SUMMARY
We describe a computational approach that integrates GRO-seq and RNA-seq data to annotate long noncoding RNAs (lncRNAs), with increased sensitivity for low abundance lncRNAs. We used this approach to characterize the lncRNA transcriptome in MCF-7 human breast cancer cells, including >700 previously unannotated lncRNAs. We then used information about the (1) transcription of lncRNA genes from GRO-seq, (2) steady-state levels of lncRNA transcripts in cell lines and patient samples from RNA-seq, and (3) histone modifications and factor binding at lncRNA gene promoters from ChIP-seq to explore lncRNA gene structure and regulation, as well as lncRNA transcript stability, regulation, and function. Functional analysis of selected lncRNAs with altered expression in breast cancers revealed roles in cell proliferation, regulation of an E2F-dependent cell cycle gene expression program, and estrogen-dependent mitogenic growth. Collectively, our studies demonstrate the use of an integrated genomic and molecular approach to identify and characterize growth-regulating lncRNAs in cancers.
Keywords: Breast cancer, Enhancer, Enhancer Transcription, Estrogen, Estrogen Receptor (ERα), Gene Regulation, GRO-seq, Long Noncoding RNA (LncRNA), Noncoding RNA, Nucleus, RNA-seq, Transcription
INTRODUCTION
Long noncoding RNAs (lncRNAs) are RNA molecules longer than 200 nucleotides that share many features with messenger RNAs (mRNAs): they are 5′ capped, polyadenylated, spliced and, in many cases, exported from the nucleus (Rinn and Chang, 2012; Sun and Kraus, 2013). The major feature distinguishing these two classes of “long” RNAs is that lncRNAs lack significant protein coding potential (Rinn and Chang, 2012; Sun and Kraus, 2013). As such, lncRNAs mediate their biological roles as RNAs, rather than as templates for protein synthesis. Recent studies based on next generation (“deep”) sequencing technologies have identified tens of thousands of lncRNAs expressed in a wide variety of cell types (Cabili et al., 2011; Derrien et al., 2012; Djebali et al., 2012; Sun and Kraus, 2014; Ulitsky and Bartel, 2013; Volders et al., 2013). The high degree of cell type-specific expression and limited evolutionary conservation of lncRNAs, however, suggests that many currently unannotated lncRNAs remain to be discovered and characterized (Guttman et al., 2011; Guttman et al., 2010; Necsulea et al., 2014; Sun et al., 2013).
The advent of next generation sequencing technologies has revolutionized the identification and annotation of lncRNAs. Common approaches include (1) large scale cloning of cDNAs, followed by deep sequencing (Carninci et al., 2005), (2) identification of previously unannotated lncRNA transcription units using characteristic histone modification signatures (i.e., those signatures associated with Pol II transcription, such as enrichment of H3K4me3 at the promoter and H3K4me36 along the gene body) (Guttman et al., 2009; Marques et al., 2013), and (3) RNA-sequencing, followed by in silico assembly of mature lncRNAs (Guttman et al., 2010; Roberts et al., 2011; Trapnell et al., 2010). These approaches have been complemented with global methods for fine mapping of the transcription start sites (TSSs) of the lncRNA genes, as well as the 5′ ends, 3′ ends, and exon/intron boundaries of the lncRNA transcripts (Guttman et al., 2010; Sun and Kraus, 2014; Trapnell et al., 2010; Ulitsky and Bartel, 2013). A wealth of information about lncRNA annotations is now available through databases including GENCODE and LNCipedia (Derrien et al., 2012; Volders et al., 2013).
Functional studies have implicated specific lncRNAs, or classes of lncRNAs, in a wide variety of biological functions in normal and disease states (Guttman et al., 2011; Huarte and Rinn, 2010; Sun et al., 2013; Sun and Kraus, 2014). Although most of the focus has been on nuclear functions of lncRNAs, including the regulation of chromatin structure and gene expression (Rinn and Chang, 2012; Sun and Kraus, 2013; Vance and Ponting, 2014; Wang and Chang, 2011), recent studies have suggested a broad range of molecular and biochemical functions of lncRNAs across the cell, including the cytoplasm (Geisler and Coller, 2013; Sun and Kraus, 2014; van Heesch et al., 2014). In the nucleus, lncRNAs function as epigenetic and transcriptional regulators by acting as scaffolds for the assembly of chromatin- and gene-regulating complexes, or guides to direct such complexes to specific sites in the genome (Rinn and Chang, 2012; Sun and Kraus, 2013; Vance and Ponting, 2014; Wang and Chang, 2011). In the cytoplasm, lncRNAs function to control mRNA processing, mRNA post-transcriptional regulation, cellular signaling, and protein activity through allosteric regulation (Geisler and Coller, 2013; Sun and Kraus, 2014).
Recent studies have implicated specific lncRNAs in the etiology of a diverse array of cancer types (Huarte and Rinn, 2010; Sun and Kraus, 2014), including hormone-responsive cancers, such as prostate cancers (e.g., the lncRNAs SChLAP1, PCAT-1, PCGEM1, and PRNCR1) (Du et al., 2013; Prensner et al., 2011; Prensner et al., 2013) and breast cancers (e.g., BCAR4) (Xing et al., 2014). Although additional studies have suggested potential roles for other lncRNAs in breast cancers (e.g., HOTAIR, GAS5, and SRA), the molecular mechanisms whereby these lncRNAs contribute to breast cancer biology are unclear (Gupta et al., 2010; Sun and Kraus, 2014). In the studies described herein, we have identified ~1900 lncRNAs in MCF-7 human breast cancer cells, including more than 700 lncRNAs not previously annotated, using a computational approach that integrates GRO-seq and RNA-seq data. Functional analyses of two of these lncRNAs, lncRNA152 and lncRNA67, suggest roles in cell proliferation and the regulation of an E2F-dependent cell cycle gene expression program, which might underlie some breast cancer phenotypes.
RESULTS
Generating a Comprehensive Catalog of LncRNAs in MCF-7 Cells Through Integrative Analysis of RNA-Seq and GRO-Seq Data
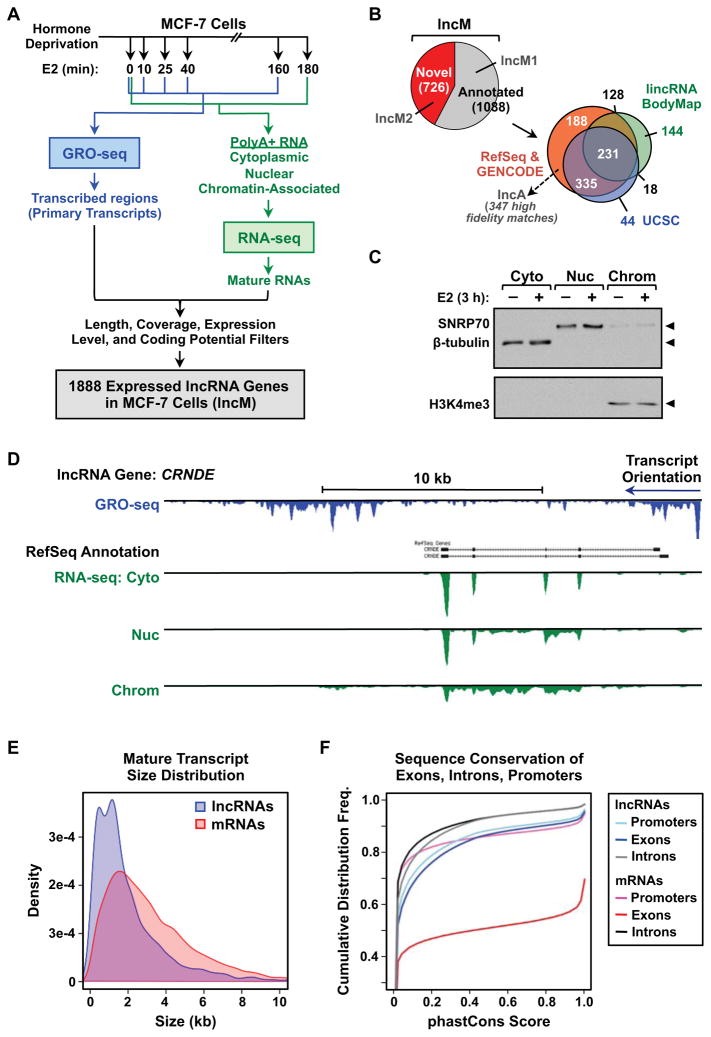
To identify and annotate lncRNAs in the estrogen receptor alpha (ERa)-positive MCF-7 breast cancer cell line, we developed a computational approach that incorporates evidence of RNA transcripts from multiple high-throughput sequencing approaches over a time course of treatment with 17β-estradiol (E2). In brief, our pipeline consists of three major parts: (1) mapping and assembly of RNA transcripts from polyadenylated (polyA+) RNA-seq (steady-state transcripts; 0 and 3 hours of E2) (Roberts et al., 2011; Trapnell et al., 2010), (2) integration of nascent RNA profiles from GRO-seq (primary transcripts or transcription units; 0, 10, 25, 40 and 160 min. of E2) (Hah et al., 2011), and (3) processing and filtering of transcripts based on length, coverage, expression levels, and coding potential (Fig. 1A; Fig. S1A). This pipeline yielded a catalog of 1,888 expressed lncRNA genes in MCF-7 cells, which we call the “lncM” set (described in detail below). Of these, 726 (38%) were not previously annotated in current lncRNA databases (e.g., RefSeq, GENCODE, UCSC, and the lincRNA BodyMap) (Fig. 1B). Furthermore, at least two thirds our unannotated lncRNAs were not identified in a recent “comprehensive” discovery study based on 7,256 RNA-seq libraries from tumors, normal tissues, and cell lines (Iyer et al., 2015).
