Summary
Mycobacterium tuberculosis M ycobacterial membrane protein Large (MmpL) proteins are important in substrate transport across the inner membrane. Herein, we show that MmpL proteins are classified into two phylogenetic clusters, where MmpL Cluster II contains three soluble domains (D1, D2, and D3) and has two full-length members, MmpL3 and MmpL11. Significantly, MmpL3 is currently the most druggable M. tuberculosis target. We have solved the 2.4 Å MmpL11-D2 crystal structure revealing structural homology to periplasmic porter subdomains of RND (multidrug) transporters. The resulting predicted Cluster II MmpL membrane topology has D1 and D2 residing, and possibly interacting, within the periplasm. Crosslinking and biolayer interferometry experiments confirm that Cluster II D1 and D2 bind with weak affinities, and guided D1-D2 heterodimeric model assemblies. The predicted full-length MmpL3 and MmpL11 structural models reveal key substrate binding and transport residues, and may serve as templates to set the stage for in silico anti-tuberculosis drug development.
Keywords: X-ray crystallography, Mycobacterium tuberculosis, tuberculosis, MmpL, crosslinking, biolayer interferometry, RND transporters, porter domain
Graphical abstract
Introduction
Mycobacterium tuberculosis (Mtb) is the pathogenic microbe responsible for the communicable disease, tuberculosis (TB), which has burdened civilization throughout history. TB continues to be a global health problem with an estimated 9 million cases and 1.4 million deaths reported in 2013 (WHO, 2014). The confluence of a progressively ineffective drug treatment regimen, emergent drug resistant strains, and AIDS/HIV synergism dictate the need to develop new treatment strategies to combat TB. Consequently, a better understanding of the complex biology of Mtb is required.
In Mtb, the MmpL (Mycobacterial membrane protein Large) protein family consists of thirteen actinobacteria-specific inner membrane proteins of approximately 1000 residues. Significantly, in the last several years, MmpL3 has become the most successful anti-TB drug target. High-throughput whole cell screens identified several potent anti-mycobacterial agents that target MmpL3, including BM212 and SQ109 (Grzegorzewicz, et al., 2012; La Rosa, et al., 2012; Owens, et al., 2013; Tahlan, et al., 2012). Moreover, mutational analyses revealed MmpL3 is essential for Mtb viability (Domenech, et al., 2005; Tullius, et al., 2011). In addition, several MmpL proteins are necessary for Mtb virulence in mice infections. MmpL4 and MmpL7 knockout mutants appear to be avirulent and have severely attenuated growth within mice lungs, and mice infected with MmpL8 and MmpL11 knockout mutants survive for considerably longer than with wild-type Mtb infection (Domenech, et al., 2005). Furthermore, a separate study suggests that MmpL5 and MmpL10 are required for Mtb survival in mice lungs (Lamichhane, et al., 2005).
MmpL proteins have been implicated in mediating substrate transport across the mycobacterial membrane. MmpL3 and MmpL11 exhibit dual roles in the export of trehalose monomycolate (TMM) for MmpL3 (Grzegorzewicz, et al., 2012; La Rosa, et al., 2012; Varela, et al., 2012) and monomeromycolyl diacylglycerol (MMDAG) and mycolate ester wax for MmpL11 (Pacheco, et al., 2013), and both have also been implicated in heme import (Tullius, et al., 2011). MmpL4 and MmpL5 have redundant functions in siderophore export, and a double MmpL4/5 mutant cannot be constructed (Wells, et al., 2013), suggesting that they are essential for siderophore-mediated iron acquisition. MmpL7 and MmpL8 have been shown to transport polyketide phthiocerol dimycocerosate and sulfolipid-1, respectively (Converse, et al., 2003; Cox, et al., 1999; Jain and Cox, 2005; Seeliger, et al., 2012). Additionally, MmpL5 and MmpL7 have been implicated in drug efflux (Lamichhane, et al., 2005; Milano, et al., 2009). These data present convincing evidence of the importance of MmpL proteins; hence, their further characterization contributes to an enhanced understanding of Mtb biology and will open up new avenues for anti-TB therapeutics.
It has been suggested that MmpL proteins belong to the Resistance-Nodulation-cell Division (RND) permease superfamily of transmembrane transporters (Domenech, et al., 2005). Inner membrane RND transporters associate with outer membrane factors, and this assembly is stabilized by periplasmic membrane fusion proteins to form a three-component efflux pump (reviewed in (Ruggerone, et al., 2013)). To-date, the five available RND transporter structures (i.e., AcrB, CusA, MexB, ZneA, and MtrD) reveal homotrimers where each monomer harbors twelve transmembrane helices (TM) with N-terminal and C-terminal periplasmic domains inserted between TM1 and TM2 and between TM7 and TM8, respectively, Figures 1A&B (Long, et al., 2010; Murakami, et al., 2002; Nakashima, et al., 2013; Pak, et al., 2013; Sennhauser, et al., 2009; Su, et al., 2012). Each periplasmic domain comprises two structurally similar porter subdomains (N-terminal porter subdomains, PN1 and PN2, and C-terminal porter subdomains, PC1 and PC2, each with a βαββαβ motif) and a docking subdomain (DN or DC), Figures 1A&B (reviewed in (Ruggerone, et al., 2013)). In all five structures, there is also an additional α-helix between TM6 and TM7 that runs almost parallel to the cytoplasmic membrane surface (Figure 1B). Furthermore, RND transporters are categorized into heavy metal efflux (HME) and hydrophilic and amphiphilic efflux (HAE) subfamilies, transporting a wide array of substrates including metals, antibiotics, detergents and dyes (reviewed in (Delmar, et al., 2014)). Driven by proton-motive-force (PMF), substrate shuttling occurs via a rotating mechanism whereby each monomer within the RND transporter homotrimer adopts a unique conformation for substrate access, binding, and release (reviewed in (Ruggerone, et al., 2013)).
Figure 1.
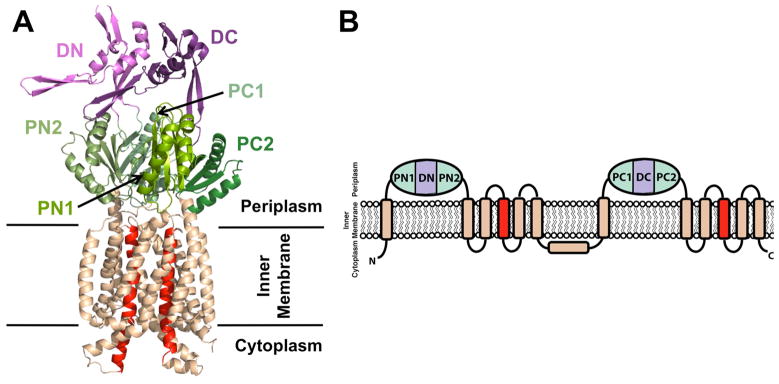
(A) Cartoon representation of monomeric CusA (PDB code: 4DNT) showing the docking, porter, and transmembrane subdomains. The N- and C-terminal docking (DN and DC) and porter (PN1, PN2, PC1, and PC2) subdomains are colored shades of purple and green, respectively, while the transmembrane subdomain is colored wheat, except for the central transmembrane helices, TM4 and TM10, which are colored red. (B) RND transporter membrane topology with two periplasmic domains, each containing two porter subdomains and one docking subdomain. An additional extracytosplasmic α-helix between TM6 and TM7 is located near the cytoplasmic membrane surface and runs almost parallel to it. Subdomain color designation is as in (A).
