Abstract
The oxygenation reactions catalyzed by cytochromes P450 (CYPs) play critical roles in plant natural products biosynthesis. At the same time, CYPs are one of most challenging enzymes to functionally characterize due to the difficulty of recombinantly expressing these membrane-associated monooxygenases. In the course of investigating rice diterpenoid biosynthesis we have developed a synthetic biology approach for functional expression of relevant CYPs in Escherichia coli. In certain cases activity was observed for only one of two closely related paralogs although it seems clear that related reactions are required for production of the known diterpenoids. Here we report that optimization of the recombinant expression system enabled characterization of not only these previously recalcitrant CYPs, but also discovery of additional activity relevant to rice diterpenoid biosynthesis. Of particular interest, CYP701A8 was found to catalyze 3β-hydroxylation of syn-pimaradiene, which is presumably relevant to momilactone biosynthesis, while CYP71Z6 & 7 were found to catalyze multiple reactions, with CYP71Z6 catalyzing the production of 2α,3α-dihydroxy-ent-isokaurene via 2α-hydroxy- ent-isokaurene, and CYP71Z7 catalyzing the production of 3α-hydroxy-ent-cassadien-2- one via 2α-hydroxy-ent-cassadiene and ent-cassadien-2-one, which may be relevant to oryzadione and phytocassane biosynthesis, respectively.
Keywords: metabolic engineering, natural products, labdane-related diterpenoids, phytoalexin, antibiotic, evolution
INTRODUCTION
The addition of oxygen catalyzed by cytochromes P450 (CYPs), particularly their ability to react with hydrophobic substrates and target chemically inert hydrogen-carbon bonds, translates to these mono-oxygenases playing critical roles in natural products biosynthesis (Hamberger and Bak 2013). For example, plants produce extensive arrays of terpenoids, whose biosynthesis most often proceeds through hydrocarbon intermediates. While C10 mono- and, to a lesser extent, C15 sesqui- terpene hydrocarbons can act as volatile signaling agents, larger terpenoids such as the C20 diterpenoids require such addition of oxygen to increase solubility and provide functional groups in order to exert biological activity.
Diterpenoids are generally derived from (E,E,E)-geranylgeranyl diphosphate (GGPP), which is cyclized and/or rearranged by class I diterpene synthases that ionize the allylic diphosphate ester bond and form varied hydrocarbon backbone structures. This can proceed directly from or after initial (bi)cyclization of GGPP catalyzed by class II diterpene cyclases. These enzymes typically form copalyl diphosphate (CPP), leading to their designation as CPP synthases (CPSs). The subsequently acting class I diterpene synthases all seem to be related to the ent-kaurene synthase (KS) required in all vascular plants for gibberellin phytohormone production and, hence, are termed KS-like (KSL). Because the hydrocarbon backbone of CPP matches that of labdanes, the resulting natural products have been termed labdane-related diterpenoids (LRDs)(Peters 2010).
The sequential activity of class II and I diterpene synthases produces multicyclic diterpenes, most often highly hydrophobic olefins. These are further elaborated to bioactive products via a range of modifications, which seems to invariably require the activity of a microsomal CYP (Zi et al. 2014a). These mono-oxygenases can catalyze not only hydrocarbon hydroxylation but also a variety of oxidative reactions such as epoxidation and alteration of carbon-carbon bonds (Mizutani and Sato 2011; Podust and Sherman 2012).
Rice (Oryza sativa) is one of most important food crops and also has become a model plant for study of LRD biosynthesis (Kitaoka et al. in press). The biochemical activity of the class II diterpene cyclases encoded by the rice CPS gene family (OsCPSs) and that of the subsequently acting class I diterpene synthases encoded by the rice KS(L) gene family have been elucidated (Schmelz et al. 2014). This work demonstrated that rice produces an array of nine different labdane-related diterpene olefins, and spurred us to develop a modular metabolic engineering system to enable biosynthesis of these compounds in Escherichia coli (Cyr et al. 2007).
Particularly with further engineering to increase flux to isoprenoid metabolism (Morrone et al. 2010b), it was possible to produce high enough concentrations of these olefins to enable their further turnover by CYPs (Kitaoka et al. in press). Although functional expression of microsomal CYPs in bacteria is traditionally considered difficult, we were able to do so via both N-terminal modification, replacing the trans-membrane helix with a leader peptide sequence (Morrone et al. 2010a; Swaminathan et al. 2009), and the critical use of synthetic genes with optimized codon usage for expression in E. coli (Wang et al. 2011). This synthetic biology approach has enabled at least partial reconstitution in E. coli of plant diterpenoid biosynthetic pathways that include CYPs (Zi et al. 2014b; Zi and Peters 2013), requiring further co-expression of a plant cytochrome P450 reductase (CPR), with incorporation of up to two CYPs to-date (Wu et al. 2013).
Through this approach we have elucidated the function of a number of rice CYPs (Wang et al. 2012a; Wang et al. 2011; Wang et al. 2012b; Wu et al. 2011; Wu et al. 2013). These studies were focused on CYPs found in two diterpenoid biosynthetic gene clusters, defined by sequentially acting OsCPSs and OsKSLs (Prisic et al. 2004; Wilderman et al. 2004), as well as a pair of CYP701A sub-family members, paralogous to the kaurene oxidase required for gibberellin biosynthesis, whose inducible transcription also suggested a role in phytoalexin biosynthesis (Itoh et al. 2004). However, in two cases activity was only observed for one of a paralogous pair of CYPs; specifically CYP99A3, but not the 83% (amino acid sequence) identical CYP99A2 (Wang et al. 2011), and CYP701A8, but not the 79% identical CYP701A9 (Wang et al. 2012b). Here we report optimization of the recombinant expression system, which enabled functional expression of these paralogs, and has revealed CYP activity that appears to be relevant to rice diterpenoid biosynthesis.