Figure 1. Integrative analysis of GRO-seq and RNA-seq data generates a comprehensive catalog of lncRNA genes in MCF-7 cells.
(A) Overview of the experimental and computational analysis pipeline for the identification of the lncRNA genes set in MCF-7 cells (lncM).
(B) Venn diagram showing the fraction of lncM genes that were previously annotated, and their overlap with annotations from RefSeq, GENCODE, UCSC Genome Browser, and the lincRNA BodyMap databases. The analysis was performed using information downloaded from the data bases in April 2014 and GENCODE ver. 19 released through the UCSC Genome Browser.
(C) Western blot showing the successful subcellular fractionation of MCF-7 cells for RNA-seq. β-tubulin (cytoplasm), SNRP70 (nucleus), and H3K4me3 (chromatin) are fraction-specific markers.
(D) Genome browser view for the locus of an annotated lncRNA gene, CRNDE, showing the RefSeq annotation, as well as GRO-seq, and fractionated RNA-seq data.
(E) Comparisons of mature transcript size distributions between lncRNA and mRNA genes assembled from cytoplasmic RNA-seq data.
(F) Cumulative distribution frequency curves showing the sequence conservation of lncRNA and mRNA genes assembled from cytoplasmic RNA-seq data.
See also Figure S1.
We sequenced polyA+ RNAs from whole cells and from each of three subcellular fractions (i.e., cytoplasmic, nucleoplasmic, and chromatin-associated; Fig. 1C) to evaluate the subcellular distribution and processing of the lncRNAs. We found that cytoplasmic lncRNAs are more completely spliced, while lncRNAs in the nucleus often contain varying amounts of unspliced introns (Fig. 1D). Thus, when annotating the lncM set, we relied on cytoplasmic RNAs (lncCyto; Fig. S1A) for the determination of exon-intron structures when possible, since it is more accurate. However, we also included transcripts that can only be assembled from nuclear RNAs, so that no lncRNAs were excluded.
In our annotation pipeline, we applied filters to ensure that the lncRNAs in the lncM set are reasonably “long,” most likely noncoding, and reliably expressed (Fig. 1A; Fig. S1A). In this regard, we (1) used a length cutoff of 200 nt for multi-exon transcripts and 1000 nt for single-exon transcripts, (2) excluded transcripts overlapping known protein coding RNAs transcribed in the same direction and eliminated transcripts with high codon substitution frequencies (i.e., phyloCSF score > 150; (Lin et al., 2011)), and (3) used a coverage threshold of 10 reads/base, with an RNA-seq FPKM > 1. Finally, we removed lncRNAs that lack evidence of a primary transcript, as determined by GRO-seq (Fig. 1A; Fig. S1A). Together, these filters helped to ensure that our annotation pipeline is sensitive and specific (Fig. S1B), and the lncM annotations are of high quality and fidelity (Fig. 1D).
Length, Exon Structure, Subcellular Distribution, and Stability of LncRNAs in MCF-7 Cells
Next, we explored the length and structure of the lncRNAs, focusing on those assembled from cytoplasmic RNAs (lncCyto; Fig. S1A), since they yielded more accurate exon-intron calls. We observed that lncRNAs have fewer distinct spliced isoforms than mRNAs (Fig. S1C), and the length of the mature lncRNAs is generally shorter, which can be attributed to a reduced number of exons per transcript (Fig. 1E; Fig S1, D and E). In addition, we observed that lncRNAs are evolutionarily less conserved compared to mRNAs, yet they display local areas of modest conservation (as measured by phastCons scores; (Siepel et al., 2005)) in the exon and promoter regions relative to the intron regions (Fig. 1F). These observations are consistent with previous reports (Derrien et al., 2012; Necsulea et al., 2014; Ulitsky and Bartel, 2013; Volders et al., 2013).
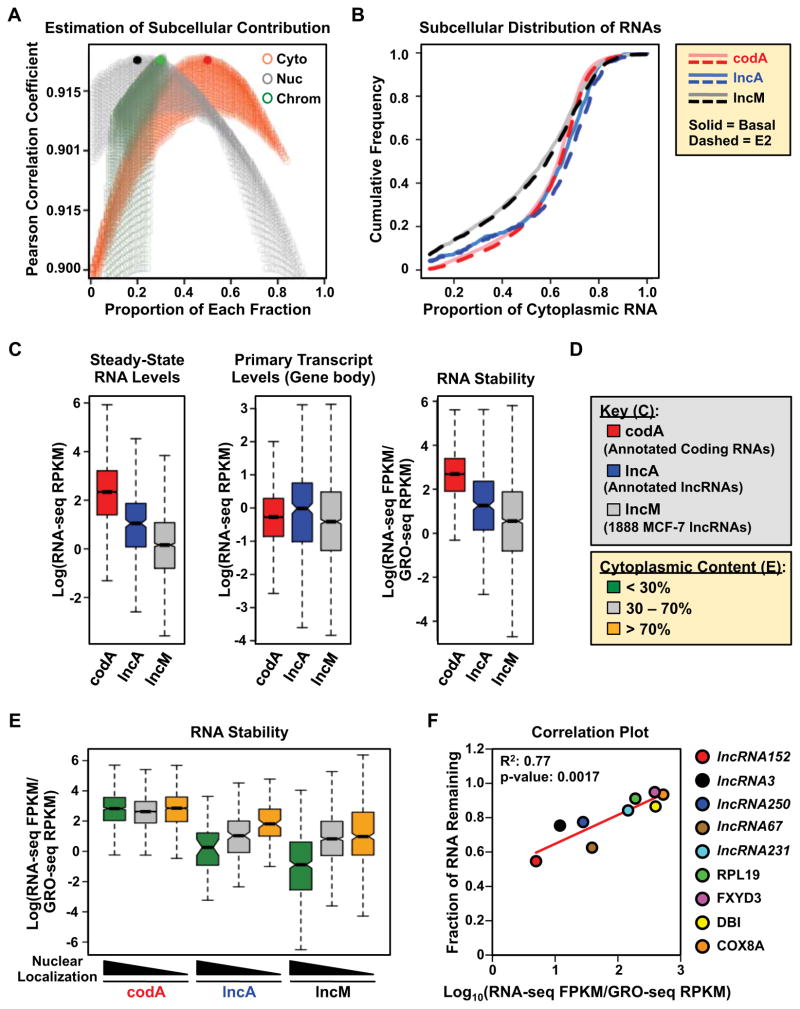
To determine the subcellular localization of lncRNAs, we calculated ratios of transcript abundance in the nuclear fractions (i.e., nucleoplasm or chromatin-associated) over the cytoplasmic fraction. These indicated a slight, but significant, enrichment of the lncM set in the nucleoplasm and on chromatin relative to protein-coding RNAs (the “codA” set) (Fig. S2A). A similar enrichment in the nucleus was not observed for previously annotated lncRNAs (the “lncA” set, a subset of lncM containing 347 lncRNAs that match perfectly with annotations in either the RefSeq or GENCODE databases; Fig. S2A), suggesting that our pipeline detects a larger fraction of nuclear lncRNAs than previous approaches. Next, we estimated the contribution of each subcellular fraction to the total RNA pool to determine the distribution of each transcript (Fig. 2A). Again, we observed that a smaller fraction of the lncM RNAs are found in the cytoplasm, compared to the codA and lncA RNAs (Fig. 2B), although a significant fraction of lncRNAs is enriched in the cytoplasm (Fig. 2, A and B).