As members of the MmpL family are large, structural and biochemical analyses of the full-length proteins has evaded the TB community thus far; however the divide and conquer strategy may prove more tractable. To this end, we present the structural characterization of a soluble domain, D2, from an MmpL Cluster II protein (depicted in Figure 3B). The 2.4 Å X-ray crystal structure of MmpL11-D2 domain reveals structural homology to the porter subdomains of RND transporters. This structure has allowed membrane topology predictions for all members of the MmpL family. Moreover, these predictions alluded to potential interactions between periplasmic domains, D1 and D2, from both MmpL11 and its closest homolog, MmpL3, and inter-domain interactions were confirmed by both affinity and crosslinking experiments. Herein, we describe the results of these studies, and discuss their implications with respect to MmpL3 and MmpL11 substrate binding and transport.
Figure 3.
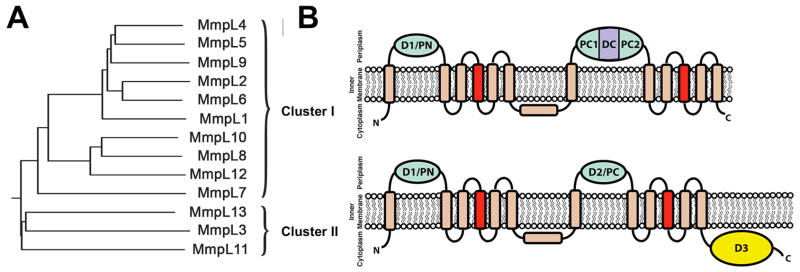
(A) Phylogenetic tree of MmpL proteins reveals two distinct clusters where the (B) predicted membrane topologies of MmpL Clusters I and II proteins are based on RND transporters. The predicted porter domains (N-terminal D1 (PN) and Cluster I C-terminal D2 (PC1 and PC2) and Cluster II C-terminal D2 (PC)) are colored green and the predicted Cluster I C-terminal docking domain (DC) is colored purple. D3 is colored yellow while the transmembrane subdomain is colored wheat, except for the central transmembrane helices, TM4 and TM10, which are colored red. The predicted additional extra-cytoplasmic α-helix located between TM6 and TM7 is shown almost parallel to the cytoplasmic membrane surface, as observed in RND transporter structures (Fig. 1B).
Results
MmpL11-D2 shares structural homology to RND transporter porter subdomains
Crystals of MmpL11-D2 were obtained using a construct that encompassed residues 390–529. MmpL11-D2 crystallized in space group C2221 with one molecule in the asymmetric unit. The mass of a single crystal was measured by MALDI-TOF to be 9556.3 Da, which corresponds to the final structural model where MmpL11-D2 is truncated at its N- and C-termini prior to crystallization.
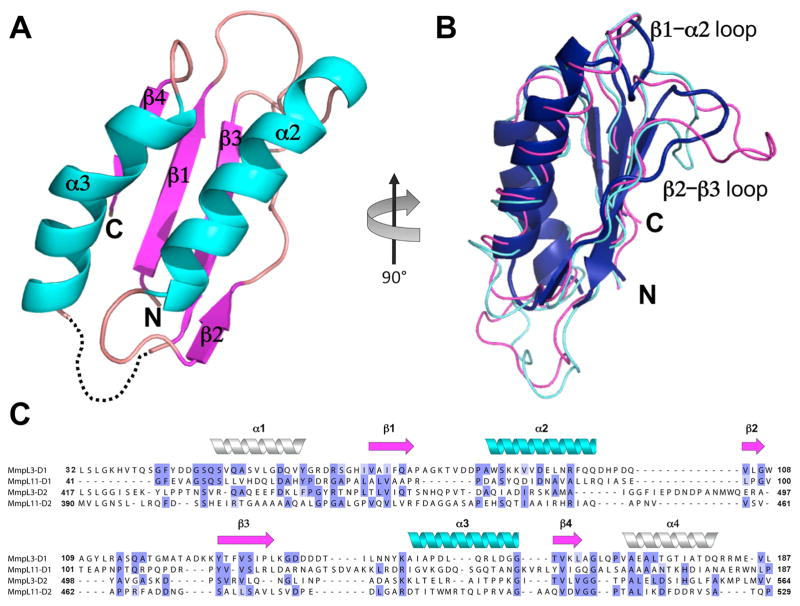
The 2.4 Å MmpL11-D2 structure reveals two anti-parallel α-helices (α2 and α3) sitting atop a four-stranded anti-parallel β-sheet, forming a βαββαβ fold enclosing a hydrophobic core of residues (Figure 2A). Notably, the three-residue β2 is interrupted with a bulge introduced by a pair of proline residues (i.e., Pro463 and Pro464), while the loop connecting β2 and β3 is stabilized by five hydrogen bonds (H-bonds) with α2 and β3 (Figure 2B). Furthermore, an extended loop region between β1 and α2 is stabilized by π-π stacking between Phe431 and His441 (Figures 2B& S1). Additionally, there are numerous H-bonds and an ion-pair between Arg430 and Asp509 stabilizing the overall globular structure, Figure S1. The observed ligands in the structure are nine iodides ions from KI and a single sulfate ion from the crystallization condition along with sixteen water molecules (Table 1).
Figure 2.
(A) Cartoon representation of MmpL11-D2 structure with missing residues 479 – 489 depicted as dashed lines. α-helices, β-strands, and loops are colored cyan, magenta, and wheat, respectively (B) MmpL11-D2 colored blue, is rotated 90° clockwise from (A) and structurally aligned with RND PC1 porter subdomains from ZneA (PDB code: 4K0E) and MexB (PDB code: 2V50), colored magenta and cyan, respectively. (C) MmpL11 and MmpL3 Cluster II D1 and D2 domain sequence alignment based on secondary structural prediction and MmpL11-D2 structure. Cylinders (α-helices) and arrows (β-sheets) colors correspond to the secondary structural elements in (A). The predicted α-helices (α1 and α4) are shown as white cylinders.
Table 1.
