Abstract
A versatile method for orchestrating the formation of side-chain-to-tail cyclic peptides from ribosomally derived polypeptide precursors is reported. Upon ribosomal incorporation into intein-containing precursor proteins, designer unnatural amino acids bearing side-chain 1,3- or 1,2-aminothiol functionalities are able to promote the cyclization of a downstream target peptide sequence via a C-terminal ligation/ring contraction mechanism. Using this approach, peptide macrocycles of variable size and composition could be generated in a pH-triggered manner in vitro, or directly in living bacterial cells. This methodology furnishes a new platform for the creation and screening of genetically encoded libraries of conformationally constrained peptides. This strategy was applied to identify and isolate a low micromolar streptavidin binder (KD = 1.1 µM) from a library of cyclic peptides produced in E. coli, thereby illustrating its potential toward aiding the discovery of functional peptide macrocycles.
Keywords: Peptide macrocyclization, Cyclic peptides, Intein, Protein splicing, Unnatural amino acid
INTRODUCTION
Macrocyclic peptides occupy a most relevant region of the chemical space and have attracted increasing attention as molecular scaffolds for addressing challenging pharmacological targets.1, 2 Important benefits arising from the cyclization of peptide sequences include pre-organization into a bioactive conformation and reduced entropic costs upon complex formation, which can result in enhanced binding selectivity and affinity, respectively, toward a target biomolecule.3–10 In addition, macrocyclization has proven to be beneficial toward improving the proteolytic stability11–13 and cell penetration ability14–16 of peptide-based molecules. These advantageous features are well reflected by the widespread occurrence of ring topologies amongst natural biologically active peptides, such as those produced by non-ribosomal peptide synthetases17, 18 (e.g. cyclosporin, polymyxin) or those belonging to the class of ribosomally synthesized and post-translationally modified peptides (RiPPs)19 (e.g., lanthipeptides20, cyclotides21, cyanobactins22).
Because of the attractive properties of macrocyclic peptides, the development of methods for the preparation of this class of compounds has attracted significant interest.23–27 Among these, strategies that rely on the cyclization of genetically encoded peptide sequences offer several advantageous features, which include the opportunity to create and evaluate large combinatorial libraries of these molecules by combining genetic randomization with high-throughput display/screening platforms.24–26 For example, biopanning of phage displayed peptide libraries cyclized via disulfide bridges28–32 or exogenous cross-linking agents33–35 has enabled the discovery of potent inhibitors of enzymes and proteins. Coupling in vitro translation methods with mRNA display, other groups have successfully isolated potent macrocyclic peptide inhibitors for a variety of other therapeutically relevant protein targets.36–41
Despite this progress, there are currently only a few methods that enable the production of macrocyclic peptides of arbitrary sequence within a cell, thereby providing the capability of coupling library production with a selection system or phenotypic screen.42–48 In this regard, a viable method is the so-called split-intein mediated circular ligation of peptides and proteins (SICLOPPS) introduced by Benkovic and coworkers,49 which enables the intracellular formation of head-to-tail cyclic peptides by exploiting the trans splicing reactivity of the natural split intein DnaE. More recently, a strategy useful for the generation in vitro and in vivo of macrocylic peptides constrained by an inter-side-chain thioether bond was reported by our group.50 While these methods provide a route to obtain head-to-tail or side-chain-to-side-chain cyclic peptides, complementary strategies to access alternative peptide macrocycle architectures would be highly desirable. For example, neither of these approaches allows the formation of side-chain-to-tail cyclic peptides, a molecular topology that is found in many bioactive peptide natural products (e.g. bacitracin A, polymixin D).
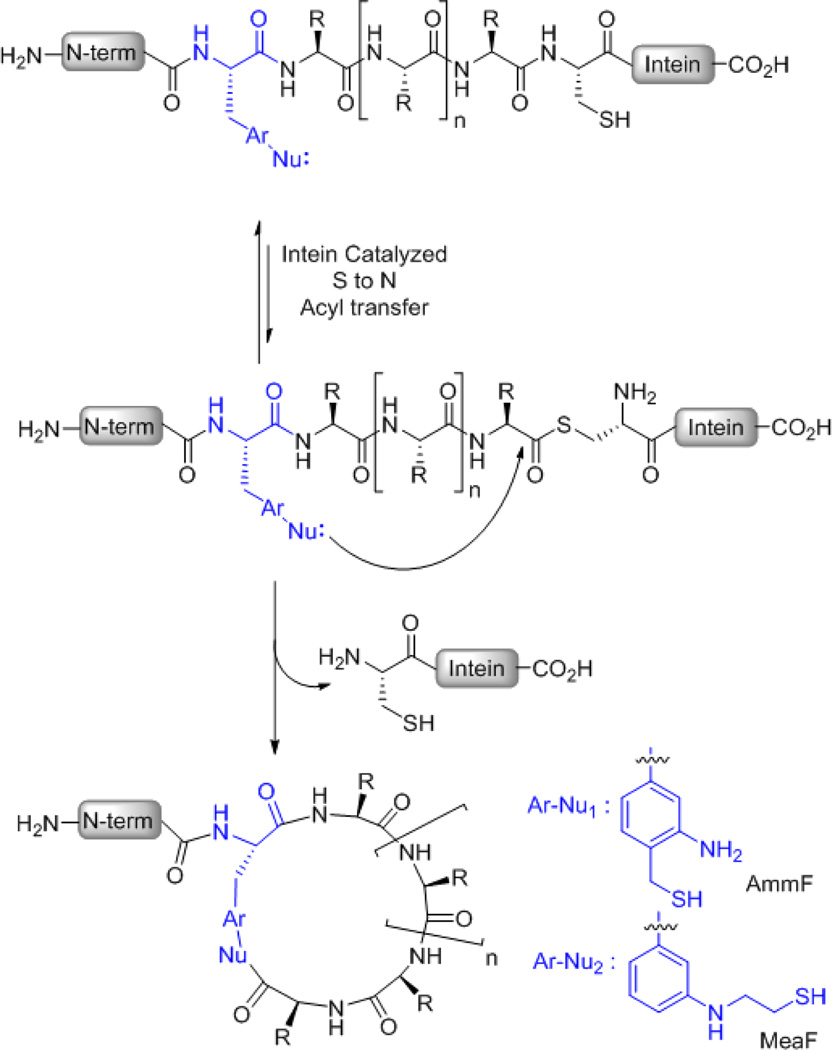
As part of our efforts toward the development of methods for the synthesis of macrocyclic organo-peptide hybrids,51–54 we recently described the ability of bifunctional oxyamine/1,3-amino-thiol aryl reagents to induce the efficient cyclization of polypeptide precursors containing a target peptide sequence flanked by a side-chain keto group and a C-terminal intein.52 Albeit two possible routes for macrocyclization are available in this system (e.g. side-chain → C-end versus C-end → side-chain ligation), our studies revealed that this process is largely driven by the intermolecular attack of the 1,3-amino-thiol aryl moiety onto the intein-catalyzed thioester bond within the polypeptide precursor.53 These mechanistic insights suggested to us the possibility of directing the biosynthesis of side-chain-to-tail macrocyclic peptides through the ribosomal expression of a precursor polypeptide in which an appropriately designed amino-thiol-based unnatural amino acid is placed upstream of an intein (Figure 1). Given the expected rate enhancement in moving from an intermolecular to an intramolecular setting, we envisioned this construct could undergo a spontaneous post-translational 'self-processing' reaction through (a) capturing of the transient intein-catalyzed thioester linkage by the side-chain thiol functionality, followed by (b) irreversible acyl shift to the neighboring amino group to give the desired macrocyclic peptide. Here, we describe the successful implementation of this design strategy and its functionality toward enabling the ribosomal synthesis of macrocyclic peptides both in vitro and in living cells. Furthermore, it is shown how this approach could be successfully applied to evolve and isolate a cyclopeptide with improved binding affinity toward a model target protein (streptavidin).
Figure 1.
Overview of the strategy for generating side-chain-to-tail macrocyclic peptides via cyclization of ribosomally derived, intein-fused precursor proteins by means of amino-thiol unnatural amino acids. 'Nu' refers to the nucleophilic 1,3- or 1,2-aminothiol moiety.