Figure 2. Nucleus-enriched lncRNAs are less stable than cytoplasm-enriched lncRNAs.
(A) Estimation of the contribution of each subcellular fraction to the total RNA content, based on the relationship a x Cyto +b x Nuc +c x Chr = Total. Pearson correlation coefficients are plotted for every pair of a x Cyto +b x Nuc +c x Chr and Total as the contribution of each fraction, a, b and c, are sampled from 0.01 to 0.99. The combination, a = 0.5; b = 0.2; c = 0.3, gives the highest correlation (solid circles).
(B) Cumulative frequency curves showing the extent of cytoplasmic localization of codA, lncA, and lncM RNAs in basal (solid) and E2-treated (dotted) conditions.
(C) Steady-state RNA levels (from RNA-seq) (left), Primary transcript levels (from GRO-seq) (middle), and relative stability (right) of annotated mRNAs (codA), annotated lncRNAs (lncA), and the lncM lncRNA set. The relative stability of RNAs is represented by the ratio of steady-state RNA levels to nascent transcript levels.
(D) Color keys for (C) through (E).
(E) Box plot showing the relative stability of codA, lncA and lncM RNAs in untreated MCF-7 cells, grouped by the extent of cytoplasmic localization.
(F) Correlation between computed RNA stability (as in panel C, right) and experimentally determined RNA stability from actinomycin D treated MCF-7 cells. Pearson correlation coefficient.
In (C) and (E), Log = the natural log (loge).
See also Figure S2.
Next, we examined the stability of the lncM and lncA lncRNAs. We observed that both have lower steady-state levels than codA mRNAs regardless of where they are localized (Fig. 2C, left and Fig. 2D; Fig. S2B), in spite of being transcribed to similar levels (Fig. 2C, middle). We devised a metric for estimating the stability of the lncM, lncA, and codA RNAs by determining the ratio of their steady-state RNA levels to their transcription levels (Fig. 2C, right). These analyses suggest that the lower steady-state levels of lncRNA compared to mRNAs are likely due to lower stability, rather than lower transcription. Interestingly, nuclear lncRNAs are significantly less stable than cytoplasmic lncRNAs (Fig. 2E; Fig. S2C), consistent with previous results (Clark et al., 2012). Furthermore, we observed that the previously unannotated lncRNAs from the lncM set (lncM2) are more nuclear and less stable than all of the other classes of RNAs that we investigated (i.e., codA, lncA, and lncM1, the latter comprising all previously annotated lncM lncRNAs; Fig. S2D). Importantly, the computed stability metric was verified experimentally for a small set of lncRNAs and mRNAs in actinomycin D stability experiments (Fig. 2F). Together, our results suggest that the lower stability of nuclear lncRNAs is why many of them have evaded earlier attempts at detection and annotation. Our approach takes advantage of GRO-seq to improve the sensitivity of detection.
Divergent and Antisense LncRNAs are Highly Transcribed and Are Dominant Contributors to the Chromatin Signatures Associated with LncRNA Gene Promoters
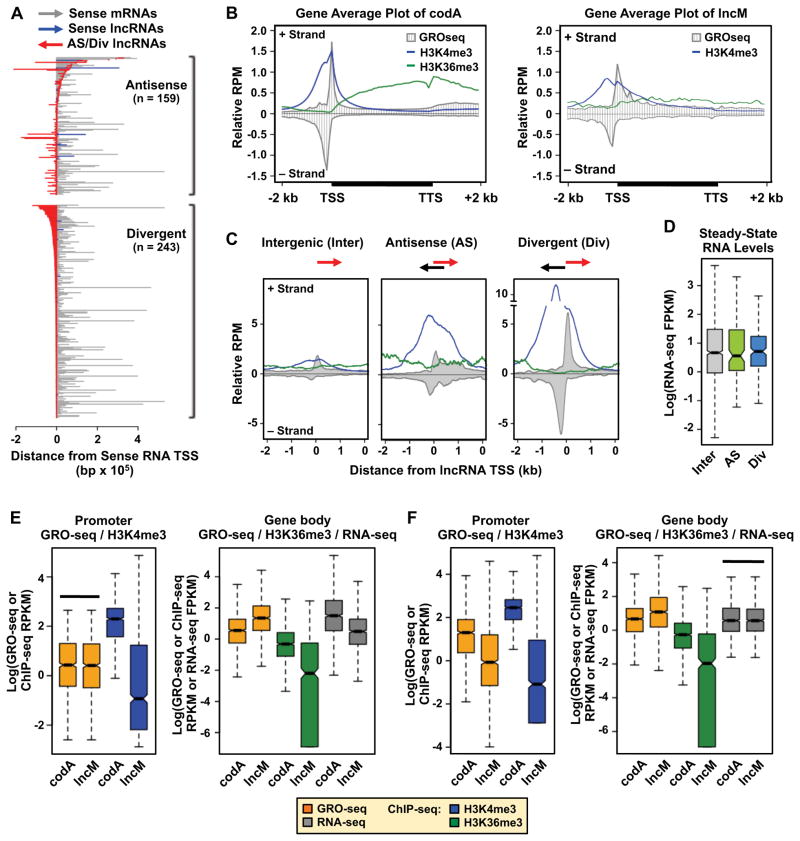
LncRNAs may be transcribed from genes that (1) do not overlap other transcription units (i.e., long intergenic noncoding RNAs, or lincRNAs), (2) run antisense to and overlap, in whole or in part, with a well-defined sense RNA gene (i.e., antisense lncRNAs), or (3) are divergent to and share a bidirectional promoter with an mRNA or a lncRNA gene (Djebali et al., 2012; Sun and Kraus, 2013). In the lncM set, we found that 1,486 (79%) are intergenic (Fig. S3A), 243 (13%) are divergent (13%), and 159 (8.4%) are antisense (Fig. 3A). Thus, a considerable fraction of lncRNAs originates from genic regions.
Figure 3. Divergent and antisense lncRNA genes are transcribed to higher levels and are more enriched for active chromatin marks than intergenic lncRNA genes, which have lower levels of promoter and gene body marks than equally expressed protein-coding genes.
(A) Graphical representation of the orientation, position, and length of antisense (top) and divergent (bottom) lncRNA genes relative to their cognate sense RNA genes.
(B) Average profiles of GRO-seq and ChIP-seq (H3K4me3 and H4K36me3) reads for codA (left) and lncM (right) genes. All gene bodies are scaled to 4 kb.
(C) Average profiles of GRO-seq and ChIP-seq (H3K4me3 and H4K36me3) reads centered on the TSSs of intergenic (Inter), antisense (AS), and divergent (Div) lncM genes.
(D) Box plot showing the steady-state RNA levels of Inter, AS, and Div lncM RNAs.
(E and F) Box plots comparing the levels of (1) active RNA Pol II (GRO-seq) and H3K4me3 (ChIP-seq) at the promoter (left), and (2) actively transcribing RNA Pol II (GRO-seq) in the gene body, H3K36me3 (ChIP-seq) in the gene body, and steady-state RNA levels (RNA-seq) (right) for selected codA genes and intergenic lncM genes. (E) Sampling of codA genes that have the same level of GRO-seq signal at the promoter as the intergenic lncM genes (box plots on the left highlighted by the solid bar above). (F) Sampling of codA genes that have the same level of steady-state RNA as the intergenic lncM genes (box plots on the right highlighted by the solid bar above).
In (D), (E), and (F), Log = the natural log (loge).
See also Figure S3.
In general, the lncM and codA gene sets show similar patterns of transcription (by GRO-seq), H3K4me3 (a mark of active promoters), and H3K36me3 (a mark of actively transcribed gene bodies) (Fig. 3B). In contrast, intergenic lncRNA genes show markedly lower levels of transcription and associated chromatin marks than genic (i.e., antisense and divergent) lncRNA genes (Fig. 3C; Fig. S3B). Thus, intergenic lncRNA genes contribute less to the overall lncM chromatin signature (Fig. 3B, right), in spite of comprising a large fraction of the lncM gene set. Interestingly, genic lncRNA genes have a much greater enrichment of promoter H3K4me3 than intergenic lncRNA genes (Fig. 3C). However, the steady-state levels of the cognate lncRNAs expressed from the genic and intergenic lncRNA genes are comparable (Fig. 3D).