MATERIALS AND METHODS
General procedures
Unless otherwise noted, chemicals were purchased from Fisher Scientific, and molecular biology reagents were purchased from Invitrogen. Gas chromatography was performed with a Varian 3900 GC with Saturn 2100 ion trap MS in electron ionization (70 eV) mode for GC-MS analyses, while an Agilent 6890N GC with flame-ionization detection was used for GC-FID analyses. Samples (1 or 5 μL) were injected in splitless mode at 50 °C and, after holding for 3 min at 50 °C, the oven temperature was increased at a rate of 14 °C/min to 300 °C, where it was held for an additional 3 min. MS data from 90 to 600 m/z were collected starting at 14 min after injection until the end of the run.
Recombinant constructs
The CYPs used here are the previously described synthetic codon-optimized N-terminally modified genes (Wang et al. 2011; Wang et al. 2012b; Wu et al. 2011), which were explicitly designed to avoid the BamHI, EcoRI, KpnI, NcoI, NdeI, NotI and XhoI restriction sites commonly found in the multiple cloning sites (MCSs) of the Duet series of dual promoter/MCS vectors (Novagen). The ability to use an NcoI or NdeI site overlapping the start codon is particularly important to provide optimal spacing relative to the Shine-Dalgarno (SD) ribosome-binding site. Accordingly, introduction of NcoI or NdeI sites at the 5′ end (overlapping the start codon), and one of the other unique restriction sites at the 3′ end by PCR amplification enables conventional “cut-and-paste” sub-cloning with retention of optimal spacing to the SD and promoter. Please note that while it was previously reported that the DEST cassette, which enables directional recombination for sub-cloning of genes from pENTR constructs, was inserted into the first MCS (MCS1) of pET-Duet1 using the NcoI and NotI restriction sites (Wu et al. 2013), it was actually inserted using the XbaI and NotI restriction sites, such that the MCS1 SD site is no longer present in the resulting pET-Duet1/DEST vector, although the promoter is retained.
The pET-Duet1/DEST/ vector was used to construct an initial set of CYP expression vectors. All the CYPs studied here (CYP71Z6, CYP71Z7, CYP99A2, CYP701A8 and CYP701A9) were each cloned into second MCS (MCS2) using 5′ NdeI and 3′ XhoI restriction sites, creating a series of pET-Duet1/DEST/CYP constructs. Subsequently, AtCPR1 (Mizutani and Ohta 1998; Urban et al. 1997) was then inserted via directional recombination into the DEST cassette, creating a series of pET-Duet1/DEST::AtCPR1/CYP co-expression constructs.
To investigate the effect of DEST cassette based sub-cloning on AtCPR1 expression, an alternative pET-Duet1/AtCPR1/CYP99A2 vector was constructed by cloning AtCPR1 into MCS1 of pET-Duet1 using 5′ NcoI and 3′ NotI restriction sites, with CYP99A2 cloned into MCS2 as described above. To provide an additional source of AtCPR1, a pCDF-Duet1/AtCPR1 vector was constructed by sub-cloning AtCPR1 into MCS2 of pCDF-Duet1 using 5′ NdeI and 3′ KpnI restriction sites. To enable co-production of the diterpene olefin substrates syn-pimaradiene or ent-isokaurene in the metabolic engineering system, the relevant class I diterpene synthase (OsKSL4 or OsKSL6, respectively) was inserted via directional recombination into a previously described pCDF-Duet1/DEST/AtCPR1 vector, creating pCDF-Duet1/DEST::OsKSL(4 or 6)/AtCPR1 vectors, an analogous construct for the production of ent-cassadiene by incorporation of OsKSL7 (i.e., creating pCDF-Duet1/DEST::OsKSL7/AtCPR1) has been previously reported (Wu et al. 2013).
Cell-free lysate preparation
CYPs preparations were carried out essentially as previously described (Swaminathan et al. 2009; Wang et al. 2012a; Wang et al. 2011; Wang et al. 2012b; Wu et al. 2011; Wu et al. 2013). Briefly, the CYPs were expressed with AtCPR1 using one of the various CYP expression vectors described above, which were generally co-transformed with the compatible pCDF-Duet1/AtCPR1 vector, into the C43 OverExpress strain of E. coli (Lucigen). Expression cultures were grown in TB medium (pH 7.5, 4 × 100 mL in 250 mL Fernbach flasks) containing appropriate antibiotics. Upon reaching an A600 of ~0.8 the cultures were shifted to 16 °C, supplemented with 5 mg/L riboflavin and 75 mg/L 5-aminolevulinic acid, and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After 72 hours the cells were harvested via centrifugation (10 min. × 5000 g), resuspended in 30 mL buffer (0.1 M Tris/HCl, pH 7.5, 0.5 mM EDTA, 20% glycerol), and twice passed through a French press homogenizer (Emulsiflex-C5: Avestin) at 1500 psi. The resulting lysates were clarified via centrifugation (20 min. × 14000 g) and the concentration of CYP in the resulting supernatant (cell-free lysate) was quantified by carbon monoxide (CO)-binding difference spectra using the standard extinction coefficient of 91 mM−1cm−1 (Omura and Sato 1964).
CYP in vitro reaction and kinetic analysis
CYP reactions were carried out in 0.9 mL buffer (0.1 M Tris/HCl, pH 7.5, 0.5 mM EDTA, 20% glycerol) containing 0.2 mM NADPH and 20 μM diterpene substrate. The assays were initiated by adding 0.1 mL cell-free lysate from recombinant cultures, and the reaction then incubated at 32 °C for 30 min. To stop the reaction the assays were incubated at 90 °C for 5 min. The reaction product was extracted with n-hexanes (3 × 2 mL). The combined organic layer was dried under a gentle stream of N2 gas and then dissolved in 100 μL n-hexane for GC-MS analysis (5 μL injection volumes).