MmpL11-D2 data collection and refinement statistics
| Data collection | |
| Wavelength (Å) | 1.54 |
| Resolution range (Å) | 53.36 - 2.4 (2.49 – 2.4) |
| Space group | C2221 |
| Unit cell (Å) | 65.60 × 91.73 × 32.85 |
| Unit cell (°) | 90 × 90 × 90 |
| Total reflections | 23972 |
| Unique reflections | 4128 |
| Multiplicity | 5.8 (6.1) |
| Completeness (%) | 99.78 (100.00) |
| Mean I/sigma (I/σ) | 20.5 (7.8) |
| Wilson B-factor (Å2) | 25.95 |
| Rmerge# | 0.109 (0.290) |
| Rpim## | 0.049 (0.133) |
| Refinement | |
| Rwork+ | 0.2077 (0.2132) |
| Rfree++ | 0.2537 (0.2519) |
| No. of atoms | 612 |
| No. of iodides | 9 |
| No. of waters | 16 |
| No. of protein residues | 80 |
| R.m.s.d., bonds (Å) | 0.003 |
| R.m.s.d., angles (°) | 0.73 |
| Ramachandran favored (%) | 96 |
| Ramachandran outliers (%) | 0 |
| B-factor (Å2) | |
| Average | 37.5 |
| Macromolecules | 37.4 |
| Solvent | 32.8 |
| Ligands | 44.6 |
| PDB code | 4Y0L |
Statistics for the highest-resolution shell are shown in parentheses.
Rmerge = Σ Σi |Ii − (I)| / Σ Σi Ii
Rpim = Σ {1/[N−1]}1/2Σi|Ii − (I)| / Σ Σi Ii
Rwork = Σ ||Fobs| − |Fcalc||/ Σ |Fobs|
Rfree was computed identically except all reflections belonged to a test set consisting of a 5% random selection of the data.
A structural homology search for MmpL11-D2 using DALI (Holm and Rosenstrom, 2010), demonstrates that the closest structural homologs are porter subdomains of RND transporters, Table S1. Thus, MmpL11-D2 was structurally aligned with AcrB (PDB code: 3W9H), CusA (PDB code: 4DNT), MexB (PDB code: 2V50), ZneA (PDB code: 4K0E), and MtrD (PDB code: 4MT1) (Bolla, et al., 2014; Nakashima, et al., 2013; Pak, et al., 2013; Sennhauser, et al., 2009; Su, et al., 2012) using RaptorX (Wang, et al., 2013). Of the approximately 1000 residues of the RND transporters, MmpL11-D2 aligns to the conserved porter subdomains, with an RMSD range of 2.0 – 2.9 Å over 69 – 74 Cα atoms. Notably, the RMSD was consistently lowest (2.0 – 2.3 Å) between MmpL11-D2 and porter subdomain, PC1. While the secondary structural elements between MmpL11-D2 and PC1 subdomains are well aligned, the β1-α2 loop and the loop connecting the three-residue β2-strand to β3 are most divergent (Figure 2B). The MmpL11-D2 β1-α2 loop tilts toward the β-sheet causing the extended β2-β3 loop to be displaced compared to that of the RND transporter, which may be a result of its interrupted β2.
MmpL3 and MmpL11 D1 and D2 have identical topologies
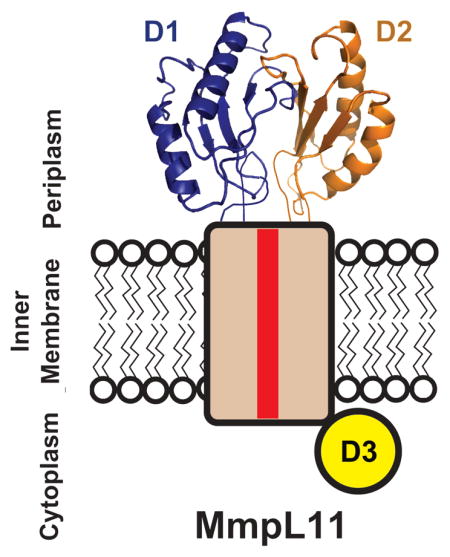
Utilizing ClustalW (Larkin, et al., 2007), the MmpL protein family phylogenetic tree reveals two distinct clusters: the majority of MmpL proteins belong in Cluster I while MmpL3, MmpL11 and MmpL13 are grouped in Cluster II (Figure 3A). A major difference is the presence of three predicted soluble domains in the MmpL3/11/13 cluster (D1, D2 and D3) whereas the other MmpL proteins have two predicted soluble domains, D1 and D2 (Figure 3B). Notably, secondary structural predictions (Cole, et al., 2008) of MmpL3/11 D1 and D2 domains are identical, suggesting that these domains harbor the conserved porter subdomain motif as observed for MmpL11-D2 (Figures 2A&C). Moreover, MmpL3/11 D1 and D2 domains each have two additional predicted α-helices, α1 and α4, which are not part of porter subdomains or observed in the MmpL11-D2 structure. Sequence alignments, as assessed by EMBOSS Needle (Rice, et al., 2000), revealed that all MmpL3/11 D1 and D2 domains share high similarities ranging from 34% between MmpL3-D1 and MmpL3-D2 domains to 13% between MmpL3-D2 and MmpL11-D2 (Figure 2C). Due to their similarity to periplasmic RND transporter porter subdomains, this would strongly suggest that MmpL3/11 D1 and D2 domains also reside in the periplasm (Figures 1B&2B). In contrast, secondary structure predictions of MmpL3/11 D3 domains are dissimilar from MmpL3/11 D1 and D2 domains and each other, where MmpL3-D3 is predicted to be largely unstructured and MmpL11-D3 predominately α-helical. Furthermore, recent in vivo fluorescence studies suggest localization of MmpL3/11 D3 domains to the cytoplasm (Carel, et al., 2014). Taken together, an MmpL3/11 topology is proposed, whereby D1 and D2 domains are periplasmic while D3 resides in the cytoplasm (Figure 3B).
Distinct from the reported RND transporter structures, the D1 and D2 periplasmic domains in MmpL3 and MmpL11 are significantly shorter, ~150 residues in MmpL3/11 as opposed to ~300 residues in RND transporters. Based on domain boundaries and structural alignments, MmpL3/11 D1 and D2 domains appear to contain a single porter subdomain βαββαβ motif with additional predicted flanking α-helices (α1 & α4, Figure 2C), whereas each RND transporter periplasmic domain contains one docking and two porter subdomains (Figure 1B) (Long, et al., 2010; Murakami, et al., 2002; Pak, et al., 2013; Sennhauser, et al., 2009). This suggests that MmpL3/11 (MmpL Cluster II proteins) belong to a new subclass of RND transporters that only contain a total of two periplasmic porter domains (D1 and D2) and a unique cytoplasmic D3 domain (Figure 3B).
