. Our data suggest that the loading protomers balance ATP turnover and dissociation from the single stranded RNA with the need to maintain RNA binding for as long as possible, thereby maximizing opportunities for loading of the third protomer to the duplex.
These observations raise the question why Ded1p has not evolved into a more potent unwinder, wherein ATP hydrolysis by the loading protomers is inhibited until unwinding occurs. This optimization for unwinding might not have occurred because Ded1p performs other functions in the cell, besides duplex unwinding as an oligomer, such as binding to and unwinding with eIF4G.
While the ATP binding state of the loading protomers appears to affect the unwinding reaction only slightly, the RNA binding state of the loading protomers to single stranded RNA or DNA strongly impacts strand separation. Unwinding involves a Ded1p oligomer even for substrates without single stranded regions, given that unwinding rate constants of blunt-end duplexes also display sigmoidal dependence on the Ded1p concentration (Yang et al., 2007a). However, the functional affinity of the Ded1p oligomer for blunt-end duplexes is significantly weaker than for those with sufficiently long unpaired RNA tails (Yang et al., 2007a). Thus, binding of unpaired RNA to the loading protomers allosterically regulates the unwinding protomer, but only when the single stranded region is in physical proximity to the duplex (Yang and Jankowsky, 2006). The interplay between the protomers and the limited coordination between the ATP-binding states of the different protomers rationalize the previously intractable connection between ATPase and unwinding activities for Ded1p, and possibly other oligomeric DEAD-box helicases.
The rate constants for ATP turnover, strand separation, and dissociation of Ded1p from the RNA with and without ATP provide direct, quantitative evidence that ATP hydrolysis is not required for strand separation by Ded1p, but linked to dissociation of the enzyme from RNA. These findings are consistent with previously published observations for Ded1p (Liu et al., 2008), and possibly other DEAD-box helicases (Chen et al., 2008; Putnam and Jankowsky, 2013b). A subset of other DEAD-box helicases might require ATP hydrolysis for strand separation (Henn et al., 2012).
Our data demonstrate that oligomerization of Ded1p provides an efficient mechanism to regulate helicase activity. The C-terminus of Ded1p serves as interface for both, oligomerization and eIF4G binding, thereby rendering both processes mutually exclusive. In the eIF4G-Ded1p complex, eIF4G replaces the loading protomers. The Ded1p-eIF4G complex displays higher RNA affinity, but lower unwinding activity than the Ded1p trimer. The C-terminus of Ded1p is a region of low complexity, with many potential sites for post-translational modifications and for interactions with other proteins. The mutually exclusive nature of Ded1p oligomerization and eIF4G binding raises the possibility that Ded1p oligomers are particularly prevalent outside of translation initiation, for example in ribosome biogenesis (Li et al., 2009).
EXPERIMENTAL PROCEDURES
Materials
RNA oligonucleotides (DHARMACON) were radiolabeled and prepared as described (Yang and Jankowsky, 2005). Sequences are given in the Supplementary Methods. Ded1p and all other protein constructs, including eIF4G/E, were expressed in E. coli and purified as described (Yang and Jankowsky, 2005)(Hilliker et al., 2011).
In vivo co-precipitation
Endogenous Ded1p was co-precipitated from S. cerevisiae (BY4741) with Ded1p or Ded1pΔC tagged with C-terminal 6xHis-Tev-Biotin-6xHis (HTB) expressed on a low copy plasmid (YcplacIII) under the Ded1p endogenous promoter. Expression levels of the tagged proteins were approximately equal to the endogenous Ded1p levels. Cells were pelleted, washed, and lysed (4°C, 50 mM NaCl, 40 mM Tris, pH 8, 0.5 mM MgCl2, 0.01% IGEPAL CA 630, 2 mM DTT, Roche protease inhibitors). The lysate was cleared by centrifugation. Lysates were incubated with Ni-NTA agarosebeads for 2h in the presence of a RNAse cocktail (Roche). Beads were washed 4 times (50 mM NaCl, 40 mM Tris, pH 8, 0.5 mM MgCl2, 0.01% IGEPAL CA 630, 2 mM DTT), samples were eluted (100 mM Tris, pH 6.8, 24% glycerol, 8% SDS, 0.02% Comassie Blue R250, 0.2 mM DTT) and resolved on a 10% Tricine-SDS Page gel. Gels were analyzed by western blot using a Ded1p antibody (α-Ded1p, polyclonal, raised against full length Ded1p, dilution 1:6,000).