Kinetic analysis was carried out essentially as described above, with cell-free lysates from recombinant cultures transformed with pCDF-Duet1/AtCPR1 and either pET-Duet1/DEST::AtCPR1/CYP99A2 or pET-Duet1/DEST::AtCPR1/CYP701A8, although the assays were only allowed to react for 1 min, diterpene substrate concentrations varied, CYP concentrations quantified and held at 15 nM for CYP701A8 and 2 nM for CYP99A2, and the final reaction volume adjusted to total 1 mL. Following the assay, 3α-hydroxy-ent-sandaracopimaradiene was added to the reaction mixture as internal standard, and relative product amount quantified by GC-FID.
Pathway reconstruction via metabolic engineering
To screen for CYP diterpenoid production in our metabolic engineering system, genes for the relevant pathway were transformed into the C41 OverExpress strain of E. coli (Lucigen). Thus, the relevant CYP was co-expressed with not only AtCPR1 using one of the pET-Duet1/DEST::AtCPR1/CYP vectors described above, but also with a GGPP synthase and the relevant CPS carried on compatible pGGxC vectors (Cyr et al. 2007), as well as the relevant OsKSL carried on one of the additionally compatible pCDF-Duet1/DEST::KSL/AtCPR1 vectors also described above. The resulting recombinant cultures were grown in TB medium (pH 7.0, 50 mL in 250 mL Fernbach flasks) containing the appropriate antibiotics. Upon reaching an A600 of ~0.8 the cultures were shifted to 16 °C, supplemented with 5 mg/L riboflavin and 75 mg/L 5-aminolevulinic acid, and induced with 1 mM IPTG. After 72 hours the resulting diterpenoids were extracted with an equal volume of n-hexanes. The organic layer was dried under a gentle stream of N2 gas and then dissolved in 1 mL n-hexanes for GC-MS analysis (1 μL injection volumes). For CYP99A2 products, the dried crude extract was methylated with diazomethane and reaction mixture was dried down again before resuspending with 1 mL n-hexanes.
Diterpenoid production and isolation
To obtain the novel CYP products culture volumes were increased (8 × 500 mL in 2.8 L Fernbach flasks). After fermentation as described above, these cultures were extracted twice with equal volumes of n-hexanes and the combined organic layer dried by rotary evaporation. The resulting residue was resuspended in 5 mL of n-hexanes and then fractioned via flash chromatography over a 4 g silica column using a Reveleris system with UV detection and automated fraction collector (Grace), with an n-hexanes to acetone step gradient (100% n-hexanes, 90% n-hexanes/acetone, and 80% n-hexanes/acetone) as the mobile phase. The mono-hydroxylated diterpenoids were eluted in the 90% n-hexanes/acetone fractions, while 2α,3α-dihydroxy-ent-isokaurene and 3α-hydroxy-ent-cassadien-2-one eluted in the 80% n-hexanes/acetone fractions. Further purification was carried out using an Agilent 1200 series HPLC instrument equipped with autosampler, fraction collector and diode array UV detection, and ZORBAK eclipse XDB-C8 column (4.6 mm × 150 mm; 5 μm), run at a 0.5 mL/min flow rate. The column was pre-equibrated with 50% acetonitrile/water (0-2 min), and eluted with 50-100% acetonitrile gradient (2-30 min), followed by a 100% acetonitrile wash (30-45 min).
Chemical structure analysis
NMR spectra for the diterpenoids were recorded at 25 °C in chloroform-d1. NMR spectra were collected using a Bruker Avance 700 spectrometer equipped with a 5-mm HCN cryogenic probe. Each purified compound was dissolved in 0.5 mL chloroform-d1. This sample was placed in a NMR microtube (Shigemi) for analysis, and chemical shifts were referenced using known chloroform-d1 signals (13C 77.0 ppm, 1H 7.24 ppm). Structural analysis was performed using 1D 1H, and 2D DQF-COSY (double-quantumfiltered correlation spectroscopy), HSQC (heteronuclear single-quantum coherence), HMBC (heteronuclear multiple bond correlation) and NOESY experiment spectra acquired at 700 MHz, and 1D 13C spectra (174 MHz) using standard experiments from the Bruker TopSpin v1.4 software. Correlations from the HMBC spectra were used to propose a partial structure and assign proton chemical shifts. The structure was then verified using HSQC to confirm chemical shift assignments.
RESULTS
Optimization reveals CYP99A2 activity
It has been previously reported on the basis of RNAi mediated co-suppression that CYP99A2 and/or 3 play a role in momilactone biosynthesis, with CYP99A2 exhibiting substantially higher expression/transcript levels than CYP99A3 (Shimura et al. 2007). We later found that CYP99A3 catalyzes conversion of the upstream syn-pimaradiene precursor to syn-pimaradien-19-oic acid that is presumably relevant to production of the eponymous 19,6-γ-lactone ring (Wang et al. 2011). However, we were unable to detect activity with CYP99A2, presumably due to incorrect folding as the characteristic peak at 450 nm was not evident in CO-binding difference spectra, and only observed the 420 nm peak associated with “incorrectly” folded CYP (Sono et al. 1996). Given the higher expression level of CYP99A2 we were interested in obtaining correctly folded enzyme to investigate the possibility that this also contributes, either separately or redundantly, to momilactone biosynthesis.