LncRNA Genes Have Lower Levels of Promoter H3K4me3 and Gene Body H3K36me3 Than mRNA Genes
Enriched H3K4me3-H3K36me3 domains have been used to identify lncRNA genes (Guttman et al., 2009; Marques et al., 2013). As noted above, lncRNA genes, in general, may have lower levels of these chromatin marks, and the H3K4me3-H3K36me3 signals from genic lncRNA genes may be influenced by overlapping promoter regions shared with divergent and antisense genes. To test this possibility, we examined the levels of promoter H3K4me3 and gene body H3K36me3 at intergenic lncM genes in comparison to codA genes that do not overlap with antisense or divergent gene loci. For this analysis, we controlled for the levels of transcription (Fig. 3E; GRO-seq) or steady-state RNA (Fig. 3F; RNA-seq) by sampling a set of codA genes that are transcribed or are expressed to similar levels as the intergenic lncM genes (Fig. 3, E and F, respectively; indicated by the black horizontal bar). When controlled either way, the intergenic lncM genes showed significantly lower levels promoter H3K4me3 and gene body H3K36me3 than the well isolated codA genes. Similar results were observed for the lncA set (Fig. S3, C and D). Differences in parameters such as transcript length, coding sequence length, and the number exons do explain the significant differences that we observed between the lncRNA genes and mRNA genes. (Fig. S3, E and F). Thus, the use of enriched H3K4me3-H3K36me3 domains to identify lncRNA genes is likely to under represent intergenic lncRNA genes.
ERα Localizes to the Proximal Promoters of E2-upregulated LncRNA Genes, Which Are Enriched in Enhancer Features
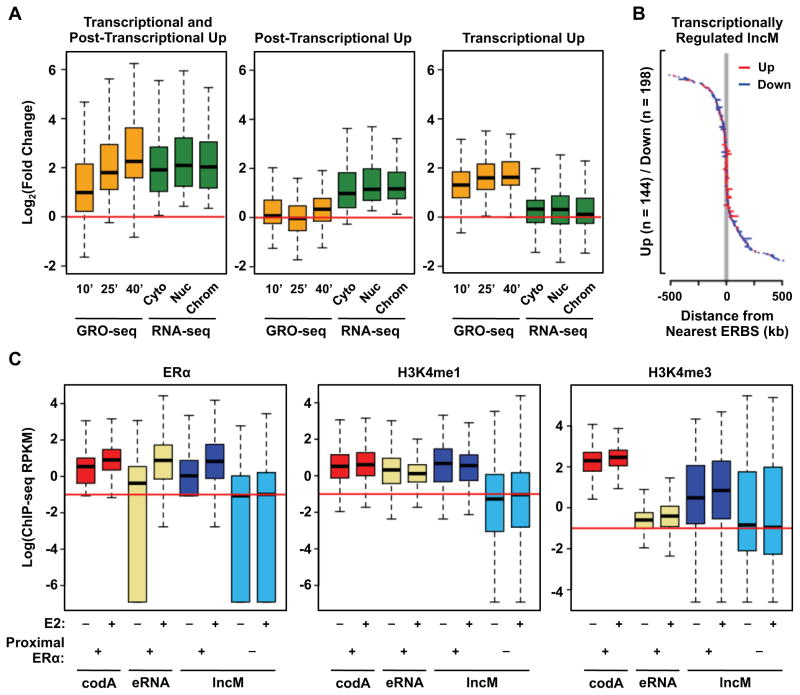
We observed that more than a quarter (531, 28%) of the lncM genes are significantly regulated by E2 (up or down regulated). By comparing regulation that was called based on GRO-seq and RNA-seq, we could distinguish between transcriptional and post-transcriptional effects of estrogen signaling (Fig. 4A; Fig. S4A). We observed changes in the steady-state levels of some lncRNAs, which were reflected in corresponding changes in the transcription of their cognate genes, while others were only affected at the transcriptional level. Not surprisingly, lncRNAs whose expression is coordinately regulated both transcriptionally and post-transcriptionally are also associated with the highest degree of regulation, either up or down (Fig. 4A; Fig. S4A).
Figure 4. ERα localizes to the promoters of a subset of lncM genes, which are enriched for enhancer features.
(A) Box plots showing the E2-induced fold changes in the expression (GRO-seq and RNA-seq) of lncM genes that are upregulated (1) both transcriptionally and post-transcriptionally, (2) post-transcriptionally only, and (3) transcriptionally only.
(B) Graphical representation of gene length and distance from the nearest ERα-binding site (ERBS) for E2-responsive, transcriptionally regulated lncM genes.
(C) Box plots comparing the levels of ERα, as well as an enhancer-associated (i.e., H3K4me1) and a promoter-associated (i.e., H3K4me3) histone marks near the TSSs of (1) codA gene promoters, (2) enhancers that produce eRNAs, and (3) lncM gene promoters, with or without nearby (proximal) ERα binding as indicated. Log = the natural log (loge).
See also Figure S4.
Interestingly, many E2-regulated lncM genes show E2-induced ERα binding at their promoters (Fig. 4B; Fig. S4B). To explore this in more detail, we measured the distance from the TSSs of transcriptionally regulated lncM genes (and codA genes) to the nearest ERα binding site (ERBS) in the E2-treated condition. Interestingly, the promoters of E2-upregulated genes were in closer proximity to an ERBS than the promoters of E2-downregulated genes (Fig. 4B; Fig. S4B), suggesting a direct involvement of ERα the E2-induced transcriptional upregulation of these genes. This result is consistent with an emerging view that lncRNAs may originate from enhancers and contribute to enhancer function (Hah et al., 2013; Orom et al., 2010; Trimarchi et al., 2014).
The coincident location of some lncRNA gene promoters with transcription factor binding sites suggests that lncRNA gene promoters may share common features with enhancers. In this regard, we observed that lncM gene promoters with proximal E2-induced ERBSs are associated with comparable levels of H3K4me1 and H3K27ac (used to define enhancers), pioneer factors, and CBP as ERBSs producing eRNAs (Figs. 4C and S4C). They are also associated with lower levels of H3K4me3 (used to define active gene promoters) than the promoters of ERα-bound protein-coding genes (Fig. 4C, right). These observations support the idea that some lncRNAs originate from enhancers and may contribute to enhancer function.
Cell Type-Specific Expression of LncRNAs Predicts the Intrinsic Molecular Subtype of Breast Cancer Cells and Suggests Unique Biological Functions for LncRNAs
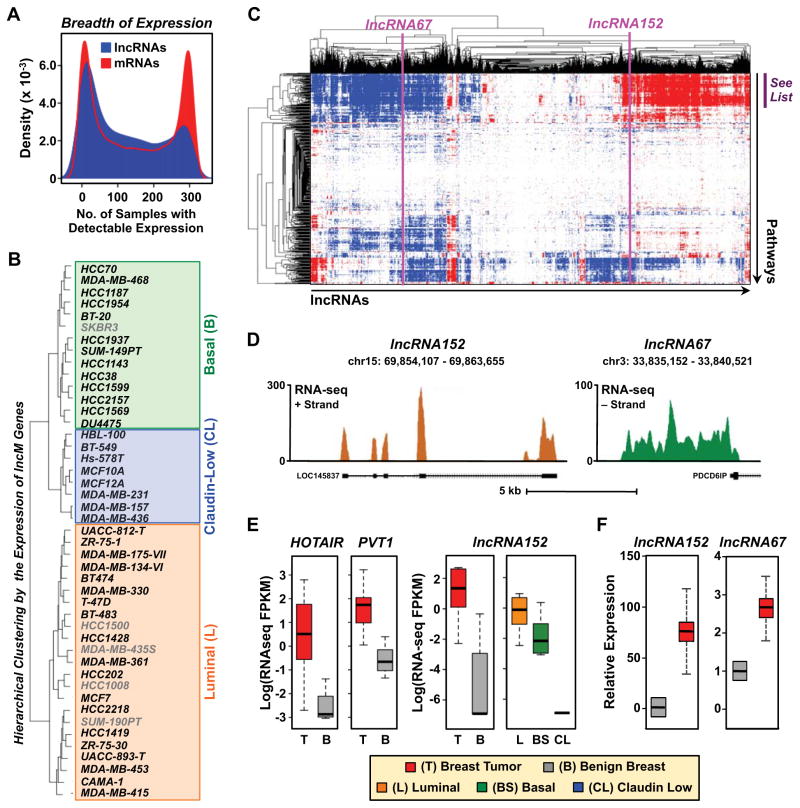
To explore the potential functions of lncRNAs in breast cancer cells, we examined the differential expression of lncRNAs and mRNAs across a panel of 304 cancerous and normal tissue samples and cell lines (Asmann et al., 2011; Kalyana-Sundaram et al., 2012). As shown previously (Rinn and Chang, 2012; Sun and Kraus, 2013; Ulitsky and Bartel, 2013), we observed that lncRNAs have a more tissue- and cell type-specific pattern of expression than mRNAs (Fig. 5A). The expression patterns of lncRNAs across the panel of cancerous and normal tissue samples and cell lines allowed accurate clustering of the samples into their respective tissue types in an unsupervised manner (Fig. S5A). Moreover, the expression patterns of the lncRNAs predicted the intrinsic molecular subtypes of a panel of 45 human breast cancer cell lines with a similar accuracy as the expression patterns of mRNAs (Fig. 5B and data not shown). Guilt-by-association (GBA) analyses (Guttman et al., 2009) suggested functions for the lncM set of lncRNAs in gene regulation and cell proliferation (with terms related to mRNA expression and processing, DNA replication, mitosis, and cell cycle) (Fig. 5C; Fig. S5B).