MmpL3 and MmpL11 D1 and D2 domains interact
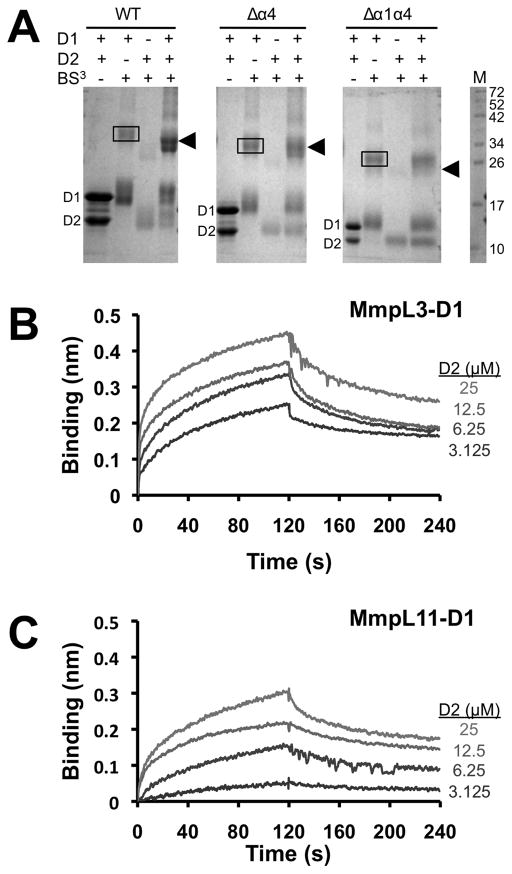
The MmpL Cluster II topology suggests that periplasmic D1 and D2 domains interact, as observed in the structures of known RND transporters (Bolla, et al., 2014; Long, et al., 2010; Murakami, et al., 2002; Pak, et al., 2013; Sennhauser, et al., 2009). To test for stable interactions between MmpL3/11 D1 and D2 domains, purified recombinant MmpL3 or MmpL11 D1 and D2 domains were mixed together and then analyzed by size exclusion chromatography. The results revealed no evidence for MmpL3/11 D1-D2 heterodimers suggesting that stable D1-D2 complexes are not formed (data not shown). To further investigate weak/transient MmpL3/11 D1-D2 interactions, the homobifunctional primary amine crosslinker BS3 was used. To this end, the abundance of lysines in MmpL3-D1 and MmpL3-D2 (nine and seven, respectively) was exploited while the BS3 crosslinking experiments could not be performed for MmpL11 due to the lack of lysines in MmpL11-D2. After combining MmpL3-D1 and MmpL3–D2 in the presence of BS3, SDS-PAGE analysis revealed the emergence of a 34.9 kDa band corresponding to the MmpL3-D1-D2 heterodimer (verified by mass spectrometry, Figure 4A – arrowhead); however, complete dimerization was not observed under conditions tested. Moreover, a prominent band corresponding to the MmpL3-D1 homodimer (38.3 kDa, verified by mass spectrometry, Figure 4A – box) is also observed, but not for that of MmpL3-D2. Thus, these results suggest that MmpL3-D1 and MmpL3-D2 may form a weak heterodimer.
Figure 4.
MmpL3 and MmpL11 D1 and D2 domains interact. (A) SDS-PAGE of MmpL3 D1 and D2 domains and their respective truncated constructs (Δα4 and Δα1α4) in the presence of BS3, suggesting that α1 helix is essential for heterodomain interaction. In all instances, the MmpL3-D1 homodimer (38.3 kDa for WT) is boxed whereas arrowheads identify the MmpL3-D1-D2 heterodimer (34.9 kDa for WT). Notably, for the Δα1α4 constructs, the heterodimer is absent. Biolayer interferometry experiments to assess interactions between (B) MmpL3 D1 (biotinylated) and D2 domains, and (C) MmpL11 D1 (biotinylated) and D2 domains. All reactions were performed at 25 °C in 20 mM sodium phosphate pH 7.4 and 150 mM NaCl. Immobilized biotinylated D1 domains were exposed to different concentrations (25 – 3.125 μM) of D2 domains, where interaction (association and dissociation) is assessed by a wavelength shift (nm).
To confirm and assess the binding affinity of the MmpL3-D1 and MmpL3-D2 interaction, biolayer interferometry was utilized. Increasing concentrations of MmpL3-D2 were titrated to biotinylated MmpL3-D1 immobilized on a streptavidin biosensor, and the association and dissociation was assessed by a shift in wavelength. This resulted in an observable but low micromolar range binding affinity (KD = 4.1 ± 0.2 μM) between MmpL3-D1 and MmpL3-D2, Figure 4B. Similar to MmpL3, biolayer interferometry reveals that the MmpL11-D1-D2 domains interact with a comparable weak KD (4.5 ± 1.1 μM), Figure 4C. These results confirm the formation of the MmpL3-D1-D2 heterodimer and demonstrate that MmpL11-D1 and MmpL11-D2 also form a heterodimer, where both interactions are in the low micromolar range.
To test the molecular determinants of MmpL3/11 D1 and D2 interactions guided by the structure of MmpL11-D2 (Figure 2A), we designed two sets of truncated variants without either the last predicted α-helix (Δα4) or the first and last predicted α-helices (Δα1α4), Figure 2C. BS3 crosslinking experiments reveal that the interaction between MmpL3-D1 and MmpL3-D2 is abrogated with the Δα1α4 domain variants whereas the interaction is restored in the Δα4 domain variants, suggesting that α1 is essential for MmpL3-D1-D2 heterodimer formation (Figure 4A). These results are supported by biolayer interferometry experiments whereby no interaction is observed between the Δα1α4 D1 and D2 variants for both MmpL3 and MmpL11. Furthermore, a similar binding affinity is obtained between the respective D1 and D2 Δα4 domains for both MmpL3 and MmpL11, implying that the α1 helix is required for D1-D2 heterodimer formation for both MmpL3 and MmpL11.
To investigate the interaction interface between MmpL3-D1 and MmpL3-D2, the SDS-PAGE protein band corresponding to the heterodimer (Figure 4A) was excised, trypsinized and analyzed with liquid chromatography tandem mass spectrometry (LC-MS/MS). Strikingly, of the seven lysines present in MmpL3-D2, only Lys504 is crosslinked to MmpL3-D1 at four distinct primary amines (i.e., N-terminus, Lys88, Lys89 and Lys125), Table S2. An intermolecular peptide is also identified between both N-termini of MmpL3-D1 and MmpL3-D2, supporting both crosslinking and biolayer interferometry data that α1 from either MmpL3/11 D1 or D2 domains, is important for domain-domain interactions.