RESULTS AND DISCUSSION
Synthesis of 1,3-aminothiol amino acid, AmmF
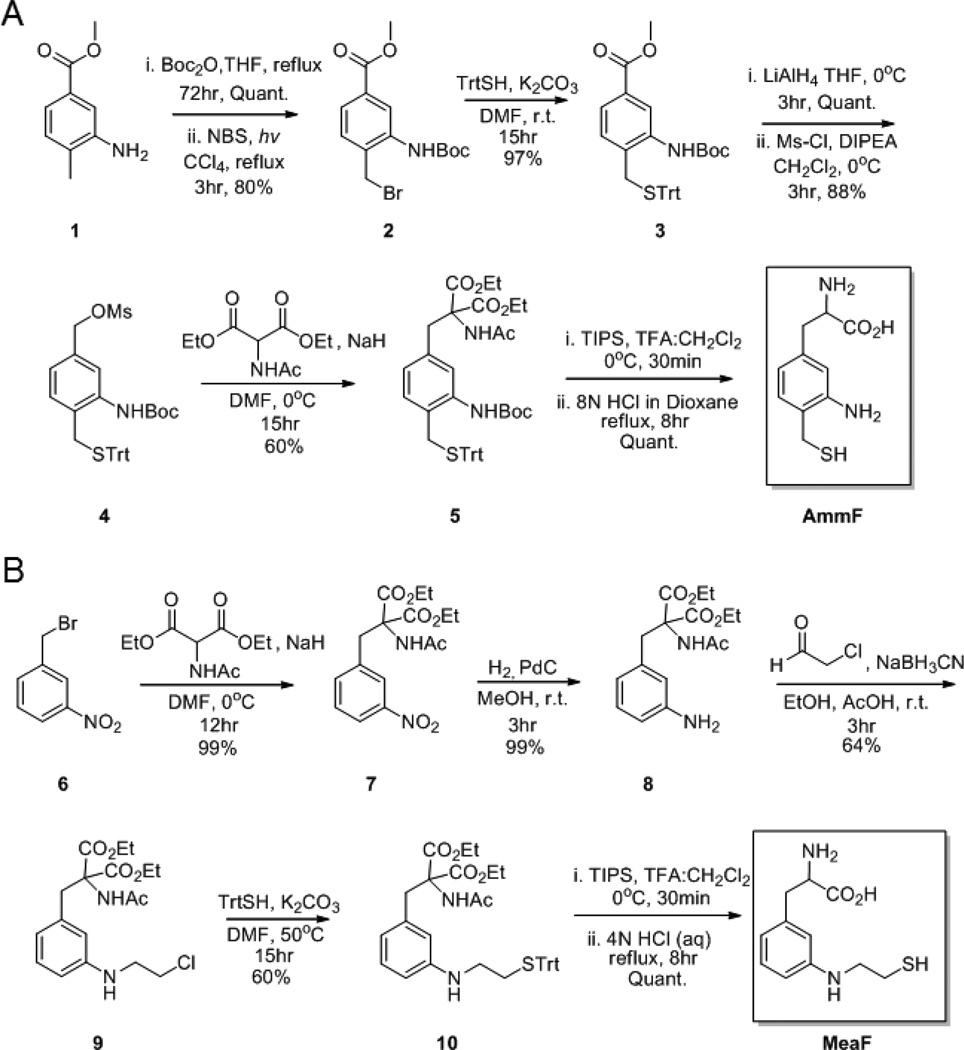
In previous work, we recognized the peculiar ability of an o-amino-mercaptomethyl-aryl (AMA) group to undergo efficient C-terminal ligation at GyrA intein thioesters under catalyst-free conditions.53 Based on this knowledge, an amino acid incorporating this reactive moiety, 3-amino-4-mercaptomethyl-phenylalanine (AmmF, Scheme 1A), was designed in order to test its ability to mediate peptide cyclization according to the strategy outlined in Figure 1. As outlined in Scheme 1A, gram-scale synthesis of AmmF was carried out starting from commercially available methyl 3-amino-4-methyl-benzoate, followed by N-Boc protection, benzylic bromination and benzylic substitution with tritylmercaptan to afford intermediate 3. Lithium aluminum hydride reduction of the methyl ester group in 3, followed by mesylation, and substitution of the mesyl group with diethylacetamidomalonate, yielded intermediate 5. Following deprotection, the desired product, AmmF, was obtained in 41% overall yield over 8 steps.
Scheme 1.
Synthetic routes for the preparation of (A) 3-amino-4-mercaptomethyl-phenylalanine (AmmF), and (B) 3-(2-mercapto-ethyl)amino-phenylalanine (MeaF).
Aminoacyl-tRNA synthetase for ribosomal incorporation of AmmF
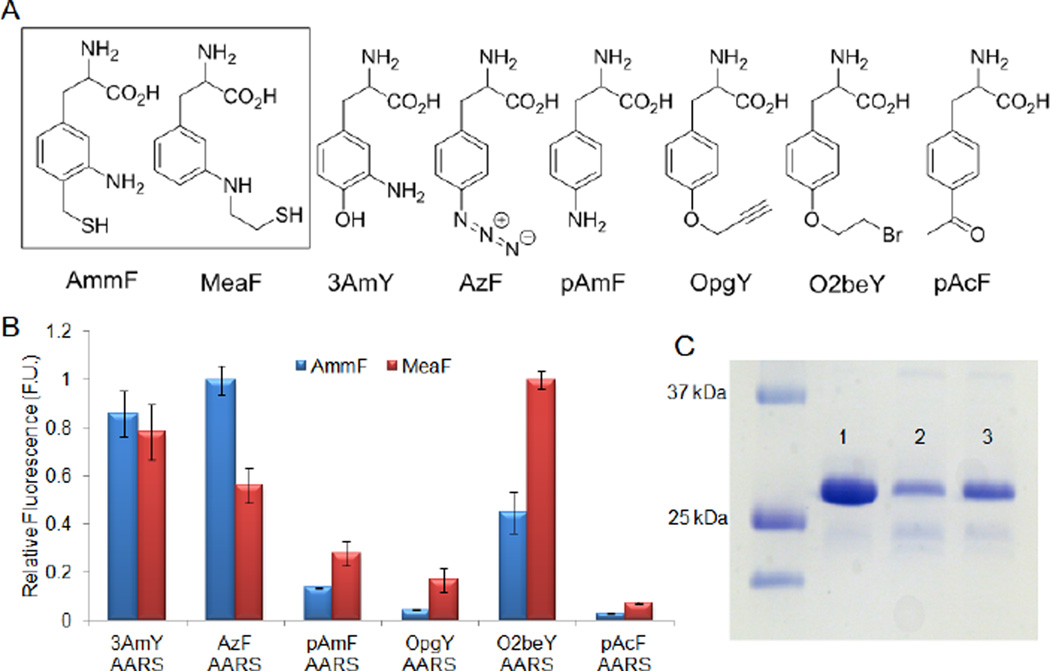
With AmmF in hand, we sought to identify an orthogonal aminoacyl-tRNA synthetase (AARS) / tRNA pair for the riboso mal incorporation of this unnatural amino acid (UAA) via amberstop codon (TAG) suppression.55 Interestingly, a number of engi neered AARS enzymes have been shown to be 'polyspecific', that is capable of charging a cognate tRNA molecule with multiple UAAs while maintaining discriminating selectivity against the natural amino acids.56, 57 We reasoned this feature could be exploited to find a suitable AARS for recognition of AmmF. To this end, we generated a panel of six previously reported AARSs derived from Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (MjTyr-RS) in combination with the cognate amber stop codon suppressor tRNA (MjtRNATyrCUA). This panel included MjTyr-RS variants selected for recognition of 3-aminotyrosine (3AmY)58, p-aminophenylalanine (pAmF)59, p-azidophenylalanine (AzF)60, p-acetylphenylalanine (pAcF)61, O-propargyltyrosine (OpgY)62, and O-(2-bromoethyl)-tyrosine (O2beY)50(Figure 2A).
Figure 2.
Orthogonal aminoacyl-tRNA synthetases for ribosomal incorporation of the amino-thiol amino acids. (A) Structures of the target amino acids, AmmF and MeaF, and other UAAs mentioned in the text. (B) Relative fluorescence measured in the YFP reporter assay for the panel of AARSs in the presence of AmmF or MeaF. Data are normalized to the highest value within each series. (C) SDS-PAGE analysis of purified wild-type YFP (lane 1) and YFP variants incorporating AmmF (lane 2) and MeaF (lane 3).