The modularity of our metabolic engineering system is derived from the use of Gateway DEST(ination) cassettes, allowing facile transfer of genes from pENTR clones into various expression constructs. To enable shuffling of multiple genes, we incorporated DEST cassettes into the Duet series of dual promoter/multiple-cloning-site (MCS) expression vectors (Cyr et al. 2007). Specifically, the DEST cassette was cloned into the first MCS (MCS1), with invariant genes, such as the GGPP synthase and cytochrome P450 reductase (CPR), then cloned into the second MCS (MCS2). To optimally space the start codon relative to a Shine-Dalgarno (SD) ribosome-binding site, genes were cloned into the SD containing pENTR/SD/d-TOPO vector. However, insertion of the DEST cassette into the Duet vectors generally left the original SD site intact, such that incorporation of genes from pENTR/SD/d-TOPO based constructs led to the presence of two SD sites, and we hypothesized that this might interfere with optimal expression.
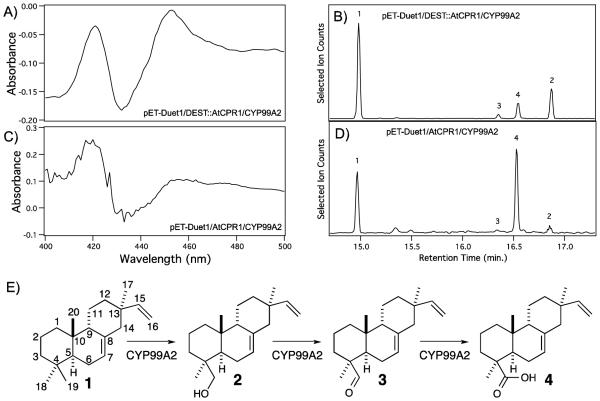
In previous work we found that CYPs seemed to be better expressed from pET derived vectors (Wu et al. 2013), so to test our hypothesis we cloned CYP99A2, with which only trace activity was previously detected (Wang et al. 2011), into the MCS2 of pETDuet1/DEST with optimal spacing to the associated SD site. Given that we have found that CPR1 from Arabidopsis thaliana (AtCPR1) was capable of reducing rice CYPs, this was inserted into the DEST cassette, constructing a pET-Duet1/DEST::AtCPR1/CYP99A2 expression vector. Strikingly, this led to expression of correctly folded CYP99A2, as determined by detection of a strong peak at 450 nm in CO-binding difference spectra with cell-free lysates from recombinant E. coli (Figure 1A). In addition, these recombinant CYP99A2 preparations are active with syn-pimaradiene, although producing largely syn-pimaradien-19-ol with smaller amounts of the further oxidized syn-pimaradien-19-al and fully oxidized syn-pimaradien-19-oic acid (Figure 1B), this still represents at least a 30-fold increase in activity relative to that previously observed.
Figure 1.
CYP99A2 catalytic activity depends on AtCPR1 expression system. A & C, CO-difference binding absorption spectra from reduced E. coli cell-free lysate following recombinant expression of CYP99A2 from either (A) pETDuet1/DEST::AtCPR1/CYP99A2 or (C) pET-Duet1/AtCPR1/CYP99A2 (i.e., without insertion of the DEST cassette and use of the Gateway cloning system). B & D, GC-MS selected ion (m/z = 257) chromatograms showing extracted product (B & D) from in vitro reactions after syn-pimaradiene was reacted with the cell-free lysates analyzed in (A) or (C), respectively (peak 1, syn-pimaradiene; peak 2, syn-pimaradien-19-ol; peak 3, syn-pimaradien-19-al; peak 4 methyl ester derivative of syn-pimaradien-19-oic acid). E, Reaction catalyzed by CYP99A2 (compounds numbered corresponding to peak labels defined above).
This improved expression system was further evaluated in the context of our metabolic engineering system. For this purpose, along with use of a previously described syn-CPP producing pGGsC construct (Cyr et al. 2007), the immediately upstream syn-pimaradiene synthase OsKSL4 was incorporated via construction of a pCDF-Duet1/DEST::OsKSL4/AtCPR1 vector. Together these led to the production of the fully oxidized syn-pimaradien-19-oic acid, although this was produced in lower yield than the initially formed syn-pimaradien-19-ol, with much smaller quantities of the intermediate syn-pimaradien-19-al observed (Supplemental Figure S1A). By contrast, with use of the originally reported pCDF-Duet1/DEST::CYP99A2/AtCPR1 (along with pGGsC and pDEST14/OsKSL4) only syn-pimaradiene is observed – i.e., no activity is evident for CYP99A2 expressed from this vector.
Kinetic analysis was conducted to evaluate CYP99A2 activity, which indicated that this exhibits a similar affinity for syn-pimaradiene as CYP99A3 (Table 1). By contrast, the lower catalytic turnover measured seems to suggest a somewhat lower catalytic efficiency for CYP99A2 relative to CYP99A3 (Table 1). However, it should be noted that the reported kcat heavily depends on the concentration of “correctly folded” CYP, which is measured using the 450 nm peak height in CO-binding difference spectra, whose relationship to observed activity has been quite variable in our hands – e.g., in certain cases activity can be observed with cell-free lysates for which no such ‘peak’ is observed (Swaminathan et al. 2009).
Table 1.
Steady-state kinetic constants against syn-pimaradiene.
| enzyme | Km (μM) | kcat (s-1) | kcat/KM (s-1μM-1) |
|---|---|---|---|
| CYP76M8*1 | 4 ± 2 | 9.0 ± 1.0 (× 10−3) | 2.3 × 10−3 |
| CYP99A2 | 1.2 ± 0.1 | 8.3 ± 0.1 | 7.0 |
| CYP99A3*2 | 2.0 ± 0.5 | 46 ± 2 | 23 |
| CYP701A8 | 27 ± 6 | 1.4 ± 0.1 | 5.2 × 10−2 |
Data from previous reports
(Wang et al. 2012a)
(Wang et al. 2011).