Discussion
Comparison of D1-D2 dimer model with CusA porter subdomain interactions
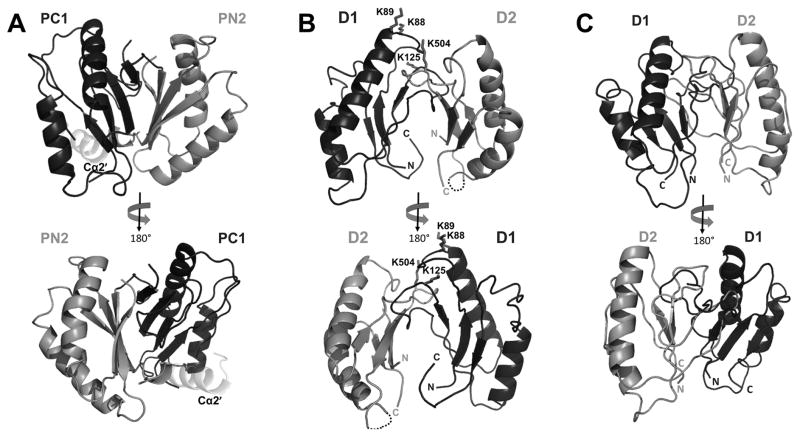
An initial heterodimer model was built based on the MmpL11-D2 structural model and the CusA porter subdomain interactions (PC1 and PN2) to satisfy the intermolecular crosslinked peptides determined for MmpL3-D1 and MmpL3-D2 (Table S2). CusA was selected due to the presence of an additional PC1 α-helix (Cα2′) that interacts with PN2 porter subdomain (Figure 5A) (Long, et al., 2010); the CusA Cα2′ helix is proposed to correspond to one of the predicted α1 helices from either MmpL3-D1 or MmpL3-D2 (Figure 2C). In short, as MmpL3/11 D1 and D2 domains are predicted to have similar porter domain structures, the MmpL11-D2 structure was threaded with the sequence of MmpL11-D1 and analogous residues within MmpL11-D1 and MmpL11-D2 were mutated to lysines based on the sequence alignment with MmpL3-D1 and MmpL3-D2 (Figure 2C). Then, the individual MmpL11-D1 and MmpL11-D2 models were oriented to bring the crosslinked lysines within the BS3 spacer arm distance of 11.4 Å (Figure 5B), using the CusA porter subdomain heterodimer as a structural template, where MmpL11-D1 corresponds to PC1 and MmpL11-D2 to PN2. Finally, the MmpL11-D1-D2 heterodimer model underwent a round of energy minimization (Figure 5B). The structure of MmpL11-D2 was further used as a template to calculate I-Tasser models for MmpL3-D1 and MmpL3-D2 (Yang, et al., 2014). Based on the MmpL11-D1-D2 heterodimer model, an MmpL3-D1-D2 heterodimer model was predicted and energy minimized, and results in a complex consistent with the crosslinking results (Figure 5C).
Figure 5.
(A) Cartoon representation of CusA PC1 and PN2 heterodimer (PDB code: 4DNT). PC1 and PN2 are colored black and dark grey, respectively, while PC1 Cα2′ is colored light grey. Cartoon representation of the heterodimer models of (B) MmpL11-D1-D2, depicting crosslinked lysine residues as sticks and (C) MmpL3-D1-D2. MmpL3/11 D1 and D2 are colored black and dark grey, respectively.
While the MmpL3/11 D1-D2 porter domain interactions are dependent on α1 helix, the CusA porter subdomain interactions are not facilitated by the Cα2′ helix. Moreover, porter interdomain interactions within RND transporters are facilitated by the formation of a β-sheet with strands donated by both porter domains (Figure 5A) and extensive interactions between docking and porter subdomains, which aid in stabilizing porter interdomain interfaces. Within the MmpL3/11 D1-D2 heterodimers, there is an absence of a stabilizing β-sheet and docking subdomains along with the absence of the second RND porter subdomain, suggesting that the α1-helix-mediated MmpL3/11 D1-D2 domain interactions evolved within the MmpL Cluster II proteins.
Implications of MmpL3/11 functions
Substrate binding
RND transporters bind their substrates or inhibitors within the same pocket through extensive PC1 subdomain interactions regardless of the vast differences in substrate sizes, ranging from 63 Da for copper to 694 Da for a pyridopyrimidine derivative inhibitor (ABI-PP) (Long, et al., 2010; Pak, et al., 2013). Strikingly, sequence alignment of the porter subdomains show that PC1 has the lowest homology within the subfamilies (56% and 25% as compared to PN1’s 75% and 37% between HAE and HME RNDs, respectively), suggesting that PC1 subdomains have evolved to confer substrate specificity. MmpL3 and MmpL11 have a variety of proposed substrates; heme (616 Da (Tullius, et al., 2011)), TMM (~1500 Da (Fujita, et al., 2005)), MMDAG and mycolate ester wax (~1300 and ~1600 Da, respectively (Pacheco, et al., 2013)); analogous to the RND transporters, they may bind these substrates primarily via their respective D1 domains. It is of interest to note that MmpL3/11 D1 domains are able to bind heme while the D2 domains do not exhibit any heme binding abilities (Tullius, et al., 2011). Additionally, it was shown that MmpL3 and MmpL11 D1 domains are able to accept heme from a proposed secreted heme transporter, Rv0203 (Owens, et al., 2013), further supporting the hypothesis that the MmpL3/11 D1 domains are mainly responsible for substrate binding. In contrast to the Cluster II MmpL proteins that only have two porter subdomains, the RND transporters have four. Thus, one may speculate that the two MmpL3/11 porter domains may allow for more flexibility to accommodate export of the larger TMM and MMDAG substrates compared to the four porter and two docking subdomains of RND transporters (Fujita, et al., 2005; Pacheco, et al., 2013).
The HAE RND sub-family porter PC1 and PN2 subdomains have numerous identical residues that interact with their respective substrates/inhibitors (Murakami, et al., 2006; Nakashima, et al., 2013); in particular, a pocket consisting of several conserved phenylalanines in AcrB and MexB PC1 subdomain is attributed to the trapping of the inhibitor, ABI-PP (Nakashima, et al., 2013), Figures S2C&D. Close inspection of the RND transporter residues involved in substrate binding reveal no obvious conserved residues in MmpL3/11 D1 or D2 domains. Instead, there is an overrepresentation of residues, especially within D1 domains (~10%), typically associated with hydrophobic substrate binding, including tyrosines, histidines, and phenylalanines (Figures S2A&B). In particular, three D1 domain conserved residues (Phe43/Phe42, Tyr61/Tyr60, and Tyr127/Tyr117 in MmpL3/11), with a particular emphasis on Tyr127/Tyr117 that is located in the vicinity of the HAE RND family substrate binding pocket, may play vital roles in substrate transport and perhaps binding of a subset of MmpL3-targeted antimycobacterial compounds (Owens, et al., 2013).