Among these, 3AmY-RS was considered a particularly promising candidate owing to the structural similarity between 3AmY and AmmF. The relative ability of the AARSs to incorporate AmmF in response to a TAG codon was established via a fluorescence assay with a Yellow Fluorescent Protein (YFP) variant containing an amber stop codon at the N-terminus. In this assay, the suppression efficiency and fidelity of each synthetase is measured based on the relative expression levels of the reporter YFP protein in the presence and in the absence of AmmF, respectively (Figure 2B). Gratifyingly, the 3AmY-RS synthetase was found capable of incorporating AmmF with good efficiency as compared to most of the other AARSs. Interestingly, the AzF-RS synthetase exhibited even better performance in that respect, as judged by the higher fluorescence signal (+15%) measured in the assay. This result is consistent with previous observations by Schultz and coworkers regarding the ability of AzF-RS to recognize various meta, para-disubstituted phenylalanine derivatives, in addition to AzF.57 Further experiments were then carried out to quantitatively assess the suppression efficiency of AzF-RS with AmmF. For this purpose, YFP(AmmF) and wild-type YFP were expressed under identical conditions and purified by Ni- affinity chromatography. Direct comparison of the expression yield (16 vs. 45 mg/L culture, respectively) indicated a suppression efficiency of 36% (Figure 2C), a value comparable to that reported for AARSs specifically evolved for the ribosomal incorporation of other UAAs.57
Characterization of AmmF-containing precursor proteins
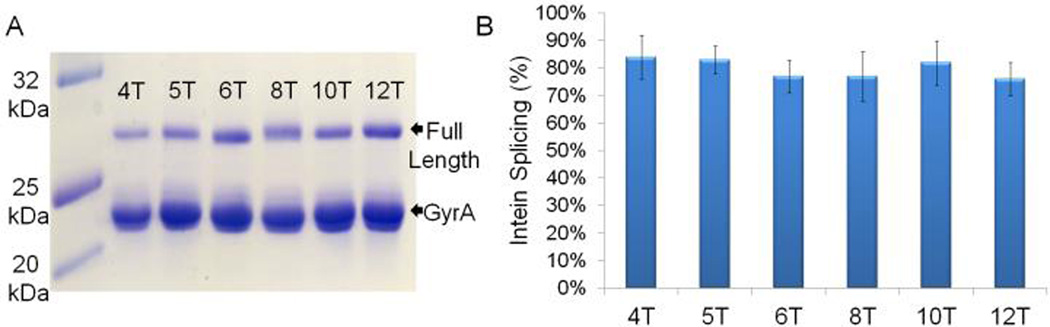
Having identified a viable AARS for AmmF incorporation, we tested the ability of this amino acid to promote peptide macrocyclization according to the scheme outlined in Figure 1. To this end, we investigated a panel of six constructs encompassing target peptide sequences of variable length (4 to 12 amino acids) framed in between AmmF and the N198A variant of GyrA mini-intein from Myobacterium xenopi63 (Entries 1–6, Table 1). These constructs would allow us to probe the formation of peptide macrocycles of varying ring size. In addition, the N-terminal chitin binding domain (CBD) would serve as a large (~8 kDa) affinity tag to facilitate both the isolation of the products formed in cells and measurement of the extent of intein cleavage via SDS-PAGE densitometry. Accordingly, the precursor proteins were produced in E. coli cells co-transformed with a pET-based plasmid encoding for the precursor protein and a pEVOL64-based plasmid for the co-expression of the AzF-RS/tRNACUA pair for the site-selective incorporation of AmmF.
Table 1.
Precursor protein constructs investigated in this study. CBD = Chitin Binding Domain; UAA = unnatural amino acid (i.e. AmmF or MeaF); GyrA = Mxa GyrA intein (N198A variant).
| Entry | Construct name |
Peptide sequence |
|---|---|---|
| 1 | CBD-4T | |
| 2 | CBD-5T | |
| 3 | CBD-6T | |
| 4 | CBD-8T | |
| 5 | CBD-10T | |
| 6 | CBD-12T | |
| 7 | MG-8T | |
| 8 | CBD-7T | |
| 9 | Strep1 | |
| 10 | Flag-Strep1 | |
| 11 | Flag-Strep2 | |
| 12 | Flag-Strep3 | |
Contrary to our expectations, only low levels of intein cleavage (20–25%) were observed for all the constructs under the applied expression conditions (12 hrs, 27°C), as determined by SDS-PAGE analysis. These values were comparable to those obtained with control constructs in which AmmF was substituted for a non-nucleophilic UAA (= pAcF). Moreover, no traces of the desired macrocyclic compounds were detected after processing the cell lysates with chitin beads.
These results were surprising, given that compounds sharing the o-amino-mercaptomethyl-aryl moiety of AmmF can induce nearly quantitative clevage of GyrA-fusion proteins (80–90%) in less than 10–12 hours.53 To investigate the basis of this unexpected lack of reactivity, the AmmF-containing constructs were isolated in full-length form using the C-terminal His tag and then incubated with thiophenol to cleave the intein and release the N-terminal CBD-containing fragment. MALDI-TOF analysis of these reactions showed the accumulation of a single product corresponding to the linear peptide with an additional mass of +26 Da for each of the constructs (Figure S1). In contrast, under identical conditions, control proteins containing pAcF instead of AmmF produced a species corresponding to the expected linear peptide with no modifications. These results pointed at a post-translational modification occurring specifically at the level of the unnatural amino acid in the AmmF-containing constructs. The observed mass difference was found to be consistent with the formation of a 2,4-dihydro-benzo[1,3]thiazine adduct (Figure 3A, species b′) resulting from a condensation/decarboxylation reaction of the AMA moiety in AmmF with pyruvate, an abundant metabolite in E. coli.65 Importantly, a similar reaction, leading to a thiazolidine adduct, was reported for polypeptides carrying an exposed N-terminal cysteine upon expression in this organism.66 To confirm the assignment, a model reaction was carried out by incubating a small-molecule surrogate of AmmF, methyl 3-amino-4-(mercaptomethyl)benzoate, with pyruvate under physiological conditions (50 mM KPi, 150 mM NaCl, pH 7.5). LC-MS analysis revealed quantitative conversion of the compound to the pyruvate condensation adduct within a short time (Figure S2). Altogether, these experiments indicated that rapid and quantitative intracellular modification of AmmF by pyruvate impedes the desired in vivo peptide cyclization process. Attempts to minimize this modification by altering the expression conditions66 failed to result in a detectable increase of the unmodified AmmF-containing protein.
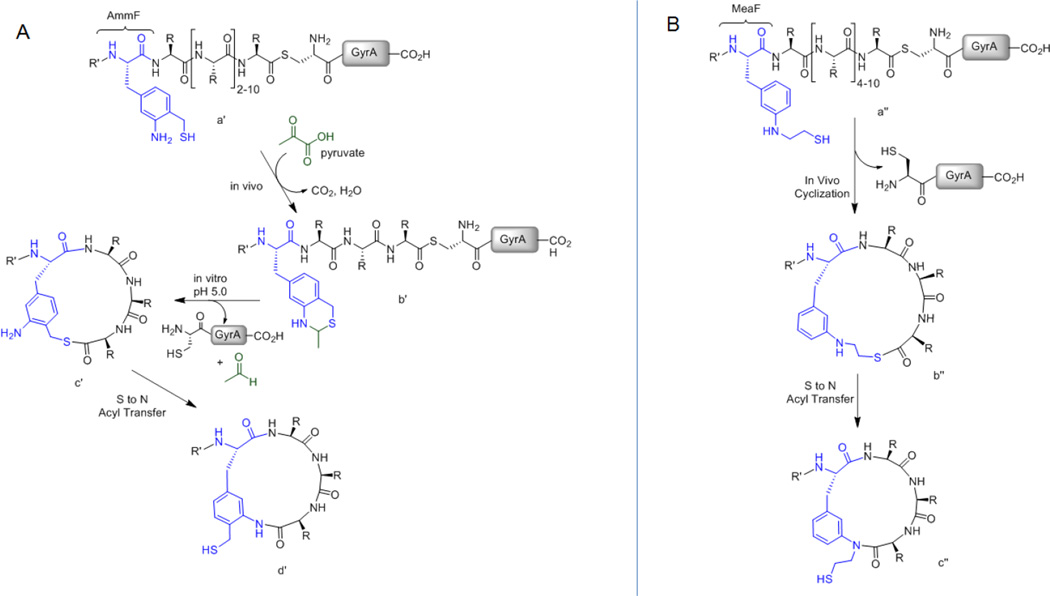
Figure 3.
Precursor polypeptides, reaction intermediates and final macrocyclic peptide products for (A) AmmF-mediated and (B) MeaF-mediated peptide cyclization. (A) Expression of the AmmF-containing precursor protein (a′) in E. coli results in the formation of the benzothiazine adduct (b′), which upon pH-induced deprotection in vitro leads to the target macrocyclic peptide (d′). (B) Expression of the MeaF-containing precursor protein (a″) in E. coli results in spontaneous, post-translational peptide cyclization to give the thiolactone intermediate (b″) and then the final macrolactam product (c″) via an S→N acyl shift rearrangement.
In vitro macrocyclization of AmmF-containing constructs
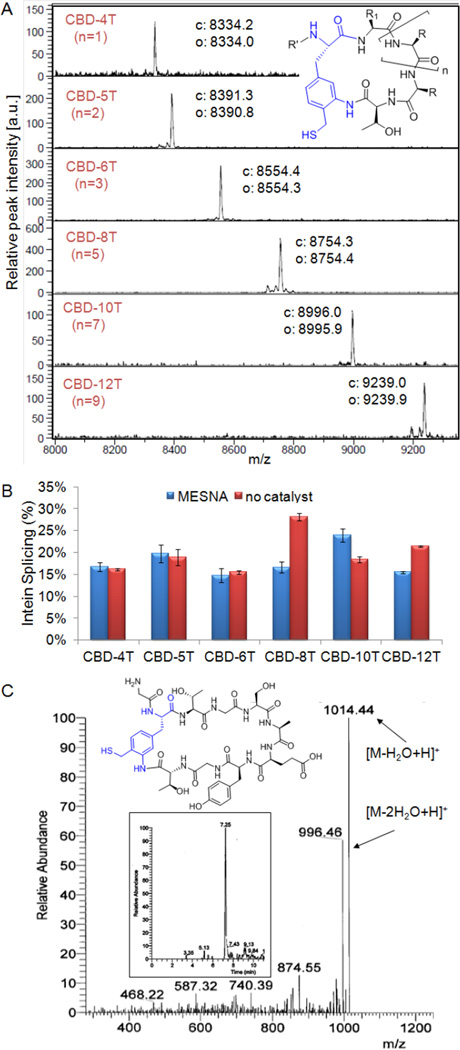
These findings prompted us to explore the possibility to execute the desired AmmF-mediated peptide macrocyclization upon chemical unmasking of its modified 1,3-amino-thiol functionality. While not directly applicable for intracellular synthesis of cyclic peptides, this approach would provide a means to trigger the formation of peptide macrocycles in a time-controlled manner in vitro. Although deprotection of cysteine-derived thiazolidines has been achieved under rather harsh, protein denaturing conditions (0.2 M methoxyamine, 6 M guanidinium chloride),67, 68 we discovered that the AmmF dihydrobenzothiazine adduct can be readily converted to the free 1,3-aminothiol counterpart by simple exposure of the protein to slightly acidic pH, under which conditions the GyrA intein remains functional. Gratifyingly, incubation of the AmmF-containing constructs at pH 5.0 and in the presence of sodium 2-sulfanylethanesulfonate (MESNA) resulted in the formation of small molecular weight products with masses corresponding to the expected CBD-fused peptide macrocycle in each case, as determined by MALDI-TOF MS (Figure S3). Notably, identical results could be achieved in the absence of any thiol catalyst, resulting in the clean formation of the desired macrocyclic lactams as the only product (Figure 4A).