Protein-protein crosslinking
Purified recombinant Ded1p (1 µM) and Ded1pΔC (1 µM) were incubated with and without 2 µM 16 bp duplex with a 3’ 25 nt single stranded RNA and 0.5 mM ADPNP/Mg2+ (Helicase reaction buffer (HRB): 40 mM Tris (pH 8.0), 50 mM NaCl, 8.3% (v/v) glycerol, 0.01% (w/v) IGEPAL CA 630, 2 mM DTT, 0.6 U/µL RNasin (Roche), and 0.5 mM MgCl2) for 60 minutes at 19°C. Formaldehyde (1% (v/v)) was added for 30 minutes at room temperature. Reactions were quenched with 0.5 mM Tris-Glycine (pH 6.8). Samples were diluted (100 mM Tris, pH 6.8, 24% glycerol, 8% SDS, 0.02% Comassie Blue R250, 0.2 mM DTT) and resolved on 10% Tricine-SDS Page gel. Gels were analyzed by western blot (α-Ded1p).
In vitro pull-down
Purified GST-Ded1p-CT (1 µM) was incubated in HRB with additional 11.6% glycerol, BSA 5 mg/mL, RNase A 20 µg/mL, RNase T1 1u/µL, and purified Ded1p or Ded1pΔC (0.5 µM) overnight (4°C). 50 µL of glutathione-sepharose resin was added, incubated for 60 min, and washed multiple times with RIPA buffer. Bound proteins were eluted with Laemmli buffer and denatured at 95°C for 5 min. Bands were resolved on 6–11.5% gradient SDS-PAGE and analyzed by Coomassie staining or western blot (α-Ded1p).
Unwinding reactions (PAGE)
Unless otherwise indicated, unwinding reactions were performed in HRB as described at 19°C (Yang and Jankowsky, 2005). ATP was added with equimolar MgCl2. For pre-steady state reactions, Ded1p was incubated with 0.5 nM radiolabled RNA for 5 min in reaction buffer, and reactions were initiated with ATP/Mg2+. Where applicable, eIF4G/E was included as described. Aliquots were removed at indicated times, reactions were stopped and applied to 15% non-denaturing PAGE. Gels were dried and RNAs visualized and quantified with a Molecular Dynamics Phosphor imager and the ImageQuant 5.2 software (Molecular Dynamics). Observed rate constants were calculated as described, taking into account simultaneously occuring strand annealing (Yang and Jankowsky, 2005). Single cycle reactions were conducted and analyzed as described above, except that the reaction was started with ATP/Mg2+ and RNA scavenger or heparin. Steady state unwinding reactions were conducted as above, but with up to 2 µM unlabeled RNA substrate in addition to the labeled RNA, and with 10 µM unlabeled top strand, to preclude re-annealing of the separated strands. Unwinding reactions were monitored by PAGE.
ATPase measurements
ATPase measurements were performed in HRB (19°C) as described (Putnam and Jankowsky, 2013a). Initial observed reaction rates (v'o) were calculated from the linear part of the plot of product vs. time.
Helicase clamping
Ded1p-RNA clamping reactions were performed for 90 min (19°C) in HRB with 0.5 nM radiolabeled RNA. After incubation, reactions were terminated by adding an equal volume of 50% glycerol, and 1 µM of a 73 nt scavenger RNA, to sequester non–stably bound protein. Protein-bound and free RNA were separated by non-denaturing PAGE (7%) at 4°C.
Sucrose gradient fractionation
Ded1p-RNA binding reactions were performed in HRB (90 µL) with 1.1 µM Ded1p and 1 mM ADPNP/Mg2+ for 60 minutes (19°C). Reactions were terminated by adding 1 µM (final) of 73 nt scavenger RNA. Samples were layered onto 4.6 ml of 6–40% sucrose gradients under the same buffer conditions. Gradients were centrifuged in a Beckman SW-55 Ti rotor at 42,000 rpm for 15 hours at 4°C. The size standards ovalbumin (3.6S, 45 kD), aldolase (7.35S, 149 kD), and catalase (11.3S, 240 kD) were mixed in equilibrium binding buffer and centrifuged in parallel with the reaction samples. Following centrifugation, samples were fractionated and radio-labeled RNA in each fraction was detected by scintillation counting (Beckman Coulter LS 6500). Size standards were examined by SDS-PAGE.
Single Molecule FRET measurements
Single-molecule FRET was measured in a home-built, total internal reflection (TIR) microscope setup (NIKON TE300 microscope, Pentamax Gen III CCD, PRINCETON INSTRUMENTS, 50 mW, frequency doubled solid-state laser, CRYSTALASER) (Fairman-Williams and Jankowsky, 2012; Yang et al., 2007b) RNA samples were immobilized in a flow-cell, coated with PEG/biotinylated PEG (Roy et al., 2008). Biotinylated RNA samples were immobilized via streptavidin (MOLECULAR PROBES). Reactions were conducted in HRB, except that 5% (v/v) glucose, glucose oxidase (SIGMA) and catalase (SIGMA) were added as oxygen-scavenging system Fluorescence and corresponding FRET values were computed as described (Yang et al., 2007b).