Given the dramatic increase in functional recombinant expression observed by directly cloning CYP99A2 into pET-Duet1, we hypothesized that increased activity might be obtained doing the same with AtCPR1 as well. Accordingly, CYP99A2 was cloned in MCS2 of pET-Duet1, and AtCPR1 into MCS1, creating a pET-Duet1/AtCPR1/CYP99A2 expression vector. This construct provided equivalent yield in the context of our metabolic engineering system (Supplemental Figure S1B). Nevertheless, while this led to observance of a predominant 420 nm peak associated with “incorrectly” folded CYP in CO-binding difference spectra with cell-free lysates from recombinant E. coli transformed with this construct (Figure 1C), these preparations actually exhibited greater activity with syn-pimaradiene relative to the pET-Duet1/DEST::AtCPR1/CYP99A2 construct, including largely producing the fully oxidized syn-pimaradien-19-oic acid (Figure 1D). This was true even when simply using equivalent volumes of cell-free lysates, and the calculated catalytic rate is significantly increased when normalized to measured content of “correctly folded” CYP.
Investigation of CYP701A8 & 9 reveals novel activity with syn-pimaradiene
From the previously reported identification of the rice kaurene oxidase, CYP701A6, and investigation of the transcriptional regulation of the small family of paralogs found in rice (CYP701A7-9 & 19), two were found to exhibit inducible transcription, CYP701A8 & 9, although transcript levels for CYP701A9 were significantly lower than those for CYP701A8 (Itoh et al. 2004). We later reported biochemical activity for CYP701A8 consistent with a role in diterpenoid phytoalexin biosynthesis, specifically C3α-hydroxylation of a number of ent-CPP derived diterpene olefins (e.g., ent-cassadiene and ent-sandaracopimaradiene) that are precursors of C3-oxy group containing natural products (Wang et al. 2012b). However, we were unable to detect activity with CYP701A9. Given that the syn-pimaradiene derived momilactones also contain a C3-oxy group, we hypothesized that CYP701A9 might catalyze the corresponding C3-hydroxylation of syn-pimaradiene. Thus, we constructed a pET-Duet1/DEST::AtCPR1/CYP701A9 vector. While no peak was visible at 450 nm in CO-binding difference spectra from an initial cell-free lysate, we also tested this in the context of our metabolic engineering system with syn-pimaradiene. While some activity was evident, only small amounts of syn-pimaradien-19-ol were produced.
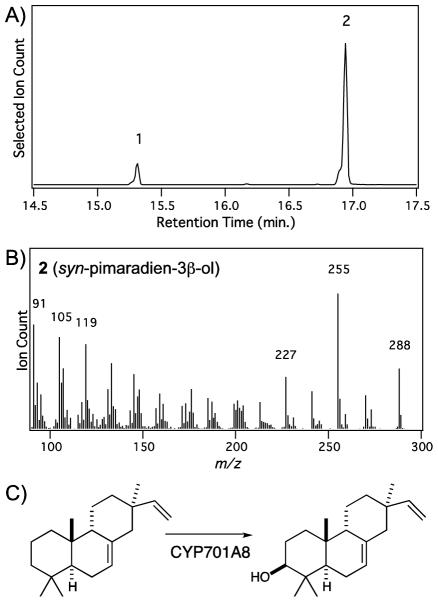
We then hypothesized that CYP701A8 might actually act on syn-pimaradiene, which might be observed upon improved recombinant expression. Accordingly, we constructed a pET-Duet1/DEST::AtCPR1/CYP701A8 vector. Strikingly, with this construct we observed significant conversion of syn-pimaradiene to a novel hydroxylated product, both in vitro and, more importantly, in the context of the metabolic engineering system as well (Figure 2). Hence, by simply scaling up culture volume, we were able to produce sufficient amounts of the novel product for NMR analysis (Supplemental Figure S2 and Supplemental Table S1). The compound was, thus, shown to be syn-pimaradien-3β-ol, consistent with the reported presence of a 3β-hydroxy group in the penultimate precursor to momilactone A (Atawong et al. 2002). However, kinetic analysis indicated that CYP701A8 exhibits a significantly lower affinity for syn-pimaradiene than CYP99A2 & 3 or CYP76M8 (Table 1).
Figure 2.
CYP701A8 catalytic activity. A, GC-MS selected ion (m/z = 272 + 288) chromatogram of extract from E. coli engineered for production of syn-pimaradiene with co-expression of CYP99A2 and AtCPR1 (peak 1, syn-pimaradiene; peak 2: syn-pimaradien-3β-ol). B, Mass spectrum for syn-pimaradien-3β-ol. C, Reaction catalyzed by CYP701A8 with syn-pimaradiene.
Clarifying CYP71Z6 & 7 activity
Previously we reported that CYP71Z6 & 7 exhibited biochemical activities consistent with roles in diterpenoid biosynthesis, with both carrying out C2-hydroxylation, of ent-isokaurene and ent-cassadiene, respectively (Wu et al. 2011). However, the amount of these products obtained was insufficient for determination of hydroxyl group configuration due to inefficient turnover of the diterpene olefin substrates, as the yield of hydroxylated compounds was only 5-15% of total diterpenoid. Hypothesizing that improved expression of CYP71Z6 & 7 might lead to higher turnover, enabling stereochemical resolution, we constructed pET-Duet1/DEST::AtCPR1/ (CYP71Z6 or CYP71Z7) vectors.