Figure 4.
pH-induced cyclization of AmmF-containing constructs in vitro. A) MALDI-TOF MS spectra of the macrocyclic peptides generated under catalyst-free conditions. The calculated ("c") and observed ("o") m/z values corresponding to the [M+H]+ adduct of the macrocylic product are indicated. R' = chitin binding domain. B) Extent of intein cleavage for constructs CBD-4T(AmmF) through CBD-12T(AmmF) at pH 5.0 after 24 hours as determined by SDS-PAGE. C) Chemical structure, extracted-ion chromatogram (insert), and MS/MS spectrum of the macrocyclic peptide obtained from pH-induced cyclization of the MG-8T(AmmF) construct. Calculated m/z values for the [M-H2O+H]+ and [M-2H2O+H]+ species are 1014.07 and 996.05, respectively.
Exposure of these macrocyclic products to iodoacetamide resulted in complete conversion to the S-alkylated adduct (+57, Figure S4), demonstrating the occurrence of the desired S→N acyl transfer rearrangement (c′ → d′ step, Figure 3A) to give rise to the hydrolytically stable, side-chain-to-C-terminus amide linkage. Although the extent of intein cleavage in these reactions was not elevated (15–28%, Figure 4B), AmmF-mediated cyclization was found to be largely independent of the target peptide length, enabling the formation of peptide macrocycles spanning from 4 to 12 amino acid residues with equal efficiency. Suspecting that steric clashes between the CBD and intein protein may disfavor attack of AmmF thiol group onto the downstream thioester (Figure 3A), an 8mer construct in which the CBD was replaced with a smaller N-terminal tail (Met-Gly) was also tested (Entry 7, Table 1). Incubation of the corresponding protein precursor at pH 5.0 resulted in the formation of the desired peptide macrocycle, whose cyclic structure was further confirmed by MS/MS analysis (Figure 4C). Importantly, the construct was determined to undergo 64% intein cleavage (vs. 28% for the CBD-fused counterpart) with the macrocycle as the only detectable product, thereby demonstrating the possibility to dramatically increase the yields of these macrocyclization reactions via the use of shorter (or more flexible) N-terminal tails.
Design and synthesis of 1,2-aminothiol unnatural amino acid for in vivo macrocycle formation
Having demonstrated the possibility to generate macrocyclic peptides in vitro by means of AmmF, we next sought an alternative aminothiol UAA to enable this process to take place in vivo. To this end, a second phenylalanine derivative was designed, namely 3-(2-mercapto-ethyl)amino-Phe (MeaF, Figure 1), which was expected to exhibit superior performance than AmmF in that respect based on the following arguments. First, the secondary amino group of MeaF was expected to disfavor condensation with pyruvate in cells, thereby preserving the side-chain aminothiol functionality required for cyclization. Second, the more acidic and less sterically hindered sulphydryl group in MeaF compared to that in AmmF (i.e. pKa of ~8.569 vs. ~9.570) would favor nucleophilic attack onto the downstream intein thioester, in particular at the near-neutral conditions of the intracellular milieu. Third, the thiolactone formed during the initial transthiosterification step was envisioned to be less strained in the presence of MeaF vs. AmmF (b″ vs. c′, Figure 3) Finally, MeaF 1,2-aminothiol moiety was expected to remain capable of undergoing an S→N acyl shift rearrangement to yield the desired N-alkyl macrolactam, as suggested by peptide ligation studies with other aminothiol compounds71.
As summarized in Scheme 1B, MeaF was prepared starting from 3-nitro-benzyl bromide via alkylation with diethylacetamidomalonate and nitro group reduction by hydrogenation with Pd/C catalyst. Reductive amination of intermediate 8 with α-chloroacetaldehyde, followed by substitution with triphenylmethylmercaptan resulted in the protected intermediate 10. Deprotection with TFA followed by decarboxylation with concentrated HCl resulted in the desired amino acid MeaF in 38% isolated yield over six steps. This route could be scaled up to the gram scale (1.5 g final product) with no significant reduction in the overall yield.
AARS for ribosomal incorporation of MeaF
The same approach described above for AmmF was applied to identify a suitable AARS for amber stop codon suppression with MeaF. Initial screening of the panel of engineered Mj AARSs showed that both pAmF-RS and AzF-RS can recognize and incorporate MeaF into the reporter YFP protein (Figure 2B). At the same time, O2beY-RS emerged as the most promising enzyme for this purpose based on the approximately 20% higher fluorescent signal in the assay (Figure 2B). This synthetase, which carries a Ala32Gly mutation in the OpgY-RS background, was engineered by our group to accommodate tyrosine derivatives containing larger alkyl substituents at the para position.50 Considering the much lower incorporation of MeaF obtained with OpgY-RS, this substitution is clearly beneficial also toward recognition of meta-substituted aromatic UAAs such as MeaF. Comparison of the expression yield of this protein (23 mg/L culture) with that of wild-type YFP (45 mg/L culture) indicated a suppression efficiency as high as 52%, that is an about 16% higher efficiency than that achieved with AmmF and AzF-RS (Figure 2C).
In vivo cyclization of MeaF-containing precursor proteins
To compare side-by-side the performance of MeaF vs. AmmF toward promoting peptide macrocyclization in vivo, the same six constructs with a 4mer to 12mer target sequence (Entries 1–6, Table 1) were expressed in E. coli in the presence of MeaF and the O2beY-RS-based suppressor system. Notably, SDS-PAGE analysis of these proteins revealed a significantly larger degree of intein cleavage (75–85%, Figure 5) as compared to the related AmmF-containing counterparts (20–25%) under identical expression conditions. Since the two sets of constructs differ only by the side-chain aminothiol functionality in the installed UAA, these results strongly suggested that the ribosomally incorporated MeaF had remained available for nucleophilic attack on the intein thioester.
Figure 5.
MeaF mediated intein cleavage. A) SDS-PAGE analysis of the precursor proteins CBD-4T(MeaF) through CBD-12T(MeaF) after expression for 12 hours in E. coli. Bands corresponding to full-length protein and cleaved GyrA intein are labeled. B) The extent of intein cleavage as measured by gel band densitometric analysis.
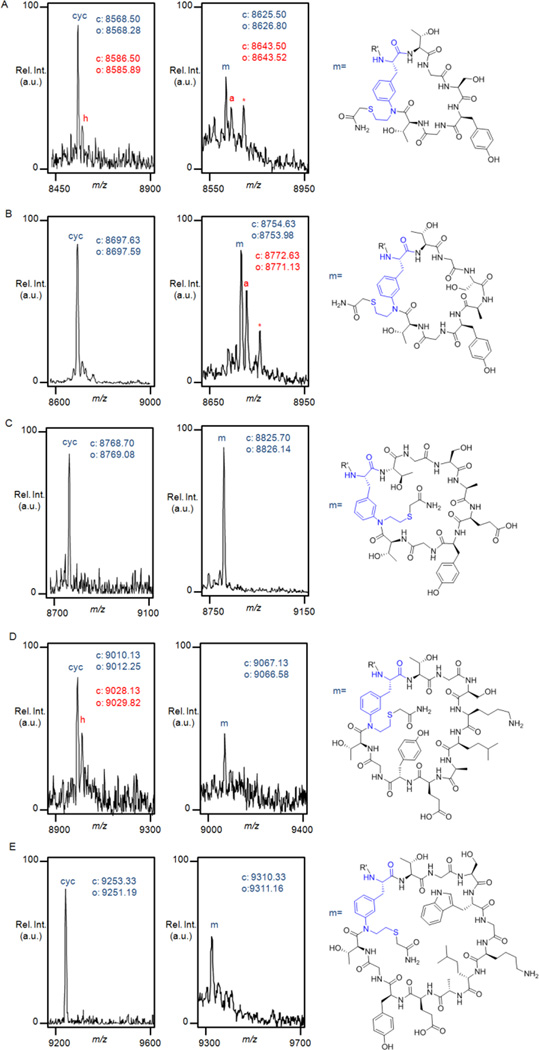
To examine the outcome of these in vivo reactions, the cleaved N-terminal fragments were isolated from the cell lysates using chitin-coated resin beads. To our delight, MALDI-TOF MS analysis of these samples revealed the occurrence of a macrocyclic product ("cyc", Figure 6) corresponding to either the thiolactone or the isobaric lactam (species b″ and c″, respectively, in Figure 3B), as the only or largely predominant product for the constructs with 6 to 12 amino acid-long target sequences. In contrast, the 4mer and 5mer constructs showed only MS signals consistent with the acyclic, hydrolyzed product (Figure S5). Although this species could arise from hydrolysis of the full-length intein-containing precursor protein directly, the higher degree of intein cleavage observed with the MeaF-containing vs. AmmF-containing constructs (Figure 5) suggests that it more likely derives from hydrolysis of the thiolactone intermediate formed after MeaF-mediated transthioesterification. Importantly, no adducts were observed for any of the constructs, indicating that the side-chain functionality of MeaF did not react with pyruvate or other intracellular metabolites.