Stopped-flow fluorescence measurements
Reactions were measured in an Applied Photophysics Pi-star fluorimeter by monitoring fluorescence change of a 2-aminopurine (2-AP) in the top strand upon strand separation. 2-AP was excited at 310 nm (4 nm slit). Emission was monitored with a 350 nm cut-off filter. Reactions were performed in HRB (19°C) with 50 nM RNA bearing the 2-AP, 0.02 mg/ml heparin as scavenger for Ded1p, and 1 µM RNA without 2-AP, to eliminate re-annealing of the labeled duplex. Ded1p was incubated with the RNA and reactions were started by rapid mixing of ATP/Mg2+ at indicated concentrations. Data were analyzed as described (Putnam and Jankowsky, 2012).
Kinetic Modeling
Kinetic modeling was performed with KINETK EXPLORER (Kintek, Austin, TX) using data obtained from unwinding experiments reported in this study and from data reported in earlier studies (Putnam and Jankowsky, 2013a). A detailed description of the data used, modeling strategy, data fits and assessment of model quality is provided in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH (GM0067700 to E.J.). We thank Dr. T. Nilsen for comments on the manuscript and Dr. M.E. Harris for use of the stopped flow fluorimeter and for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.A.P., Z.G. and E.J. designed the study. A.A.P., Z.G., F.L., H.J., Q.Y., performed the experiments and purified proteins. A.A.P. performed the kinetic modeling. A.A.P., Z.G. and E.J. wrote the paper.
References
- Beckham C, Hilliker A, Cziko AM, Noueiry A, Ramaswami M, Parker R. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol Biol Cell. 2008;19:984–993. doi: 10.1091/mbc.E07-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernoult-Lange M, Baconnais S, Harper M, Minshall N, Souquere S, Boudier T, Benard M, Andrey P, Pierron G, Kress M, et al. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA. 2012;18:1702–1715. doi: 10.1261/rna.034314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Jankowsky E. Unwinding initiation by the viral RNA helicase NPH-II. J Mol Biol. 2012;415:819–832. doi: 10.1016/j.jmb.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firczuk H, Kannambath S, Pahle J, Claydon A, Beynon R, Duncan J, Westerhoff H, Mendes P, McCarthy JE. An in vivo control map for the eukaryotic mRNA translation machinery. Mol Syst Biol. 2013;9:635. doi: 10.1038/msb.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin JW, Hu YX, McKay DB. Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J Mol Biol. 2010;402:412–427. doi: 10.1016/j.jmb.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, Bradley MJ, De La Cruz EM. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu Rev Biophys. 2012;41:247–267. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- Jamieson DJ, Rahe B, Pringle J, Beggs JD. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991;349:715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- Jarmoskaite I, Bhaskaran H, Seifert S, Russell R. DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA. Proc Natl Acad Sci U S A. 2014;111:E2928–E2936. doi: 10.1073/pnas.1404307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schnier J, Slonimski PP. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Putnam AA, Jankowsky E. DEAD-box helicases form nucleotide-dependent, long-lived complexes with RNA. Biochemistry. 2014;53:423–433. doi: 10.1021/bi401540q. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012;490:121–125. doi: 10.1038/nature11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, Russell R, Lambowitz AM. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci USA. 2011;108:12254–12259. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Ogilvie VC, Wilson BJ, Nicol SM, Morrice NA, Saunders LR, Barber GN, Fuller-Pace FV. The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res. 2003;31:1470–1480. doi: 10.1093/nar/gkg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Potratz JP, Cannon B, Simpson ZB, Ziehr JL, Tijerina P, Russell R. DEAD-box helicase proteins disrupt RNA tertiary structure through helix capture. PLoS Biol. 2014;12:e1001981. doi: 10.1371/journal.pbio.1001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam A, Jankowsky E. Analysis of duplex unwinding by RNA helicases using stopped-flow fluorescence spectroscopy. Methods Enzymol. 2012;511:1–27. doi: 10.1016/B978-0-12-396546-2.00001-2. [DOI] [PubMed] [Google Scholar]
- Putnam AA, Jankowsky E. AMP sensing by DEAD-box RNA helicases. J Mol Biol. 2013a;425:3839–3845. doi: 10.1016/j.jmb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam AA, Jankowsky E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim Biophys Acta. 2013b;1829:884–893. doi: 10.1016/j.bbagrm.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, G W, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. Crystal structure and nucleotide binding of the Thermus thermophilus RNA helicase Hera N-terminal domain. J Mol Biol. 2006;361:731–743. doi: 10.1016/j.jmb.2006.06.065. [DOI] [PubMed] [Google Scholar]
- Sharma D, Jankowsky E. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol. 2014;49:343–360. doi: 10.3109/10409238.2014.931339. [DOI] [PubMed] [Google Scholar]
- Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007a;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Yang Q, Fairman ME, Jankowsky E. DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J Mol Biol. 2007b;368:1087–1100. doi: 10.1016/j.jmb.2007.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.