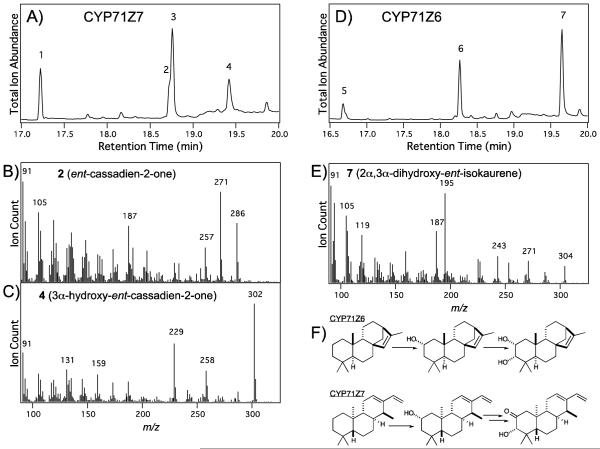
Use of the relevant construct led to improved activity for CYP71Z7 against ent-cassadiene in the metabolic engineering system, with observation of two new products beside the previously reported ent-cassadien-2-ol, such that oxygenated diterpenoids were predominant (Figure 3A). The apparent molecular ions for the new compounds suggested that these were an ent-cassadienone (m/z = 286) and further hydroxylated derivative (m/z = 302), respectively (Figure 3B & C). Again, by simply scaling up culture volume, we were able to produce sufficient amounts of both the 2-hydroxy and novel dienonol products for complete NMR analysis (Supplemental Figures S3 & S4 and Supplemental Tables S2 & S3). The 2-hydroxy group was, thus, shown to occupy the α position – i.e., CYP71Z7 produces 2α-hydroxy-ent-cassadiene – consistent with the presence of a 2α-hydroxy group in the derived phytocassanes A & B. In addition, the novel dienonol compound was found to be 3α-hydroxy-ent-cassadien-2-one, comprising a subset of the oxy groups found in phytocassane D (3α-hydroxy-ent-cassadien-2,11-dione).
Figure 3.
CYP71Z6 and CYP71Z7 catalytic activities. A & D, GC-MS chromatograms of cell-free lysate from E. coli engineered for production of either (A) ent-cassadiene with co-expression of CYP71Z7 and AtCPR1 (peak 1, ent-cassadiene; peak 2, ent-cassadien-2-one; peak 3; 2α-hydroxy-ent-cassadiene, peak 4, 3α-hydroxy-ent-cassadien-2-one), or (D) ent-isokaurene with co-expression of CYP71Z6 and AtCPR1 (peak 5, ent-isokaurene; peak 6, 2α-hydroxy-ent-isokaurene; peak 7, 2α,3α-dihydroxy-ent-isokaurene). B, C & E, Mass spectra from (B) ent-cassadien-2-one, (C) 3α-hydroxy-ent-cassadien-2-one, or (E) 2α,3α-dihydroxy-ent-isokaurene. F, Reactions catalyzed by CYP71Z6 & CYP71Z7 (as indicated).
Similarly, use of the relevant construct also led to improved activity for CYP71Z6 against ent-isokaurene in the metabolic engineering system, with even more predominant yield of hydroxylated diterpenoids (Figure 3D). Again, in addition to the previously observed 2-hydroxy product, a novel compound was observed whose apparent molecular ion (m/z = 304) suggested it was a dihydroxyl derivative (Figure 3E). By scaling up the cultures we were able produce sufficient amounts of both the 2-hydroxy and novel diol product for NMR analysis (Supplemental Figures S3 & S5 and Supplemental Table S4). The 2-hydroxy was, thus, shown to occupy the α position – i.e., CYP71Z6 produces 2α-hydroxy-ent-isokaurene. In addition, the novel diol was found to be 2α,3α-dihydroxy-ent-isokaurene.
DISCUSSION
Optimization of CYP expression
Functional recombinant expression of eukaryotic microsomal CYPs in E. coli is generally considered challenging. In previous work we have reported that the use of N-terminally modified and synthetic (codon-optimized) CYP genes leads to successful recombinant expression in many, but not all cases (Kitaoka et al. in press). Here we report further optimization, through vector choice and sub-cloning procedure to achieve ideal spacing to a single (i.e., non-redundant) Shine-Dalgarno ribosome-binding site, which has not only enabled functional expression of previously recalcitrant CYPs (i.e., CYP99A2 and CYP701A9), but also detection of new activity with previously investigated CYPs (i.e., CYP71Z6 & 7 and CYP701A8).
CYPs in momilactone biosynthesis
The momilactones contain a minimum of three oxygenated carbons (C3, C6 and C19). We have previously reported that CYP76M8 will carry out C6β-hydroxylation of syn-pimaradiene, consistent with the 19,6β-configuration of the eponymous lactone ring (Wang et al. 2012a). Here we report that CYP99A2 acts as a C19-oxidase with syn-pimaradiene (Figure 1). Although this activity is redundant with that previously reported for CYP99A3 (Wang et al. 2011), the higher transcript expression level for CYP99A2 nevertheless suggests that this may be relevant. More critically, as reported here CYP701A8 can act as a syn-pimaradiene C3β-hydroxylase (Figure 2), with such configuration consistent with previous work on momilactone biosynthesis (Atawong et al. 2002).
While still targeting C3, the change in stereospecificity of CYP701A8 with syn-pimaradiene relative to ent-CPP derived olefins is intriguing. Perhaps the most obvious similarity around the targeted position is the presence of co-axial methyl groups at C4 and C10. However, structural examination of the ent-CPP derived diterpenes with which the 3α position is targeted (e.g., ent-cassadiene) versus that of syn-pimaradiene for which the 3β position is hydroxylated reveals differences even in the relative orientation of these groups (Figure 4A).