Figure 6.
MALDI-TOF spectra of the macrocyclization products obtained from the MeaF-containing constructs as isolated from E. coli cells after 12 hours (left panel) and 24 hours (right panel), using pH 5.0 buffer and pH 8.0 buffer containing iodoacetamide, respectively. (A) CBD-6T(MeaF); (B) CBD-7T(MeaF); (C) CBD-8T(MeaF); (D) CBD-10T(MeaF); and (E) CBD-12T(MeaF). See also Figures S5–S6. "cyc" = macrocyclic product in either thiolactone or lactam form; "h" = hydrolyzed product, "m" = macrolactam (S-carboxyamidomethyl adduct), "a" = acyclic side product (S-carboxyamidomethyl adduct), * = acetylated "a". The calculated ("c") and observed ("o") m/z values corresponding to the [M+H]+ adducts are indicated and color coded.
To assess the occurrence and extent of S→N acyl shift rearrangement in the 6mer to 12mer macrocyclic products, the corresponding cell lysate samples were first treated with iodoacetamide under alkaline conditions (pH 8) prior to chitin-affinity purification. Through this procedure, the macrolactam is converted to its corresponding S-carboxyamidomethyl adduct ('m', Figure S6) whereas the thiolactone is hydrolysed and alkylated, thus appearing as the S-alkylated adduct of the linear acyclic peptide ('a'). These experiments showed that the 6mer, 10mer, and 12mer-based macrocycles were present exclusively or mostly (~90% for 12mer) in the thiolactone form. As an exception, the 8mer-based macrocycle was found to have undergone partial rearrangement, being present in about 40% as the desired lactam product and the remaining 60% as thiolactone as estimated based on the intensity of the corresponding MS signals (Figure S6C) and assuming the two species share similar ionization properties. Collectively, these results demonstrated the ability of MeaF to efficiently mediate the first step (i.e. intramolecular transthioesterification) of the envisioned macrocyclization process within 12 hours. The partial rearrangement of the 8mer-based construct also suggested that the second step (S,N acyl shift) is comparatively much slower. This finding is consistent with the slow kinetics of the S,N acyl transfer observed for 1,2-aminothiol moieties containing secondary amines in the context of peptide ligation studies71.
Given the remarkable stability in vivo of the thiolactone intermediate for the 6mer to 12mer constructs, we reasoned that longer culture times could result in the intracellular formation of the desired macrolactam product. Accordingly, cell cultures were grown for 24 hours prior to isolation and analysis of the spliced products using the procedure described above. Gratifyingly, these experiments showed a dramatic increase in the S,N acyl transfer rearrangement for all the constructs. Indeed, the 6mer-based macrocycle was found to consist for the most part of the desired lactam product (~60%, Figure 6A). Even better results were achieved with the 8mer, 10mer, and 12mer-based macrocycles, which were isolated exclusively in the lactam form (Figure 6, C-E). These results were corroborated by parallel tests in which the peptides were isolated via chitin affinity immediately after cell lysis under alkaline conditions (pH 8.0). Under these conditions, MS analysis revealed a macrocycle : hydrolyzed product ratio (corresponding to the lactam : thiolactone ratio) comparable or identical to the 'm' : 'a' ratio obtained after incubation with iodoacetamide, thus further confirming the occurrence of the S,N acyl transfer step inside the cells.
An intriguing aspect emerging from these studies concerns the effect of the target peptide length on the outcome of MeaF-mediation peptide cyclization in vivo. Our results indicate that 4mer and 5mer sequences undergo MeaF-induced transthioesterification, but the resulting thiolactones are apparently susceptible to hydrolysis before the irreversible S,N acyl transfer process can take place. In contrast, both of these steps occur efficiently in the context of longer peptide sequences (6–12 amino acids). Possibly, a higher ring strain in the thiolactone intermediate and/or slower kinetics for the subsequent ring contraction step may be at the basis of the higher susceptibility of the shorter constructs toward hydrolysis. In the future, it will be interesting to determine whether alternative aminothiol UAAs and/or alterations within the peptide sequence (e.g. at the 'Intein-1' site) can provide access also to these smaller rings. Another implication of the results above is that 7mer peptide sequences should also be amenable to cyclization. To test this hypothesis, the construct corresponding to Entry 8 in Table 1 was prepared and expressed. As anticipated, this construct underwent extensive cyclization in vivo (~75–80% intein cleavage), resulting in the formation of the desired CBD-fused peptide macrocycle as the major product (~60:40 lactam:thiolactone ratio, Figure 6B). Altogether, these studies demonstrated the functionality of the methodology outlined in Figure 3B to orchestrate the spontaneous, intracellular formation of side-chain-to-tail peptide macrocycles in E. coli.
Generation of a streptavidin-binding macrocyclic peptide in living cells
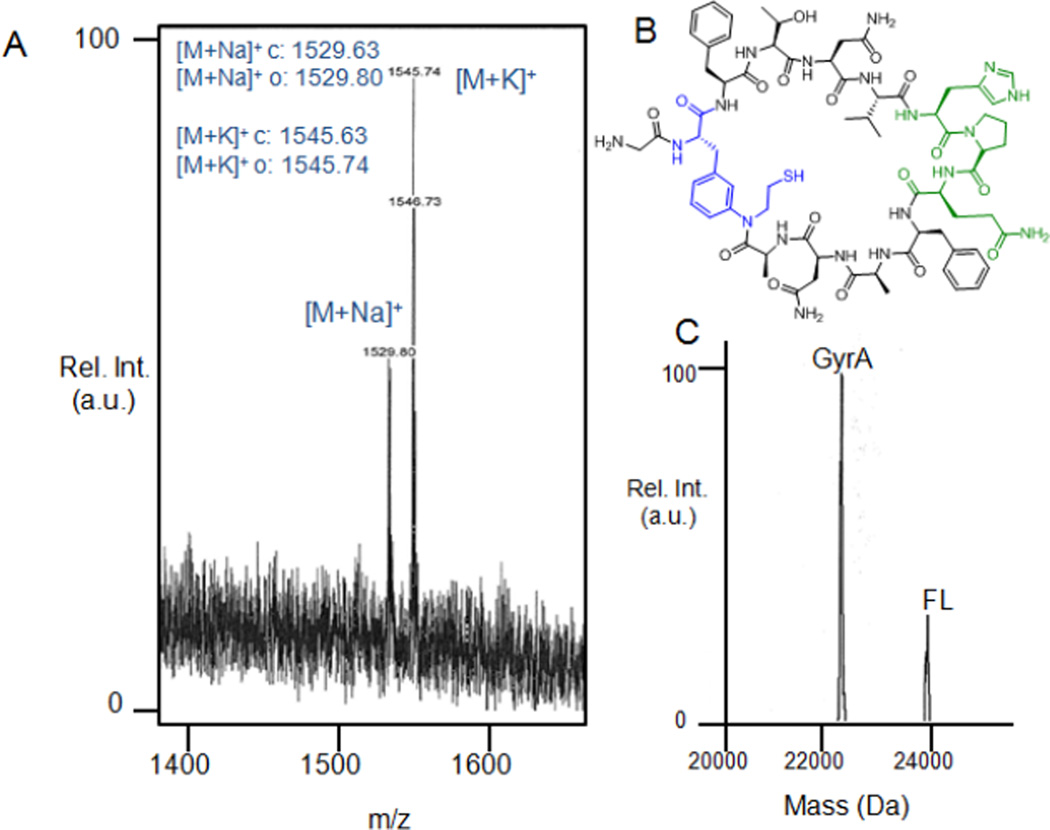
To demonstrate the production of a bioactive macrocyclic peptide in living cells, an additional construct (Entry 9, Table 1) was designed based on a streptavidin-binding head-to-tail cyclopeptide reported by Benkovic and coworkers.72 In this case, the precursor protein consists of a short N-terminal tail (Met-Gly), followed by MeaF and a 11mer peptide sequence encompassing a histidine-proline-glutamine (HPQ) motif known to interact with streptavidin.73,74 After expression of this construct in E. coli, cell lysates were passed over streptavidin-coated beads to isolate any streptavidin-bound material. Remarkably, analysis of the eluate revealed the presence of the desired macrocyclic peptide (cyclo(Strep1)) as the only product, as shown in the MS spectrum in Figure 7A. The lack of detectable amounts of the acyclic by-product, which would be also captured by this procedure as determined via control experiments, indicated that MeaF-mediated cyclization had occurred with high efficiency, that is outcompeting hydrolysis of the full-length construct and/or of the thiolactone intermediate. Furthermore, the precursor protein was determined to have undergone >75% cleavage (Figure 7C), a value comparable to that observed for target sequences of comparable length in the CBD-fused format. Based on the expression level of the isolated GyrA protein and extent of in vivo cyclization, the yield of the Strep1-derived cyclopeptide was estimated to be around 0.85 mg L−1.