Proton-motive-force
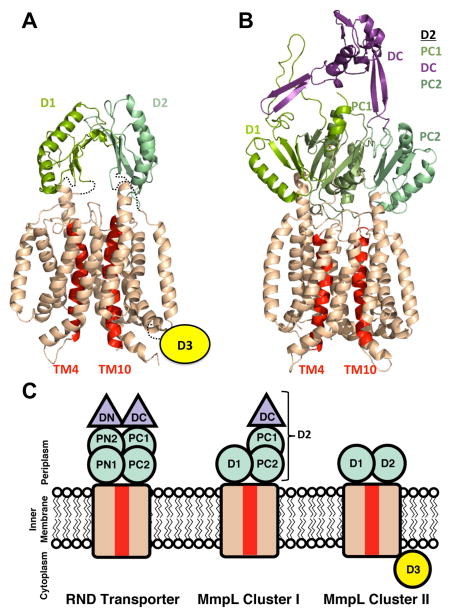
To-date, all RND transporters are reported to utilize PMF to provide the necessary energy for substrate transport. Charged residues have been implicated to play critical roles in PMF. In particular, a conserved aspartate, located in the middle of TM4, has been shown to be essential for transporter function (Franke, et al., 2003; Goldberg, et al., 1999; Guan and Nakae, 2001; Janganan, et al., 2013; Murakami, et al., 2002; Pak, et al., 2013). The mechanism for Cluster II MmpL substrate export and perhaps import is unknown although recent inhibitory Mtb compounds, such as BM212 and SQ109, were suggested to non-specifically target MmpL3 by dissipating its electrochemical proton gradient (Li, et al., 2014). More importantly, a recent report demonstrated that Corynebacterium glutamicum CmpL4, the closest homolog of Cluster II MmpL13, is dependent on PMF (Yang, et al., 2014), making a convincing argument that MmpL3 and MmpL11 also rely on a coupled proton gradient for substrate transport. MmpL3 and MmpL11 Phyre2 models (Kelley and Sternberg, 2009) were analyzed for the conservation of proton relay network residues. Within TM4, MmpL3 Asp251 and MmpL11 Asp248 correspond to the conserved essential aspartate required for PMF (Figure 6A). Furthermore, the corresponding PMF-associated TM10 residue found in CusA, AcrB, MtrD, and MexB (Janganan, et al., 2013; Long, et al., 2010; Murakami, et al., 2002; Sennhauser, et al., 2009) is present in MmpL3 and MmpL11 (Asp640 and Asp609, respectively) whereas TM11 contains positively charged residues (MmpL3 Arg672 and MmpL11 Arg641) analogous to CusA Lys984 (Long, et al., 2010).
Figure 6.
Phyre2 models of (A) MmpL11 with additional restraints from the crosslinking results and (B) MmpL4. The transmembrane domains are colored wheat, except for TM4 and TM10, which are colored red. The different periplasmic porter subdomains are in shades of green and the proposed MmpL4 docking domain is colored purple. The Cluster II MmpL (MmpL11) cytoplasmic D3 domain is signified by a yellow circle. (C) A cartoon representing the domain architecture of RND transporters, MmpL Cluster I and II proteins. Subdomain color designations are as in (A) and (B).
Cluster I and II MmpL periplasmic domains share similar motifs
The previously published structural model for Cluster II MmpL3/11 is reminiscent of the RND superfamily and is similar to our updated MmpL3/11 topology and model supported by structural and biochemical data (Figures 3B&6A) (Li, et al., 2014). The Phyre2 structural prediction (Kelley and Sternberg, 2009) for Cluster I MmpL4 reveals a similar topology of two periplasmic domains, each inserted between TM1/2 and TM7/8, respectively (Figure 6B). Cluster I MmpL D1 domains are predicted to have identical porter βαββαβ motifs as Cluster II MmpL D1 and D2 domains (Figure S3). In contrast, Cluster I MmpL D2 domains are much larger than Cluster II MmpL D2 domains, consisting of approximately 350 residues. This larger Cluster I MmpL D2 domain is predicted to correspond to an RND docking and two porter subdomains, as observed within domains of RND transporters (Figure 6C). Finally, Cluster II MmpL proteins contain cytoplasmic D3 domains, whereas Cluster I MmpL proteins do not (Figure 6). Thus, the Cluster I MmpL model is more similar to RND transporters than the Cluster II MmpL model, and contains docking domains that may play roles in protein interactions with accessory and outer membrane channel proteins, as observed for RND transporters (Du, et al., 2014; Su, et al., 2011).
Significance
Structural information on biologically important Mtb MmpL proteins has remained elusive. Herein, we report the structure of an MmpL periplasmic domain, MmpL11-D2, and provide a first glimpse of periplasmic inter-domain interactions (D1-D2) within MmpL3 and MmpL11. These analyses are of significance as MmpL3 is currently the most promising anti-tuberculosis drug target. Furthermore, we have demonstrated the diversity of the periplasmic domain architecture within the RND transporter superfamily. The canonical RND transporters have an elaborate six-subdomain superstructure, the Cluster I MmpL proteins have a pared down assembly and the Cluster II MmpL proteins appear to have the minimal components required for substrate transport within this RND superfamily (Figure 6C). Many outstanding questions remain regarding the structural characterization of MmpL proteins. All known RND transporter structures are organized as homotrimers implying that the functional oligomeric state of MmpL proteins are also trimeric. Additionally, the orientation of the periplasmic domains D1 and D2, and the roles each domain plays in facilitating substrate transport need further investigation. Finally, the structure/function of D3, which is unique for Cluster II MmpL proteins, is still unresolved. To fully understand these new subclasses of RND transporters, full-length structures of both Cluster I and II MmpL proteins are necessary.
Experimental Procedures
Domain cloning, expression and purification
DNA sequences of MmpL3 residues 32 – 187 and 419 – 560 and MmpL11 residues 41 – 187 and 390 – 529, corresponding to D1 and D2 domains (Table S3), were PCR-amplified from Mtb genomic DNA and cloned into pET28a (Novagen) using NdeI and HindIII for all domains except for MmpL11-D1, where BamHI and XhoI were used (Fermentas Scientific), as outlined in Table S3. All domain constructs, which encode for fusion proteins with N-terminal His6, were overexpressed and purified with the following protocol. Expression plasmids encoding individual domains were transformed into BL21-Gold (DE3) cells and grown at 37 °C in LB medium containing 30 μg/mL kanamycin. Protein expression was induced when cells reached OD600 of 0.8 by the addition of 1 mM IPTG and cells were harvested after 4 hours by centrifugation at 5100 rpm for 20 minutes, followed by resuspension in 50 mM Tris, pH 7.4, 350 mM NaCl and 10 mM imidazole. Cells were then lysed by sonication after addition of egg hen lysozyme (5 mg, Sigma) with phenylmethylsulfonyl fluoride (40 μM, Sigma) and the cell lysate centrifuged at 14000 rpm for 20 minutes. The supernatant was filtered using a 0.45 μm membrane, loaded onto a Ni2+-charged HisTrap column (GE Healthcare), and eluted with a linear imidazole gradient. Fractions containing D1 or D2 domains (between 100 – 250 mM imidazole) were visualized by SDS-PAGE, pooled and concentrated using an Amicon centrifugal filter (10 kD cut-off, Millipore). Further purification was achieved by size exclusion chromatography (SEC) using an S75 gel filtration column (GE Healthcare) pre-equilibrated with 50 mM Tris pH 7.4, 150 mM NaCl, yielding nearly 100% homogeneous protein. Cleavage of the His6-tag was conducted in cleavage buffer (50 mM Tris pH7.4, 150 mM NaCl, 10 mM CaCl2) by the addition of 1 mL thrombin-agarose suspension (Sigma). After an overnight incubation at 4 °C, the thrombin-agarose was removed on a glass frit. Each domain was further purified over an S75 SEC column pre-equilibrated with 50 mM Tris pH 7.4, 150 mM NaCl to separate it from the His6-tag.