Figure 5. Differential expression and guilt-by-association analyses connect lncRNA expression to breast cancer biology.
(A) Density plot showing the breadth of expression of lncM lncRNAs and annotated mRNAs across a panel of 304 tissue samples and cell types.
(B) Differential expression analysis of lncRNAs predicts the intrinsic molecular subtype of breast cancer. Hierarchical clustering of 45 breast cancer cell lines into intrinsic molecular subtypes [luminal (L), basal (B), and claudin-low (CL)] based on the differential expression of lncM lncRNAs.
(C) “Guilt-by-association” analysis showing that lncM lncRNAs are associated with cancer-related molecular pathways. The list of pathways demarcated by the purple bar can be found in Figure S6B.
(D) Genome browser views for the loci encoding lncRNA152 (LOC145837) (left) and lncRNA67 (right) showing the mature RNA start and stop sites (mapped by 5′ and 3′ RACE, respectively), cytoplasmic RNA-seq data from MCF-7 cells, and RefSeq annotations.
(E) Box plots comparing the expression of lncRNAs in breast tumors (T, red), benign breast tissues (B, grey), and breast cancer cell lines representing different intrinsic molecular subtypes (Luminal, L; Basal, BS; Claudin low, CL: (left) HOTAIR, PVT1 and (right) lncRNA152. Log = the natural log (loge).
(F) Relative expression of lncRNA152 and lncRNA67 in ERα+ invasive ductal breast carcinomas (red) and benign breast tissues (grey) as determined by RT-qPCR. β-actin mRNA was used as an internal control. Each bar represents the mean + SEM, n = 12. Carcinoma is significantly different than benign breast (Student’s t-test; p-value < 0.05).
See also Figure S5.
Selected LncRNAs Are Required for Breast Cancer Cell Growth and Cell Cycle-Related Gene Expression
We selected two lncRNAs from the lncM set for further functional studies based on their expression patterns, regulation, and association with cell cycle terms in the GBA analyses: (1) lncRNA152 (a.k.a. LOC145837, RP11-279F6.1, or DRAIC (Sakurai et al., 2015)) a previously annotated, but poorly characterized, lncRNA and (2) lncRNA67, a previously unannotated lncRNA (Fig. 5, C and D; Fig. S5C). Like HOTAIR and PVT1 (Fig. 5E, left), two breast cancer-associated lncRNAs that have been characterized previously (Guan et al., 2007; Gupta et al., 2010), lncRNA152 and lncRNA67 show elevated expression in breast tumors compared to benign breast tissue (Fig. 5E, right and 5F) and less aggressive breast cancer cell lines (Fig. S5D). More specifically, lncRNA152 shows elevated expression in luminal (low risk) and basal breast cancers compared to claudin-low breast cancers (Fig. 5E, right), as well as elevated expression in prostate tumors compared to benign prostate tissue (Fig. S5E).
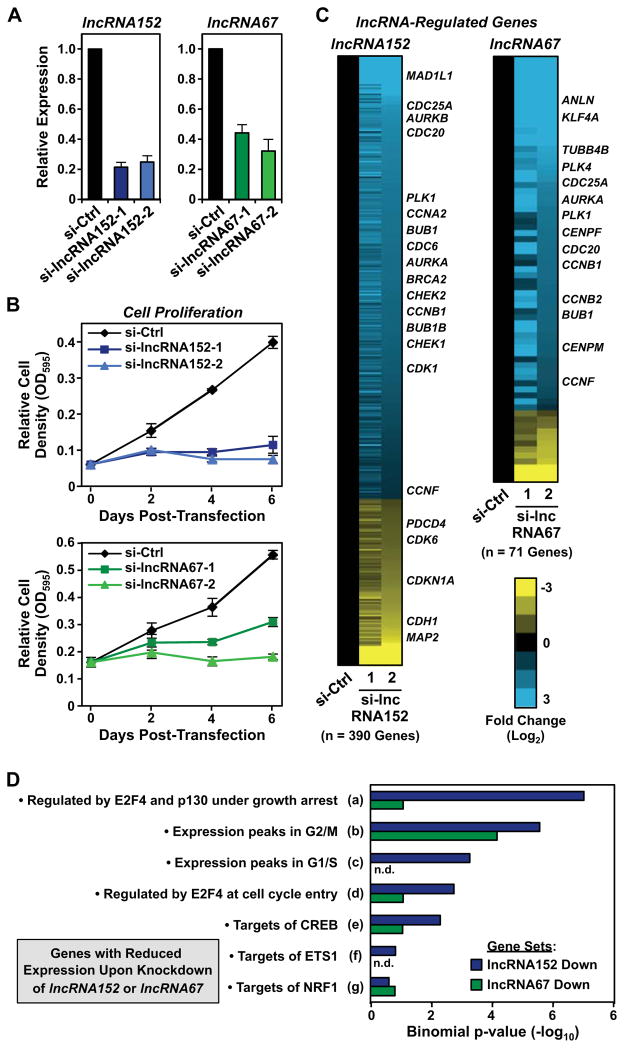
In cell proliferation assays, siRNA-mediated knockdown of lncRNA152 or lncRNA67 (Fig. 6A) dramatically inhibited the growth of MCF-7 cells (Fig. 6B). Knockdown of lncRNA152 also inhibits the growth of LNCaP prostate cancer cells (Sakurai et al., 2015). These results are unlikely to be due to off-target effects of the siRNAs because: (1) similar results were observed when comparing to multiple different control siRNAs (Fig. S6A), (2) three additional siRNAs for each lncRNA showed the same effects on cell growth (not shown), (3) similar effects of the siRNAs on cell growth were observed in another lncRNA152- and lncRNA67-expressing cell line (T47D), but not in a non- or low-expressing cell line (MDA-MB-231) (Fig. S6, B and C), and (4) Dox-induced ectopic expression of lncRNA152 or lncRNA67 increased the growth of MCF-7 cells and partially rescued the effects of siRNA-mediated knockdown (Fig. S6D).
Figure 6. LncRNA152 and lncRNA67 are required for cell cycle-related gene expression programs and the growth of breast cancer cells.
(A) siRNA-mediated knockdown of lncRNA152 (left) or lncRNA67 (right) in MCF-7 cells using two independent siRNA oligos. The expression of lncRNA152 and lncRNA67 after knockdown was monitored by RT-qPCR. β-actin mRNA was used as an internal control. Each bar represents the mean + SEM, n = 3.
(B) Analysis of the growth of MCF-7 cells after control (si-Ctrl) or siRNA-mediated knockdown of lncRNA152 (top) and lncRNA67 (bottom) (si-lncRNA152 and si-lncRNA67, respectively) over 6 day time course post-transfection. Each point represents the mean ± SEM, n = 3.