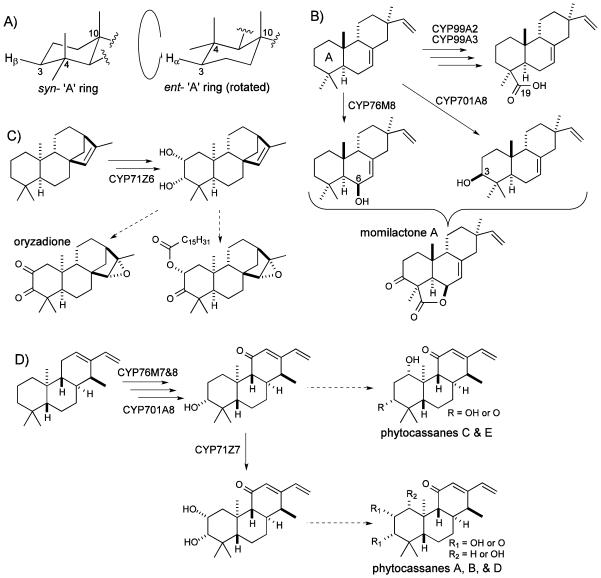
Figure 4.
Putative roles for CYP activities reported here in rice diterpenoid biosynthesis. A, Difference in configurations of ‘A’ rings from syn- or ent-CPP derived diterpenes, with regards to hydrogen targeted by CYP701A8 (note that the ent- ‘A’ ring has been “flipped over” relative to the standard orientation, and the syn- ‘A’ ring). B, Three CYP activities against syn-pimaradiene targeting all three oxygenated positions common to the momilactones. C, Putative role of CYP71Z6 in production of oryzadione and related fatty acid ester derivative. D, Role of CYP71Z7 in phytocassane biosynthetic network.
Regardless of the underlying structure-function relationships, with identification of CYP701A8 as a C3β-hydroxylase, CYPs targeting all three positions relevant for momilactone biosynthesis are now known (Figure 4B). However, in each case the relevant activity has only been observed with the common substrate syn-pimaradiene, leaving the order in which these CYPs act unclear. From the measured kinetic constants it seems likely that CYP701A8 acts later given its substantially higher KM, but such comparison still leaves open the relative order in which CYP99A2 & 3 versus CYP76M8 act, as these have approximately equivalent KM values for syn-pimaradiene (Table 1). Intriguingly, in addition to acting as a C19 oxidase, CYP99A2 also seems to produce small amounts of syn-pimaradien-3β-ol (data not shown), but does not further elaborate this, consistent with addition of the 3β-hydroxy group at a later step in momilactone biosynthesis.
CYP71Z6 & 7 activity in diterpenoid biosynthesis
We have previously suggested that the C2-hydroxylase activity of CYP71Z6 with ent-isokaurene might be relevant to orzyalide biosynthesis (Wu et al. 2011). This hypothesis was based in part on the assumption that the resulting compound was 2β-hydroxy-ent-isokaurene, corresponding to the previous isolation of 2β,3α-dihydroxy-15,16-epoxy-ent-kaurane from rice (Watanabe et al. 1992). However, as shown here CYP71Z6 actually produces 2α-hydroxy-ent-isokaurene instead (Figure 3). While this difference in configuration does not rule out a role for CYP71Z6 in oryzalide biosynthesis (e.g., the observed rice metabolite may not serve as an intermediate in such biosynthesis), it does leave it in question. On the other hand, a related compound with a C2α ester linked fatty acid has been reported (Kono et al. 2004), indicating that 2α-hydroxylation is relevant to rice diterpenoid biosynthesis (Figure 4C). Moreover, the further hydroxylation catalyzed by CYP71Z6 to 2α,3α-dihydroxy-ent-isokaurene is potentially relevant to biosynthesis of oryzadione (2,3-diketo-15,16-epoxy-ent-kaurane).
The production of 2α-hydroxy-ent-cassadiene by CYP71Z7 is more clearly relevant to phytocassane biosynthesis, as phytocassanes A & B both contain a 2α-hydroxy substituent. In addition, the further oxidation of this hydroxyl to a keto by CYP71Z7 also may be relevant to the production of phytocassane D, which contains a 2-oxo group. However, the 3α-hydroxylation subsequently catalyzed by CYP71Z7 seems unlikely to be relevant given the much higher affinity exhibited by CYP701A8 (KM ~200 versus 4 µM, respectively), which also catalyzes 3α-hydroxylation of ent-cassadiene (Wang et al. 2012a; Wu et al. 2011). Indeed, it seems likely that CYP71Z7 acts at a later stage of phytocassane biosynthesis (Figure 4D), as RNAi mediated knock-down of the encoding transcript suppresses accumulation of the C2-oxygenated (i.e., A, B & D), while increasing that of the other phytocassanes (Okada 2011).
Insight into CYP paralogs
Given the previously reported higher expression level of CYP99A2 relative to its closely related paralog CYP99A3 (Shimura et al. 2007), we were particularly interested in characterizing the biochemical activity of the previously recalcitrant CYP99A2. This was enabled by the optimized recombinant expression approach developed here, which revealed potential redundancy of the CYP99A2 & 3 paralogs in momilactone biosynthesis. By contrast, with the similarly closely related paralogs CYP701A8 & 9, our results raise the possibility that CYP701A9 has lost catalytic activity and may be in the process of becoming a pseudo-gene, which would be consistent with the previously observed low expression level (Itoh et al. 2004).