Truncated constructs are outlined in Table S3. Expression and purification for the truncated domains proceed as with the full-length domain.
Crystallization, data collection, structure determination and refinement of MmpL11-D2 (residues 390 – 529)
Purified MmpL11-D2 (residues 390 – 529) was concentrated to 10 mg/mL in 50 mM Tris pH 7.4, 150 mM NaCl for crystallization trials. Several MmpL11-D2 (residues 390 – 529) crystals grew after two years in 0.1 M MES pH 6.7, 2 M MgSO4. Crystals were soaked, for two minutes, in 0.5 M KI dissolved in mother liquor containing 20% glycerol and a diffraction dataset was collected at 100 K. A single-wavelength anomalous dispersion (SAD) dataset, which diffracted to 2.4 Å, was collected from an iodide-soaked crystal (λ= 1.54 Å) with unit dimensions of 65.6 Å × 91.7 Å × 32.9 Å and 1 molecule per asymmetric unit in space group C2221. The images were indexed, integrated and reduced using iMOSFLM (Battye, et al., 2011). Data collection statistics are summarized in Table 1. The initial phase and model were determined by SAD using phenix.Autosol (Adams, et al., 2010). The final model was determined using reiterative rounds of model building with phenix.Autobuild (Adams, et al., 2010) followed by manual building through Coot (Emsley, et al., 2010) and refinement with phenix.refine (Adams, et al., 2010). The final model contained MmpL11-D2 residues 424–511; however no electron density was observed for residues 479–489 that correspond to a loop region. The stereochemistry and geometry of MmpL11-D2 was validated with program Molprobity (Chen, et al., 2010) with final refinement parameters summarized in Table 1. All molecular graphics were prepared with PyMOL (DeLano, 2002).
Crosslinking experiments
Crosslinking experiments with BS3 (Pierce, Inc.) were performed with 50 μM proteins in 20 mM sodium phosphate pH 7.4, 150 mM NaCl. Briefly, 50 μM MmpL3 D1 and D2 domains, as well as the two sets of truncated constructs (i.e., Δα1α4 and Δα4) were incubated together in the absence or presence of 10-fold molar excess BS3 on ice for 2 hrs. Reactions were quenched by the addition of 1M Tris pH 7.4 to a final concentration of 50 mM. SDS-PAGE was performed to assess the formation of crosslinked products.
Mass spectrometry
Excised SDS-PAGE bands corresponding to the potential crosslinked heterodimer were in-gel digested with trypsin, as described (Tokhtaeva, et al., 2015). nLC-MS/MS with Collision Induced Dissociation (CID) was performed on an Orbitrap XL (Thermo Fisher, Waltham, MA) integrated with an Eksigent nano-LC. A prepacked reverse-phase column (Acutech Scientific C18 with a dimension of 75 μm × 20 cm containing resin (Biobasic C18, 5-μm particle size, 300-Å pore size, Acutech Scientific, San Diego, CA) was used for peptide chromatography and subsequent CID analyses. ESI conditions using the nano-spray source (Thermo Fisher) for the Orbitrap were set as follows: capillary temperature of 220 °C, tu be lens 110 V and a spray voltage of 2.3 kV. The flow rate for reverse-phase chromatography was 0.5 μl/min for loading and 400 nl/min for analytical separation (buffer A: 0.1% formic acid, 3% ACN; buffer B: 0.1% formic acid, 100% ACN). Peptides were resolved by the following gradient: 0–40% buffer B over 180 min, and then returned to 0% buffer B for equilibration of 20 min. The Orbitrap was operated in data-dependent mode with a full precursor scan at high-resolution (60,000 at m/z 400) and ten MS/MS experiments at low resolution on the linear trap while the full scan was completed. For CID the intensity threshold was set to 5000, where mass range was 350–2000. Spectra are searched using Protein Prospector software (http://prospector2.ucsf.edu/prospector/mshome.htm) in which results with p<0.05 (95% confidence interval) were considered significant and indicating identity. Spectra for crosslinked peptides with score differences greater than 5 were examined manually.
Biolayer interferometry experiment
MmpL3 and MmpL11 D1 and D2 domain binding affinities were determined by biolayer interferometry (BLItz; ForteBio Inc.). All binding reactions were performed at 25 °C in 20 mM sodium phosphate pH 7.4 and 150 mM NaCl. Biotinylated D1 and D2 domains (NHS-PEG4-Biotin, ThermoScientific) were immobilized on streptavidin biosensors and exposed to different concentrations (25 – 3.125 μM) of interacting domains, as well as EC869 CdIO11 (Morse, et al., 2012) as a negative control. A buffer reference was subtracted from all binding curves before curve fitting. Curve fitting and data processing were performed using BLItz Pro software (ForteBio Inc.).
Model building
To build the MmpL3 D1 and D2 heterodimer model, I-Tasser models for the individual domains were first determined based on the MmpL11-D2 structure (Yang, et al., 2014). Both domain models were then oriented according to the CusA structure (PDB code: 4DNT) (Su, et al., 2012) where D1 and D2 were aligned to the PC1 and PN2 porter subdomains, respectively. The heterodimer models were energy minimized with Yasara force fields (Krieger, et al., 2009).
Supplementary Material
Highlights.
MmpL3 and MmpL11 contain two periplasmic domains (D1,D2) and a cytoplasmic one (D3)
The structure of MmpL11-D2 is homologous to RND transporter porter subdomains
MmpL3/11 D1 and D2 bind with weak affinities
MmpL3/11 D1-D2 models were built based on cross-linking data and RND transporters
Acknowledgments
This work was supported by grant AI095208 (C.W.G.) and P30DK063491 (J. P. W.) from the National Institute of Health. We thank the staff at Stanford Synchrotron Radiation Lightsource (SSRL) and Advanced Light Source (ALS) at Berkeley National Laboratories for their invaluable help in data collection. ALS is supported in part by the U.S. DOE Contract DE-AC02-05CH11231, and University of California Office of the President, Multicampus Research Programs and Initiatives grant MR-15-328599. The SSRL Structural Molecular Biology Program is supported by the U.S. DOE Office of Biological and Environmental Research and by the NIH-GMS (including P41GM103393). We also thank Robert Morse, Angelina Iniguez, Yama Latif, Mario Garcia, Daniel Yabuno, Katie Nguyen and Shivani Desai for technical assistance. Additionally, we thank Heidi Contreras, Robert Stroud, Tom Poulos and Cedric Owens for critical reading of the manuscript.