(C) siRNA-mediated knockdown of lncRNA152 (left) or lncRNA67 (right) alters gene expression in MCF-7 cells. Heat maps showing the relative expression lncRNA-regulated genes and their associated fold changes (log2) in expression upon knockdown of lncRNA152 or lncRNA67 calculated from RNA-seq FPKM values.
(D) Transcription factor target analysis using the GREAT analysis tool performed on the high-confidence set of RefSeq genes downregulated upon siRNA-mediated knockdown of lncRNA152 or lncRNA67. The full gene set descriptors can be found in Figure S7A.
See also Figure S6.
To determine the roles of lncRNA152 and lncRNA67 in the control of gene expression, we performed RNA-seq on control and lncRNA knockdown MCF-7 cells. These analyses also allowed us to validate the specificity of siRNA oligos targeting each lncRNA of interest. We derived high-confidence regulated RefSeq gene lists by filtering the Cuffdiff-called regulated gene lists with a fold cutoff of either 2^(0.8) or 2^(−0.8). Knockdown of lncRNA152 or lncRNA67 significantly altered the expression of 390 and 71 genes, respectively (Fig. 6C; Fig. S6E). For both lncRNA152 and lncRNA67, 70 to 80% of the affected genes had reduced expression upon lncRNA knockdown, suggesting primarily positive roles for the lncRNAs in gene regulation. In both cases, the regulated gene set contains many genes involved in cell cycle regulation, consistent with the cell growth phenotypes observed. Analysis of the downregulated genes in more details using GREAT (Genomic Regions Enrichment of Annotation Tools) (McLean et al., 2010) revealed an enrichment for genes whose expression peaks at key points in the cell cycle and are likely targets of the E2F4 transcription factor (Fig. 6D; Fig. S6F).
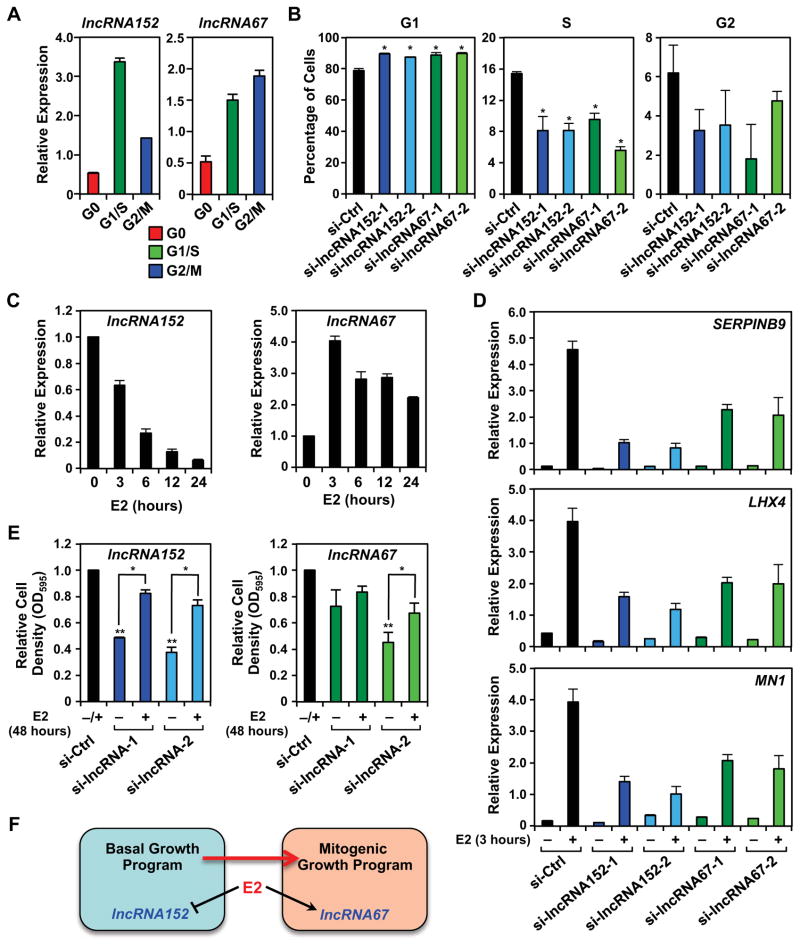
Finally, we explored the biology of lncRNA152 and lncRNA67 in cell cycle control and estrogen-dependent signaling. We observed that the expression of both lncRNA152 and lncRNA67 varies dramatically during the cell cycle, with both showing significant expression during G1/S in MCF-7 cells (Fig. 7A). Additionally, knockdown of either lncRNA promoted accumulation of the cells in G1 phase, with a corresponding reduction in S phase, as assessed by FACS (Fig 7B), consistent with their expression patterns. Since the mitogenic effects of estrogen in breast cancers are mediated, in part, by control of the cell cycle, we examined the interplay between estrogen signaling and lncRNA152 and lncRNA67. Interestingly, estrogen differentially regulated the expression of these lncRNAs, with lncRNA152 downregulated and lncRNA67 upregulated by estrogen treatment in MCF-7 cells (Fig. 7C). In both cases, the effects of estrogen were mediated at the transcriptional level, as determined by GRO-seq (Fig. S7). However, knockdown of either lncRNA inhibited the expression of a subset of estrogen-regulated genes (Fig. 7D). Interestingly, estrogen treatment partially rescued the effects of lncRNA152 knockdown on the growth of MCF-7 cells (Fig. 7E, left), but was less effective at doing so with lncRNA67 knockdown (Fig. 7E, right). Collectively, these results indicate that lncRNA152 and lncRNA67 are required for breast cancer cell growth and cell cycle-related gene expression. Our results suggest that lncRNA152 may be more important for basal growth, whereas lncRNA67 may be more important for estrogen-stimulated growth (Fig. 7F).
Figure 7. LncRNA152 and lncRNA67 regulate the cell cycle and estrogen-dependent signaling in breast cancer cells.
(A) Analysis of lncRNA152 (left) and lncRNA67 (right) expression in MCF-7 cells throughout the cell cycle. Total RNA was isolated from synchronized MCF-7 cells (G0, serum withdrawn; G1/S, double thymidine block/hydroxyurea; G2/M, nocodazole) and analyzed by RT-qPCR. β-actin mRNA was used as an internal control. Each bar represents mean + SEM, n = 3.
(B) siRNA-mediated knockdown of lncRNA152 or lncRNA67 alters cell cycle profile in MCF-7 cells, as assessed by propidium iodide staining and FACS analysis. Asterisks represent significant differences versus the control knockdown (si-Ctrl) (Student’s t-test, p-value < 0.05).
(C) Relative expression of lncRNA152 (left) and lncRNA67 (right) in estrogen-withdrawn MCF-7 cells following a time course of E2 treatment as determined by RT-qPCR. Each bar represents the mean + SEM, n = 3. * p-value < 0.05, Student’s t-test.
(D) Effect of lncRNA152 or lncRNA67 knockdown on the expression of estrogen-regulated genes in MCF-7 cells as determined by RT-qPCR with RPL19 mRNA as an internal control. Forty-eight hours after transfection of the siRNAs, the cells were treated for 3 hours with E2 and total RNA was collected. Each point represents the mean ± SEM, n = 3. Each bar represents the mean + SEM, n = 3.
(E) Effect of lncRNA152 or lncRNA67 knockdown on the growth of MCF-7 cells as determined by crystal violet staining. Twenty-four hours after transfection of the siRNAs, the cells were treated for 3 days with E2 and the cell density was determined. Each point represents the mean ± SEM, n = 3. Asterisks, p-value < 0.05 (**, relative to si-Ctrl; *, relative to –E2).
(F) Model depicting the unique roles of lncRNA152 and lncRNA67 in basal and E2-dependent mitogenic growth.
See also Figure S7.
DISCUSSION
In this study, we describe a robust and accurate genomic and computational pipeline for annotating lncRNAs. Using this approach, we identified 1888 lncRNAs in MCF-7 human breast cancer cells, more than 700 of which were not previously annotated. Functional analyses of two specific lncRNAs, lncRNA152 and lncRNA67, show how interplay between lncRNA-mediated regulatory pathways and the estrogen signaling pathway can control gene expression programs driving both basal and mitogenic growth of breast cancer cells.