Figure 7.
Isolation of streptavidin-binding peptide macrocycle from E. coli. (A) MALDI-TOF spectrum and (B) chemical structure of the cyclo(Strep1) as isolated from E. coli cells using streptavidin-coated beads. The HPQ motif is highlighted in green. (C) LC-MS spectrum of the Ni-affinity purified full-length precursor protein ('FL') and spliced GyrA intein (GyrA) illustrating the extent of intein cleavage occurred in vivo.
Isolation of improved streptavidin-binding peptide from macrocyclic peptide library
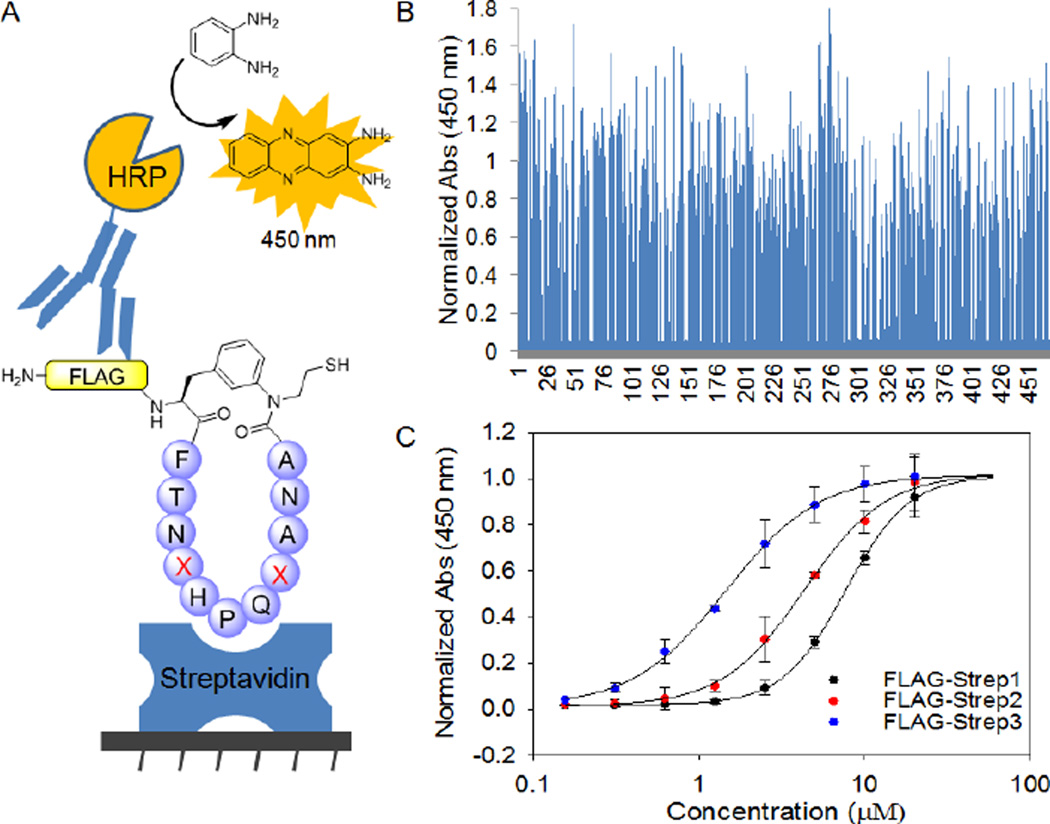
To begin to explore the potential utility of this methodology toward the discovery of macrocyclic peptides with novel or improved function, cyclo(Strep1) was subjected to mutagenesis in search for cyclopeptide variants with enhanced affinity toward streptavidin. To this end, we availed of insights from the available crystal structure of streptavidin bound to a HPQ-containing peptide isolated by phage display.75 Inspection of this complex revealed that whereas the key HPQ motif inserts into the biotin binding site of streptavidin, the amino acid residues flanking this region also lie within short distance from the protein surface (< 6 Å, Figure S7). In addition, these residues are likely to influence the β-turn conformation of the HPQ sequence, thereby affecting the affinity of the peptide ligand for the protein. Accordingly, a library of cyclic peptides was generated by randomizing the positions flanking the HPQ tripeptide in the Strep1 target sequence (V18, F22) using the degenerate codon NDC (12 codons/12 amino acids). In the corresponding precursor proteins, a FLAG peptide (sequence: DYKDDDDK) was placed upstream of the UAA and the semi-randomized target sequence in order to obtain macrocyclic peptides with an N-terminal affinity tag useful for detection purposes. As a reference, a construct encoding for a FLAG-tagged version of cyclo(Strep1) was also prepared (Entry 10, Table 1).
To screen the library, an ELISA-like assay was implemented and validated using a synthetic biotinylated FLAG peptide. As schematically illustrated in Figure 8A, this assay relies on capturing the streptavidin-binding peptide on a streptavidin-coated plate, followed by colorimetric detection and quantification of the amount of bound peptide by means of an anti-FLAG antibody-horseradish peroxidase (HRP) conjugate. After expression and cell lysis in a 96-well plate format, 480 recombinants from the Flag-Strep1(V18NDC/F22NDC) library were screened using this assay. This experiment led to the identification of ten variants displaying a significantly enhanced response (>1.5 fold) as compared to the reference Flag-cyclo(Strep1) peptide (Figure 8B). Re-screening of these ‘hits’ yielded two most promising variants, named Flag-Strep2 and Flag-Strep3, which exhibited >2-fold higher signal in the immunoassay over that of the parent cyclic peptide (Figure S8). Upon sequencing, Flag-Strep2 was found to carry a single mutation (F22S), whereas both of the HRP flanking positions were mutated in Flag-Strep3 (V18Y, F22D) (Entries 11–12, Table 1). Interestingly, many of the other improved variants contain a Tyr or Phe residue in position 18 (4/10) and a polar residue (H, D, S, R) in position 22 (6/10), thus defining a consensus for these sites. Efficient production in vitro of the two best cyclic peptides, Flag-cyclo(Strep2) and Flag-cyclo(Strep3), confirmed that both sequences undergo MeaF-mediated cyclization, leading to the expected macrocycles as the only product (Figure S9). These cyclic peptides, along with the parent cyclic peptide Flag-cyclo(Strep1), were then tested in the assay to measure their relative affinity for streptavidin (Figure 8C). From the resulting dose-response curve, the parent cyclic peptide was found to bind streptavidin with an equilibrium dissociation constant (KD) of 7.7 µM. Importantly, both of the evolved macrocyclic peptides showed significantly (2- to 7-fold) improved affinity for this protein, with measured KD values of 4.2 µM (Flag-cyclo(Strep2)) and 1.1 µM (Flag-cyclo(Strep3). Altogether, these studies provides a proof-of-principle demonstration of the utility of the present methodology in aiding the discovery of macrocyclic binders against a target protein of interest.
Figure 8.
Selection of improved streptavidin-targeting cyclic peptides. (A) Schematic representation of the in vitro assay used to screen the Strep1-derived library and measure KD for peptide binding to streptavidin. HRP: horseradish peroxidase. (B) Results from the primary screening of the Flag-Strep1(V18NDC/F22NDC) library in 96-well format. The absorbance values are normalized to that of the reference cyclic peptide Flag-cyclo(Strep1). (C) Streptavidin binding curves for isolated Flag-cyclo(Strep1) (KD = 7.7 ± 0.6 µM), Flag-cyclo(Strep2) (KD = 4.2 ± 0.1 µM), and Flag-cyclo(Strep3) (KD = 1.1 ± 0.2 µM)
CONCLUSIONS
In summary, we have developed a novel and versatile methodology useful for generating peptide macrocycles from ribosomally produced polypeptides in vitro and in vivo. This approach leverages the ability of two genetically encodable aminothiol amino acids, AmmF and MeaF, to induce a side-chain→C-end peptide cyclization via an intein-mediated intramolecular transthioesterification followed by a ring contraction through an S,N acyl transfer rearrangement. Whereas the AmmF-based strategy (Figure 3A) provides a means to generate macrocyclic peptides in vitro in a pH-controlled manner, the MeaF-based strategy (Figure 3B) offers the opportunity to achieve the spontaneous cyclization of peptide sequences of variable length (6mer to 12mer) and composition directly in living bacterial cells. As such, it provides a new approach, complementary in scope to those previously reported,49, 50 to direct the biosynthesis of cyclic peptides of completely arbitrary sequence. Compared to SICLOPPS, for example, the present method offers the versatility to access the desired peptide macrocycle in a broader range of molecular arrangements (i.e. cyclic, lariat, or C-terminally fused to a carrier protein) according to the nature of the N-terminal tail. In this study, this structural feature was leveraged to couple in-cell library generation with a high throughput in vitro assay to achieve the affinity maturation of a streptavidin-targeting cyclic peptide. Beyond that, we expect this methodology to make possible the screening of genetically encoded cyclopeptide libraries in a wide range of other formats, which include not only intracellular selection42–44, 48 or reporter systems45, but also two-hybrid76 or surface display77 systems. Whereas the molecular size of the macrocycles currently accessible with this method (i.e. 900–1,600 Da) seems appropriate for targeting extended biomolecular interfaces characterizing protein-protein interactions,28–31, 78 future studies will explore whether this strategy can be further evolved to provide access to smaller peptide ring structures.