Supplementary Material
Acknowledgments
Funding: This work was supported by grants from the USDA (AFRI-NIFA 2014-67013-21720) and NIH (GM076324), and funds from Iowa State University, to R.J.P.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
REFERENCES
- Atawong A, Hasegawa M, Kodama O. Biosynthesis of rice phytoalexin: enzymatic conversion of 3ß-hydroxy-9ß-pimara-7,15-dien-19,6ß-olide to momilactone A. Biosci Biotechnol Biochem. 2002;66(3):566–570. doi: 10.1271/bbb.66.566. [DOI] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ. A Modular Approach for Facile Biosynthesis of Labdane-Related Diterpenes. J Am Chem Soc. 2007;129:6684–6685. doi: 10.1021/ja071158n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Bak S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368(1612):20120426. doi: 10.1098/rstb.2012.0426. doi:10.1098/rstb.2012.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M. A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol. 2004;54:533–547. doi: 10.1023/B:PLAN.0000038261.21060.47. [DOI] [PubMed] [Google Scholar]
- Kitaoka N, Lu X, Yang B, Peters RJ. The application of synthetic biology to elucidation of plant terpenoid metabolism. Mol Plan. doi: 10.1016/j.molp.2014.12.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono Y, Kojima A, Nagai R, Watanabe M, Kawashima T, Onizawa T, Teraoka T, Watanab M, Koshino H, Uzawa J, Suzuki Y, Sakurai A. Antibacterial diterpenes and their fatty acid conjugates from rice leaves. Phytochemistry. 2004;65(9):1291–8. doi: 10.1016/j.phytochem.2004.03.016. doi:10.1016/j.phytochem.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ohta D. Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 1998;116:357–367. doi: 10.1104/pp.116.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Sato F. Unusual P450 reactions in plant secondary metabolism. Arch Biochem Biophys. 2011;507(1):194–203. doi: 10.1016/j.abb.2010.09.026. doi:10.1016/j.abb.2010.09.026. [DOI] [PubMed] [Google Scholar]
- Morrone D, Chen X, Coates RM, Peters RJ. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem J. 2010a;431(3):337–344. doi: 10.1042/BJ20100597. doi:10.1042/BJ20100597. [DOI] [PubMed] [Google Scholar]
- Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, Peters RJ. Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol. 2010b;85:1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K. The biosynthesis of isoprenoids and the mechanisms regulating it in plants. Biosci Biotechnol Biochem. 2011;75(7):1219–25. doi: 10.1271/bbb.110228. [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsome II. Solubilization, purification, and properties. J Biol Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- Peters RJ. Two rings in them all: The labdane-related diterpenoids. Nat Prod Rep. 2010;27(11):1521–1530. doi: 10.1039/c0np00019a. doi:10.1039/C0NP00019A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust LM, Sherman DH. Diversity of P450 enzymes in the biosynthesis of natural products. Nat Prod Rep. 2012;29(10):1251–66. doi: 10.1039/c2np20020a. doi:10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ. Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 2004;136(4):4228–4236. doi: 10.1104/pp.104.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 2014;79(4):659–678. doi: 10.1111/tpj.12436. doi:10.1111/tpj.12436. [DOI] [PubMed] [Google Scholar]
- Shimura K, Okada A, Okada K, Jikumaru Y, Ko K-W, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N, Koga J, Nojiri H, Yamane H. Identification of a biosynthetic gene cluster in rice for momilactones. J Biol Chem. 2007;282:34013–34018. doi: 10.1074/jbc.M703344200. [DOI] [PubMed] [Google Scholar]
- Sono M, Roach MP, Coulter ED, Dawson JH. Heme-Containing Oxygenases. Chem Rev. 1996;96(7):2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell. 2009;21:3315–3325. doi: 10.1105/tpc.108.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272(31):19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Okada K, Yamazaki K, Wu Y, Swaminathan S, Yamane H, Peters RJ. Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J Biol Chem. 2012a;287(9):6159–6168. doi: 10.1074/jbc.M111.305599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Peters RJ. CYP99A3: Functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J. 2011;65(1):87–95. doi: 10.1111/j.1365-313X.2010.04408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Wu Y, Peters RJ. CYP701A8: A rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 2012b;158(3):1418–1425. doi: 10.1104/pp.111.187518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kono Y, Uzawa J, Teraoka T, Hosokawa D, Suzuki Y, Sakurai A, Teraguchi M. Structures of Oryzalic Acid B and Three Related Compounds, a Group of Novel Antibacterial Diterpenes, Isolated from Leaves of a Bacterial Leaf Blight-Resistant Cultivar of Rice. Biosci Biotechnol Biochem. 1992;56:113–117. [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 2004;135(4):2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hillwig ML, Wang Q, Peters RJ. Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization of CYP71Z6 & 7. FEBS Lett. 2011;585:3446–3451. doi: 10.1016/j.febslet.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang Q, Hillwig ML, Peters RJ. Picking sides: Distinct roles for CYP76M6 and -8 in rice oryzalexin biosynthesis. Biochem J. 2013;454(2):209–216. doi: 10.1042/BJ20130574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ. To Gibberellins and Beyond! Surveying the Evolution of (Di)Terpenoid Metabolism. Annu Rev Plant Biol. 2014a;65:259–286. doi: 10.1146/annurev-arplant-050213-035705. doi:10.1146/annurev-arplant-050213-035705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Matsuba Y, Hong Y, Jackson A, Pichersky E, Tantillo DJ, Peters RJ. Biosynthesis of lycosantalonol, a cis-prenyl derived diterpenoid. J Am Chem Soc. 2014b;136:16951–16953. doi: 10.1021/ja508477e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Peters RJ. Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae. Org Biomol Chem. 2013;11(44):7650–2. doi: 10.1039/c3ob41885e. doi:10.1039/c3ob41885e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.