A LncRNA Annotation Pipeline that Combines Data from Transcription and Steady-State RNA Analyses
In our lncRNA annotation pipeline, we used GRO-seq to define the transcription units/primary transcripts and RNA-seq to define the exons. Our approach provides advantages over previous approaches that rely solely on (1) steady-state RNA levels from RNA-seq (Guttman et al., 2010; Roberts et al., 2011; Trapnell et al., 2010) or (2) promoter and gene body histone modification patterns (i.e., H3K4me3 and H3K36me, respectively) (Guttman et al., 2009; Marques et al., 2013) to identify the lncRNA transcripts for the following reasons. First, lncRNA genes, as a group, are transcribed to similar levels as mRNA genes, but lncRNAs, as a group, are considerably less stable than mRNAs (Fig. 2, C through E; Fig. S2). Thus, GRO-seq, which measures transcription, rather than steady-state RNA levels (Hah et al., 2011), provides a sensitive approach for identifying lncRNA transcripts that might otherwise go undetected. This includes the previously unannotated lncM2 set, which contains lncRNAs that are enriched in the nucleus and are generally less stable (Fig. S2, C and D). Second, intergenic lncRNA genes, which represent the majority of lncRNA genes, have significantly lower levels promoter H3K4me3 and gene body H3K36me3 than mRNA genes (Fig. 3C). This makes many lncRNA genes difficult to detect using patterns of H3K4me3 and H3K36me3 enrichment. Thus, integrated analyses combining multiple genomic and bioinformatic approaches represent the most robust means of detecting and annotating lncRNAs.
LncRNA Genes and Transcripts: From the Nucleus to the Cytoplasm
LncRNA genes show a variety of orientations, locations, and distributions with respect to mRNA genes and other lncRNA genes (e.g., antisense, divergent, and intergenic), similar to those observed for mRNA genes (Sun and Kraus, 2013). Interestingly, we observed that some lncRNA genes have TSSs located proximal to ERa binding sites, which are enriched for features indicative of active enhancers (e.g., H3K4me1, H3K27ac, CBP, pioneer factors) (Figs. 4C and S4C). Previous studies have shown that lncRNAs originating from enhancers may contribute to enhancer function, acting as “ncRNA-activating (ncRNA-a)” lncRNAs {Orom, 2010 #86;Trimarchi, 2014 #93}. The presence of enhancer features in the promoter regions of lncRNA may dictate a distinct mode for regulating gene expression compared to lncRNA genes, whose promoters are enriched in chromatin features more typical of promoter regions.
Our cell fractionation experiments allowed us to track the subcellular distribution of mature lncRNAs. We observed a greater enrichment of the lncM lncRNA set in the nucleoplasm and on chromatin compared to the cytoplasm, although a significant fraction of lncRNAs are enriched in the cytoplasm. Enrichment in the nucleus was not observed for previously annotated lncRNAs (the lncA set), suggesting that our pipeline detects a larger fraction of nuclear lncRNAs than previous approaches. This is likely due to the fact that nuclear lncRNAs, as a group, are significantly less stable than cytoplasmic lncRNAs (Fig. 2E; Fig. S2; (Clark et al., 2012)) and the greater sensitivity of GRO-seq in detecting less stable lncRNAs. The computed stability metric that we developed based on RNA-seq/GRO-seq ratios, which we validated experimentally (Fig. 2F), should be a useful tool for exploring lncRNA stability in future studies.
These observations raise some important questions for future studies. For example, how are lncRNAs directed to particular subcellular compartments? What are the molecular mechanisms underlying the reduced stability of lncRNAs compared to mRNAs, especially nuclear lncRNAs? What are the unique features and functions of cytoplasmic lncRNAs and nuclear lncRNAs? In spite of the focus on nuclear lncRNAs in the literature, recent studies have begun to elucidate the functions of cytoplasmic lncRNAs (Geisler and Coller, 2013; Sun and Kraus, 2014; van Heesch et al., 2014). However, more studies are needed in this area.
Breast Cancer-Associated LncRNAs Regulate Cell Cycle Gene Expression to Control Cell Proliferation
As shown previously and illustrated herein, lncRNAs exhibit a high level of tissue- and cell type-specific expression compared to mRNAs (Rinn and Chang, 2012; Sun and Kraus, 2014; Ulitsky and Bartel, 2013). Importantly, the differential expression patterns of lncRNAs carry useful information about tissue and cell identity, as well as the intrinsic molecular subtypes of breast cancers (Fig. 5B). The latter suggests that lncRNA expression patterns may have potential utility as diagnostic or prognostic indicators in breast cancer patients.
We have determined the roles of two lncRNAs in the growth of breast cancer cells, using MCF-7 cells as a model: (1) lncRNA152 (a.k.a. LOC145837, RP11-279F6.1, or DRAIC (Sakurai et al., 2015)) a previously annotated, but poorly characterized lncRNA and (2) lncRNA67, a previously unannotated lncRNA. Both of these lncRNA show elevated levels of expression in breast cancers compared to benign breast tissue (Fig. 5, E and F) and are associated with gene expression, RNA processing, DNA replication, and the cell cycle in guilt-by-association analyses (Fig. 5C). Functional analyses (i.e., using siRNA-mediated knockdown or ectopic expression) revealed key roles for lncRNA152 and lncRNA67 in breast cancer cell proliferation (Fig. 6B), the expression of genes involved in cell cycle regulation (Fig. 6C), and cell cycle progression (Fig. 7B). The lncRNA152- and lncRNA67-regulated gene sets are enriched for genes whose expression peaks at key points in the cell cycle and are likely targets of the E2F4 transcription factor (Fig. 6D). The results from all of these assays are consistent, pointing to a role for lncRNA152 and lncRNA67 in the control of cell proliferation by regulating the cell cycle. In addition, the expression and functions of lncRNA152 and lncRNA67 interface with the estrogen signaling pathway (Fig. 7, C through E). Collectively, our results indicate that both lncRNA152 and lncRNA67 are required for breast cancer cell growth and cell cycle-related gene expression, but that lncRNA152 may be more important for basal growth, whereas lncRNA67 may be more important for estrogen-stimulated growth (Fig. 7F).
EXPERIMENTAL PROCEDURES
Additional details on the experimental procedures can be found in the Supplemental Materials.
Cell culture and treatments
MCF-7 cells were maintained in MEM medium with 5% calf serum. For experiments involving estrogen treatment, the cells were grown for at least 3 days in phenol red-free medium and then treated with ethanol (vehicle) or 17β-estradiol (E2; 100 nM) as indicated.
Cell fractionation, RNA isolation, and polyA+ RNA-seq
Two biological replicates of 107 MCF-7 cells were subjected to cell fractionation into cytoplasmic, nuclear, and chromatin fractions. Total RNA was isolated from each fraction using the PARIS kit (Ambion). Total RNA was also isolated from unfractionated MCF-7 cells using the RNeasy kit (QIAgen). The RNA collected from each subcellular fraction, as well as the unfractionated MCF-7 cells, was processed for whole genome polyadenylated RNA sequencing (polyA+ RNA-seq).
The total RNA samples were subjected to enrichment of polyA+ RNA using Dynabeads Oligo(dT)25 (Invitrogen) as described previously (Zhong et al., 2011). Strand-specific RNA-seq libraries were prepared from the polyA+ RNA as described previously (Zhong et al., 2011). The RNA-seq libraries were sequenced using an Illumina HiSeq 2000 as follows: (1) fractionated RNA samples were sequenced using paired-end methodology with a length of 100 nt (PE100) and (2) unfractionated RNA samples were sequenced using single-end methodology with a length of 50 nt (SE100).
Computational pipeline for annotation of lncRNAs
We developed a computational pipeline to annotate lncRNAs using RNA-seq and GRO-seq data, which includes the following steps: (1) RNA-seq read mapping to the human genome (NCBI 37, hg19) using the spliced read aligner TopHat version ver. 2.0.4 (Kim et al., 2013); (2) Transcriptome assembly using Cufflinks ver. 2.0.2 (Trapnell et al., 2010), applying a minimal read coverage threshold (>10 reads per base) and a size selection filter (>200 bp; >1 kb for single exon transcripts); (3) Merging of filtered transcripts into two distinct, non-overlapping sets using Cuffmerge: a cytoplasmic set and a nuclear set, with the latter containing lncRNAs from both the nucleoplasmic and chromatin-associated fractions; (4) Filtering transcripts versus known annotations (RefSeq or in GENCODE ver. 12) and classifying them based on gene location and orientation; (5) Filtering transcripts lacking evidence of a primary transcript using published GRO-seq data sets from control and E2-treated MCF-7 cells (Hah et al., 2011); (6) Filtering transcripts based on a coding potential threshold using PhyloCSF (Lin et al., 2011), excluding transcripts from our lncRNA catalog with a PhyloCSF score greater than 150; and (7) Filtering transcripts based on a transcript abundance threshold (FPKM >1).