EXPERIMENTAL SECTION
Cloning and plasmid construction
Genes encoding for the precursor proteins were placed under an ITPG-inducible T7 promoter in pET22 vectors (Novagen). Construction of the plasmids for expression of constructs CBD-4T through CBD-12T was described previously.52 Construct 9 of Table 1 was prepared following an identical procedure. Construct 10 of Table 1 was prepared by stepwise assembly PCR using FLAG-HPQ-GyrA_for1/3, FLAG-HPQ-GyrA_for2/3, and FLAG-HPQ-GyrA_for3/3 (Table S1) as forward primers, T7_Term_long_rev as reverse primer, and pET22b_CBD-12T as template. The PCR product (0.68 Kbp) was cloned into the Nde I / Xho I cassette of pET22b(+) vector. The FLAG-Strep1-derived library was prepared through randomization of position V18 and F22 in pET22b_FLAG-Strep1 with NDC degenete codons (= 12 codons encoding for C, D, F, G, H, I, L, N, R, S, V, Y). The corresponding plasmid library was prepared by overlap extension PCR using primers HPQ_SOE(A)_for, HPQ_SOE(A)_rev, HPQ(NDC)_SOE(B)_for, and HPQ(NDC)_SOE(B)_rev for fragment generation and then primers HPQ_SOE(A)_for and T7m _Term_long_rev for amplification of the final gene (1.35 Kbp), which was then cloned into the Xba I / Xho I cassette of pET22b(+) vector. The library was transformed into E. coli DH5α cells and a plasmid stock was generated by pooling > 1,000 c.f.u.
Synthesis of AmmF and MeaF
Detailed synthetic procedures and characterization data for AmmF and MeaF as provided as Supplemental Information.
Fluorescence-based assay
E. coli BL21(DE3) cells were co-transformed with a pET22_YFP(stop) plasmid50 encoding for MetGly(amber stop)YFP-His6 and a pEVOL plasmid encoding for the appropriate aminoacyl-tRNA synthetase. Cells were then grown in LB media containing ampicillin (50 mg/L) and chloramphenicol (26 mg/L) at 37°C overnight. The overnight cultures were used to inoculate 96-deep well plates containing minimal M9 media. At an OD600 of 0.6, cell cultures were induced by adding arabinose (0.06%), IPTG (0.2 mM), and either AmmF or MeaF at a final concentration of 2 mM. After overnight growth at 27°C, the cell cultures were diluted (1:1) with phosphate buffer (50 mM, 150 mM NaCl, pH 7.5) and fluorescence intensity (λex = 514 nm; λem = 527 nm) was determined using a Tecan Infinite 1000 plate reader. Cell cultures containing no unnatural amino acid were included as controls. Each sample was measured in triplicate.
Protein expression and purification
Proteins were expressed in E. coli BL21(DE3) cells co-transformed with the plasmid encoding for the biosynthetic precursor and pEVOL_AzF or pEVOL_O2beY vectors. After overnight growth, cells were used to inoculate M9 medium (0.4 L) supplemented with ampicillin (50 mg L−1), chloramphenicol (34 mg L−1), and 1 % glycerol. At an OD600 of 0.6, protein expression was induced by adding L-arabinose (0.05%), IPTG (0.25 mM), and AmmF or MeaF (2 mM). Cultures were grown for an additional 12–24 h at 27 °C and harvested by centrifugation at 3,400 g. Frozen cells were resuspended in Tris buffer (50 mM; pH 7.4) containing 300 mM NaCl and 20 mM imidazole and lysed by sonication. Protein purification by Ni-NTA affinity chromatography was carried out using a Tris buffer (50 mM, pH 7.4, NaCl 150 mM) containing 50 mM and 300 mM imidazole for protein loading and elution, respectively. Protein samples were concentrated in potassium phosphate buffer (50 mM, NaCl 150 mM, pH 7.5), and stored at −80 °C.
In vitro macrocyclization reactions
Reactions were carried out incubating the AmmF-containing precursor protein (50 µM) in potassium phosphate buffer (50 mM, NaCl 150 mM, pH 5.0) in the presence of TCEP (10 mM). For the thiol-catalyzed reactions, 10 mM MESNA was also added. After 1.5 h, the pH of the solution was adjusted to 5.0. Intein cleavage was monitored and quantified by SDS-PAGE and densitometry analysis of the gel bands using ImageJ software. The low MW products (8–10 kDa) of the reactions were analyzed by MALDI-TOF using a Brüker Autoflex II mass spectrometer.
In vivo macrocyclization reactions
The AmmF-containing precursor proteins were expressed in E. coli BL21(DE3) cells (50 mL culture) as described above. After 12 and 24 hours after induction, cells were harvested by centrifugation (3,400 g), re-suspended in 800 µL of potassium phosphate buffer (50 mM, NaCl 150 mM) at either pH 5.0 or 8.0, followed by lysis via sonication. The samples at pH 5.0 were used for detection of the thioester intermediate and they were processed immediately. The samples at pH 8.0 were incubated with iodoacetamide (50 mM) for 3 hours at room temperature prior to further processing. For protein isolation, the cell lysate samples were clarified by centrifugation (20,100 g) and passed over chitin-coated beads (New England Biolabs), the beads were washed with the lysis buffer, and the chitin-bound proteins were eluted with 70% acetonitrile in water (1 mL) followed by MALDI-TOF MS analysis.
Isolation of streptavidin binding peptide
The Strep1(MeaF) protein construct was expressed in E. coli BL21(DE3) cells (50 mL culture) for 24 hours at 27 °C as described above. After harvesting, cells were re-suspended in 800 µL of potassium phosphate buffer (50 mM, NaCl 150 mM, 7.5), lysed via sonication, followed by centrifugation at 20,100 g. The clarified cell lysate was passed over streptavidin beads (Pierce) and the immobilized peptide was eluted with 70% acetonitrile in water and analyzed by MALDI-TOF MS.
Library Screening
Recombinants from the FLAG-Strep1(V18NDC/F22NDC) library were expressed in E. coli BL21(DE3) cells in 96 deep well plates (1 mL culture) for 24 hours at 27 °C. After harvesting, cells were re-suspended in 400 µL of TBS (Tris 50 mM, NaCl 150 mM, 7.4), and incubated for 1 h at 37 °C in the presence of lysosyme, DNase, and MgCl2 to lyse the cells, followed by centrifugation at 3,400 g. The clarified cell lysate (200 µL) was transferred to NeutrAvidin coated 96-well plates (Pierce) and incubated for 2 h at room temperature. The plates were washed three times with 200 µL TBS + 0.5% Tween 20. Next, a solution of anti-FLAG-antibody HRP conjugate (100 µL, 1:2500 dilution) was added to each well and incubated for 1 h. The plates were washed three times with 200 µL TBS + 0.5% Tween 20. 100 µL of SigmaFast OPD solution was added to each well and the absorbance at 450 nm was measured after 20 minutes. The signals were normalized to the signal corresponding to the cells containing the FLAG-Strep1 cyclic peptide. The ten most active variants (>1.5 rel. act.) were re-expressed and screened again in triplicate using the same procedure (Figure S8).