Footnotes
Author contributions
N.C. and C.W.G. conceived the experiments; N.C., R.T., Y.L., J.C., and G.B. performed the experiments; N.C., J.W., and C.W.G analyzed the data; N.C. and C.W.G. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolla JR, Su CC, Do SV, Radhakrishnan A, Kumar N, Long F, Chou TH, Delmar JA, Lei HT, Rajashankar KR, et al. Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS One. 2014;9:e97903. doi: 10.1371/journal.pone.0097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carel C, Nukdee K, Cantaloube S, Bonne M, Diagne CT, Laval F, Daffe M, Zerbib D. Mycobacterium tuberculosis proteins involved in mycolic acid synthesis and transport localize dynamically to the old growing pole and septum. PLoS One. 2014;9:e97148. doi: 10.1371/journal.pone.0097148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse SE, Mougous JD, Leavell MD, Leary JA, Bertozzi CR, Cox JS. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System on World Wide Web. 2002 http://www.pymol.org.
- Delmar JA, Su CC, Yu EW. Bacterial multidrug efflux transporters. Annu Rev Biophys. 2014;43:93–117. doi: 10.1146/annurev-biophys-051013-022855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P, Reed MB, Barry CE., 3rd Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Naka T, Doi T, Yano I. Direct molecular mass determination of trehalose monomycolate from 11 species of mycobacteria by MALDI-TOF mass spectrometry. Microbiology. 2005;151:1443–1452. doi: 10.1099/mic.0.27791-0. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Pribyl T, Juhnke S, Nies DH. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. The Journal of biological chemistry. 1999;274:26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Kordulakova J, et al. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol. 2012;8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Nakae T. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J Bacteriol. 2001;183:1734–1739. doi: 10.1128/JB.183.5.1734-1739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Cox JS. Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e2. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janganan TK, Bavro VN, Zhang L, Borges-Walmsley MI, Walmsley AR. Tripartite efflux pumps: energy is required for dissociation, but not assembly or opening of the outer membrane channel of the pump. Molecular microbiology. 2013;88:590–602. doi: 10.1111/mmi.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins. 2009;77(Suppl 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa V, Poce G, Canseco JO, Buroni S, Pasca MR, Biava M, Raju RM, Porretta GC, Alfonso S, Battilocchio C, et al. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob Agents Chemother. 2012;56:324–331. doi: 10.1128/AAC.05270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane G, Tyagi S, Bishai WR. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun. 2005;73:2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li K, Schurig-Briccio LA, Feng X, Upadhyay A, Pujari V, Lechartier B, Fontes FL, Yang H, Rao G, Zhu W, et al. Multitarget drug discovery for tuberculosis and other infectious diseases. J Med Chem. 2014;57:3126–3139. doi: 10.1021/jm500131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Upadhyay A, Fontes FL, North EJ, Wang Y, Crans DC, Grzegorzewicz AE, Jones V, Franzblau SG, Lee RE, et al. Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014 doi: 10.1128/AAC.03229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano A, Pasca MR, Provvedi R, Lucarelli AP, Manina G, Ribeiro AL, Manganelli R, Riccardi G. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis (Edinb) 2009;89:84–90. doi: 10.1016/j.tube.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, Goulding CW. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21480–21485. doi: 10.1073/pnas.1216238110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500:102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- Owens CP, Chim N, Goulding CW. Insights on how the Mycobacterium tuberculosis heme uptake pathway can be used as a drug target. Future Med Chem. 2013;5:1391–1403. doi: 10.4155/fmc.13.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens CP, Chim N, Graves AB, Harmston CA, Iniguez A, Contreras H, Liptak MD, Goulding CW. The Mycobacterium tuberculosis secreted protein Rv0203 transfers heme to membrane proteins MmpL3 and MmpL11. The Journal of biological chemistry. 2013;288:21714–21728. doi: 10.1074/jbc.M113.453076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco SA, Hsu FF, Powers KM, Purdy GE. MmpL11 protein transports mycolic acid-containing lipids to the mycobacterial cell wall and contributes to biofilm formation in Mycobacterium smegmatis. The Journal of biological chemistry. 2013;288:24213–24222. doi: 10.1074/jbc.M113.473371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak JE, Ekende EN, Kifle EG, O’Connell JD, 3rd, De Angelis F, Tessema MB, Derfoufi KM, Robles-Colmenares Y, Robbins RA, Goormaghtigh E, et al. Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18484–18489. doi: 10.1073/pnas.1318705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Ruggerone P, Murakami S, Pos KM, Vargiu AV. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr Top Med Chem. 2013;13:3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- Seeliger JC, Holsclaw CM, Schelle MW, Botyanszki Z, Gilmore SA, Tully SE, Niederweis M, Cravatt BF, Leary JA, Bertozzi CR. Elucidation and chemical modulation of sulfolipid-1 biosynthesis in Mycobacterium tuberculosis. The Journal of biological chemistry. 2012;287:7990–8000. doi: 10.1074/jbc.M111.315473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennhauser G, Bukowska MA, Briand C, Grutter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. Journal of molecular biology. 2009;389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Su CC, Long F, Lei HT, Bolla JR, Do SV, Rajashankar KR, Yu EW. Charged amino acids (R83, E567, D617, E625, R669, and K678) of CusA are required for metal ion transport in the Cus efflux system. Journal of molecular biology. 2012;422:429–441. doi: 10.1016/j.jmb.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011;470:558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE, 3rd, et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokhtaeva E, Capri J, Marcus EA, Whitelegge JP, Khuzakhmetova V, Bukharaeva E, Deiss-Yehiely N, Dada LA, Sachs G, Fernandez-Salas E, et al. Septin dynamics are essential for exocytosis. The Journal of biological chemistry. 2015;290:5280–5297. doi: 10.1074/jbc.M114.616201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius MV, Harmston CA, Owens CP, Chim N, Morse RP, McMath LM, Iniguez A, Kimmey JM, Sawaya MR, Whitelegge JP, et al. Discovery and characterization of a unique mycobacterial heme acquisition system. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5051–5056. doi: 10.1073/pnas.1009516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela C, Rittmann D, Singh A, Krumbach K, Bhatt K, Eggeling L, Besra GS, Bhatt A. MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem Biol. 2012;19:498–506. doi: 10.1016/j.chembiol.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ma J, Peng J, Xu J. Protein structure alignment beyond spatial proximity. Sci Rep. 2013;3:1448. doi: 10.1038/srep01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RM, Jones CM, Xi Z, Speer A, Danilchanka O, Doornbos KS, Sun P, Wu F, Tian C, Niederweis M. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 2013;9:e1003120. doi: 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The World Health Organization (WHO) Global tuberculosis report 2014. 2014 http://www.who.int/tb/publications/global_report/en/
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2014;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lu S, Belardinelli J, Huc-Claustre E, Jones V, Jackson M, Zgurskaya HI. RND transporters protect Corynebacterium glutamicum from antibiotics by assembling the outer membrane. Microbiologyopen. 2014;3:484–496. doi: 10.1002/mbo3.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.