Additional analyses of lncRNAs
After annotating the lncRNAs, we performed a variety of additional analyses to characterize the lncRNAs as a class of RNAs, including (1) Estimation of sequence conservation using phastCons scores (Siepel et al., 2005), extracted from the vertebrate phastCons 46-way alignment (UCSC Genome Browser), setting the region from −1000 bp to −1 bp relative to the TSS as the promoters; (2) Estimation of the contribution of each subcellular fraction to the total population of polyA+ RNAs using the relationship a x Cyto + b x Nuc + c x Chrom = Total (where Cyto, Nuc, Chrom, are the FPKM values of each transcript in the specified fraction; a, b, and c indicate their corresponding contributions; and Total is the estimated total FPKM); (3) Estimation of transcript stability, calculated as the ratio of RNA-seq FPKM over GRO-seq RPKM for each lncRNA and mRNA transcript; (4) Determination of regulation at the transcriptional level (GRO-seq) versus the steady-state RNA level (RNA-seq) using the Bioconductor package edgeR with a 5% false discovery rate (FDR) for GRO-seq data (Hah et al., 2011; Robinson et al., 2010) and Cuffdiff with a 5% FDR for RNA-seq data (Trapnell et al., 2013); and (5) Determination of the breadth and specificity of lncRNA and mRNA expression using RNA-seq datasets from 135 tumor tissues, 27 benign tissues, 109 tumor cell lines, and 22 benign cell lines of the breast, prostate, stomach, melanocytes, pancreas, bladder, kidney, salivary gland, lymphoid and myeloid tissue (Kalyana-Sundaram et al., 2012), as well as three additional breast cancer cell lines and eight benign breast tissue samples (Asmann et al., 2011).
Analysis of histone modification, coregulator, and transcription factor signatures
Published GRO-seq and ChIP-seq (H3K4me3, H3K36me3, H3K4me1, H3K27ac, ERa, CBP, FOXA1, and AP2 ) data sets from untreated or E2-treated MCF-7 cells were used to explore the enrichment of histone modifications, coregulators, transcription factors, and transcription at specific loci. Metagene plots were generated as described (Hah et al., 2011). Boxplot representations were generated using the boxplot function in R.
Guilt-by-association analyses
Guilt-by-association analyses were performed as described previously (Guttman et al., 2009). Briefly, the expression (based on RNA-seq) of each lncRNA in the lncM set across a panel of 304 tissues and cell lines was correlated with the expression of each mRNA. Each lncRNA was then associated with the entire list of mRNAs, ranked by their correlation with the lncRNA. Gene set enrichment analysis (GSEA) on the ranked list of mRNAs was used to associate the lncRNAs with pathways and signatures that were significantly enriched.
Functional analyses of lncRNAs in MCF-7 cells
Transient RNAi-mediated knockdown of lncRNAs in MCF-7 cells was performed by transfection of (1) custom-designed siRNAs targeting selected lncRNAs or (2) a commercially available control siRNA (Sigma, MISSION siRNA universal negative control) using Lipofectamine RNAiMAX reagent (Life Technologies). Forty-eight hours post transfection, the cells were collected for RT-qPCR and RNA-seq. RT-qPCR detection of lncRNAs and mRNAs in total RNA was performed as described previously (Hah et al., 2013; Sun et al., 2012) using gene-specific primers.
RNA-seq after lncRNA knockdown using polyA+ RNA was performed as described above using the dUTP method (Zhong et al., 2011). Differentially-regulated RefSeq mRNAs were called by Cuffdiff, using a 5% FDR. Fold changes in expression were represented in heatmaps using Java Treeview. Transcription factor target analysis was performed using GREAT (McLean et al., 2010) on the regulated mRNA set.
RNA stability analyses
The stability of lncRNAs and mRNAs was determined (1) experimentally by treating MCF-7 cells by with 2.5 μg/mL actinomycin D (Sigma) for four hours and then monitoring RNA levels by RT-qPCR and (2) computationally by taking the log10(RNA-seq FPKM/GRO-seq RPKM).
Cell proliferation assays and cell cycle analyses
After transient RNAi-mediated knockdown of specific lncRNAs as described above, MCF-7 cells were grown for the number of days indicated and stained with 0.1% crystal violet in 200 mM phosphoric acid, and washed and destained with 10% acetic acid. The acetic acid destain was collected and read at absorbance 595 nm. Expression of lncRNAs throughout the cell cycle was determined for G0 (serum withdrawn), G1/S (double thymidine block/hydroxyurea), and G2/M (nocodazole) synchronized MCF-7 cells. FACS analysis was performed on siRNA-transfected cells fixed in 85% ethanol and stained with propidium iodide.
Data sets
The RNA-seq data sets generated for this study, as well as a detailed list of the lncRNA annotations, can be accessed through GEO using the series accession number GSE63189.
Supplementary Material
Highlights.
A sensitive approach for annotating lncRNAs that integrates GRO-seq and RNA-seq data
Identification of lncRNAs in breast cancer cells, including >700 not yet annotated
Differences between lncRNA and mRNA gene promoters; similarities with enhancers
LncRNAs 152 and 67 control the cell cycle and gene expression in breast cancer cells
Acknowledgments
We thank (1) Arul Chinnaiyan and E. Aubrey Thompson for RNA-seq data sets, (2) the Tissue Management Resource at UT Southwestern’s Harold C. Simmons Comprehensive Cancer Center for RNA from breast cancer samples, (3) Rosemary Conry, Balaji Parameswaran, Tulip Nandu, and Anusha Nagari for technical assistance, and (4) members of the Kraus lab for helpful discussions about this project and constructive comments on this manuscript. This work was supported by a predoctoral fellowship from the American Heart Association to M.S., a postdoctoral fellowship from the Susan G. Komen Breast Cancer Foundation to S.S.G., and grants from the Cancer Prevention and Research Institute of Texas (CPRIT; RP130607) and the NIH/NIDDK (DK058110) to W.L.K.
Footnotes
AUTHOR CONTRIBUTIONS
W.L.K. conceived the project based on previous studies in the lab from Hah et al. (2011) and secured funding to support the project. M.S. and S.S.G. grew, treated, and collected the cells for RNA-seq, performed the subcellular fractionation of RNAs, and carried out the 5′ and 3′ RACE analyses. M.S. developed and executed the computational pipeline for annotating lncRNAs, analyzed all of the genomic data, and generated a list of candidate lncRNAs for further analysis. S.S.G. generated the RNA-seq libraries, selected specific lncRNAs for further analysis, and performed all of the functional analyses of lncRNA152 and lncRNA67. D.S.K. performed the actinomycin D RNA stability experiments. M.S. and S.S.G. made the figures and wrote an initial draft of the paper. W.L.K. edited the figures and text to generate the final version of the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asmann YW, Hossain A, Necela BM, Middha S, Kalari KR, Sun Z, Chai HS, Williamson DW, Radisky D, Schroth GP, et al. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res. 2011;39:e100. doi: 10.1093/nar/gkr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, Asangani IA, Kothari V, Prensner JR, Lonigro RJ, et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. 2012;149:1622–1634. doi: 10.1016/j.cell.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol. 2013;14:R131. doi: 10.1186/gb-2013-14-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013 doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Reon BJ, Anaya J, Dutta A. The lncRNA DRAIC/PCAT29 locus constitutes a tumor-suppressive nexus. Mol Cancer Res. 2015;13:828–838. doi: 10.1158/1541-7786.MCR-15-0016-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Isaacs GD, Hah N, Heldring N, Fogarty EA, Kraus WL. Estrogen regulates JNK1 genomic localization to control gene expression and cell growth in breast cancer cells. Mol Endocrinol. 2012;26:736–747. doi: 10.1210/me.2011-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Kraus WL. Minireview: Long noncoding RNAs: new “links” between gene expression and cellular outcomes in endocrinology. Mol Endocrinol. 2013;27:1390–1402. doi: 10.1210/me.2013-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Kraus WL. From discovery to function: the expanding roles of long non-coding RNAs in physiology and disease. Endocr Rev. 2014:er20141034. doi: 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, et al. LncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc. 2011;2011:940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.