KD determination for streptavidin binding
The cyclic peptides Flag-cyclo(Strep1), Flag-cyclo(Strep2), and Flag-cyclo(Strep3) were obtained via cyclization of the corresponding full-length precursor proteins in TBS in the presence of TCEP (20 mM) and thiophenol (10 mM). The full-length precursor proteins were obtained via expression in E. coli BL21(DE3) cells for 7 h at 22 °C, followed by Ni-affinity purification as described above. In each case, the macrocyclization reaction proceeded quantitatively yielding the cyclic peptide as the only product as determined by SDS-PAGE and MS (Figure S9). A solution of each of the macrocyclic peptide (50 µL) was added to NeutrAvidin coated plates at varying concentration (0.156 – 20 µM) and assayed in duplicate as described above. KD values were calculated with SigmaPlot via fitting of the dose-response curves using a 1:1 binding model.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by the National Science Foundation grant CHE-1112342 and in part by National Institute of Health grant R21 CA187502. N.T.J. and L.J.P. acknowledges the NSF REU program for financial support. MS instrumentation was supported by the NSF grants CHE-0840410 and CHE-0946653. We also thank Dr. Kestutis Bendinskas (SUNY Oswego) for providing access to Bruker Autoflex II MALDI-TOF mass spectrometer.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Synthetic procedures, additional MS spectra and SDS-PAGE gels. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery--an underexploited structural class. Nat. Rev. Drug Discov. 2008;7:608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
- 2.Marsault E, Peterson ML. Macrocycles Are Great Cycles: Applications, Opportunities, and Challenges of Synthetic Macrocycles in Drug Discovery. J. Med. Chem. 2011;54:1961–2004. doi: 10.1021/jm1012374. [DOI] [PubMed] [Google Scholar]
- 3.Al-Obeidi F, Castrucci AM, Hadley ME, Hruby VJ. Potent and prolonged acting cyclic lactam analogues of alpha-melanotropin: design based on molecular dynamics. J. Med. Chem. 1989;32:2555–2561. doi: 10.1021/jm00132a010. [DOI] [PubMed] [Google Scholar]
- 4.Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 5.Dechantsreiter MA, Planker E, Matha B, Lohof E, Holzemann G, Jonczyk A, Goodman SL, Kessler H. N-Methylated cyclic RGD peptides as highly active and selective alpha(V)beta(3) integrin antagonists. J. Med. Chem. 1999;42:3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 6.Graciani NR, Tsang KY, McCutchen SL, Kelly JW. Amino acids that specify structure through hydrophobic clustering and histidine-aromatic interactions lead to biologically active peptidomimetics. Bioorg. Med. Chem. 1994;2:999–1006. doi: 10.1016/s0968-0896(00)82048-9. [DOI] [PubMed] [Google Scholar]
- 7.Fasan R, Dias RL, Moehle K, Zerbe O, Vrijbloed JW, Obrecht D, Robinson JA. Using a beta-hairpin to mimic an alpha-helix: cyclic peptidomimetic inhibitors of the p53-HDM2 protein-protein interaction. Angew. Chem. Int. Ed. 2004;43:2109–2112. doi: 10.1002/anie.200353242. [DOI] [PubMed] [Google Scholar]
- 8.Dias RL, Fasan R, Moehle K, Renard A, Obrecht D, Robinson JA. Protein ligand design: from phage display to synthetic protein epitope mimetics in human antibody Fc-binding peptidomimetics. J. Am. Chem. Soc. 2006;128:2726–27232. doi: 10.1021/ja057513w. [DOI] [PubMed] [Google Scholar]
- 9.Cardoso RM, Brunel FM, Ferguson S, Zwick M, Burton DR, Dawson PE, Wilson IA. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J. Mol. Biol. 2007;365:1533–1544. doi: 10.1016/j.jmb.2006.10.088. [DOI] [PubMed] [Google Scholar]
- 10.Henchey LK, Porter JR, Ghosh I, Arora PS. High Specificity in Protein Recognition by Hydrogen-Bond-Surrogate alpha-Helices: Selective Inhibition of the p53/MDM2 Complex. Chembiochem. 2010;11:2104–2107. doi: 10.1002/cbic.201000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satoh T, Li S, Friedman TM, Wiaderkiewicz R, Korngold R, Huang Z. Synthetic peptides derived from the fourth domain of CD4 antagonize off function and inhibit T cell activation. Biochem. Biophys. Res. Commun. 1996;224:438–443. doi: 10.1006/bbrc.1996.1045. [DOI] [PubMed] [Google Scholar]
- 12.Fairlie DP, Tyndall JDA, Reid RC, Wong AK, Abbenante G, Scanlon MJ, March DR, Bergman DA, Chai CLL, Burkett BA. Conformational selection of inhibitors and substrates by proteolytic enzymes: Implications for drug design and polypeptide processing. J. Med. Chem. 2000;43:1271–1281. doi: 10.1021/jm990315t. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Liao W, Arora PS. Enhanced metabolic stability and protein-binding properties of artificial alpha helices derived from a hydrogen-bond surrogate: application to Bcl-xL. Angew. Chem. Int. Ed. 2005;44:6525–6529. doi: 10.1002/anie.200501603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudmundsson OS, Vander Velde DG, Jois SD, Bak A, Siahaan TJ, Borchardt RT. The effect of conformation of the acyloxyalkoxy-based cyclic prodrugs of opioid peptides on their membrane permeability. J. Pept. Res. 1999;53:403–413. doi: 10.1034/j.1399-3011.1999.00077.x. [DOI] [PubMed] [Google Scholar]
- 15.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J. Am. Chem. Soc. 2006;128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- 17.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 18.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 19.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 21.Craik DJ, Conibear AC. The chemistry of cyclotides. J. Org. Chem. 2011;76:4805–4817. doi: 10.1021/jo200520v. [DOI] [PubMed] [Google Scholar]
- 22.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat. Chem. Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White CJ, Yudin AK. Contemporary strategies for peptide macrocyclization. Nat. Chem. 2011;3:509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- 24.Frost JR, Smith JM, Fasan R. Design, synthesis, and diversification of ribosomally derived peptide macrocycles. Curr. Opin. Struct. Biol. 2013;23:571–580. doi: 10.1016/j.sbi.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Smith JM, Frost JR, Fasan R. Emerging strategies to access Peptide macrocycles from genetically encoded polypeptides. J. Org. Chem. 2013;78:3525–3531. doi: 10.1021/jo400119s. [DOI] [PubMed] [Google Scholar]
- 26.Passioura T, Katoh T, Goto Y, Suga H. Selection-based discovery of druglike macrocyclic peptides. Annu. Rev. Biochem. 2014;83:727–752. doi: 10.1146/annurev-biochem-060713-035456. [DOI] [PubMed] [Google Scholar]
- 27.Aboye TL, Camarero JA. Biological synthesis of circular polypeptides. J. Biol. Chem. 2012;287:27026–27032. doi: 10.1074/jbc.R111.305508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrighton NC, Farrell FX, Chang R, Kashyap AK, Barbone FP, Mulcahy LS, Johnson DL, Barrett RW, Jolliffe LK, Dower WJ. Small peptides as potent mimetics of the protein hormone erythropoietin. Science. 1996;273:458–464. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 29.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 30.Livnah O, Stura EA, Johnson DL, Middleton SA, Mulcahy LS, Wrighton NC, Dower WJ, Jolliffe LK, Wilson IA. Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2.8 Å. Science. 1996:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 31.Skelton NJ, Chen YM, Dubree N, Quan C, Jackson DY, Cochran A, Zobel K, Deshayes K, Baca M, Pisabarro MT, Lowman HB. Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein I. Biochemistry. 2001;40:8487–8498. doi: 10.1021/bi0103866. [DOI] [PubMed] [Google Scholar]
- 32.Pasqualini R, Koivunen E, Ruoslahti E. A peptide isolated from phage display libraries is a structural and functional mimic of an RGD-binding site on integrins. J. Cell Biol. 1995;130:1189–1196. doi: 10.1083/jcb.130.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinis C, Rutherford T, Freund S, Winter G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 2009;5:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 34.Angelini A, Cendron L, Chen SY, Touati J, Winter G, Zanotti G, Heinis C. Bicyclic Peptide Inhibitor Reveals Large Contact Interface with a Protease Target. ACS Chem. Biol. 2012;7:817–821. doi: 10.1021/cb200478t. [DOI] [PubMed] [Google Scholar]
- 35.Jafari MR, Deng L, Kitov PI, Ng S, Matochko WL, Tjhung KF, Zeberoff A, Elias A, Klassen JS, Derda R. Discovery of Light-Responsive Ligands through Screening of a Light-Responsive Genetically Encoded Library. ACS Chem. Biol. 2014;9:443–450. doi: 10.1021/cb4006722. [DOI] [PubMed] [Google Scholar]
- 36.Yamagishi Y, Shoji I, Miyagawa S, Kawakami T, Katoh T, Goto Y, Suga H. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem. Biol. 2011;18:1562–1570. doi: 10.1016/j.chembiol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi Y, Morimoto J, Suga H. In vitro selection of anti-Akt2 thioether-macrocyclic peptides leading to isoform-selective inhibitors. ACS Chem. Biol. 2012;7:607–613. doi: 10.1021/cb200388k. [DOI] [PubMed] [Google Scholar]
- 38.Morimoto J, Hayashi Y, Suga H. Discovery of macrocyclic peptides armed with a mechanism-based warhead: isoform-selective inhibition of human deacetylase SIRT2. Angew. Chem. Int. Ed. 2012;51:3423–3427. doi: 10.1002/anie.201108118. [DOI] [PubMed] [Google Scholar]
- 39.Schlippe YVG, Hartman MCT, Josephson K, Szostak JW. In Vitro Selection of Highly Modified Cyclic Peptides That Act as Tight Binding Inhibitors. J. Am. Chem. Soc. 2012;134:10469–10477. doi: 10.1021/ja301017y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann FT, Szostak JW, Seebeck FP. In vitro selection of functional lantipeptides. J. Am. Chem. Soc. 2012;134:8038–8041. doi: 10.1021/ja302082d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawakami T, Ishizawa T, Fujino T, Reid PC, Suga H, Murakami H. In Vitro Selection of Multiple Libraries Created by Genetic Code Reprogramming To Discover Macrocyclic Peptides That Antagonize VEGFR2 Activity in Living Cells. ACS Chem. Biol. 2013 doi: 10.1021/cb300697h. [DOI] [PubMed] [Google Scholar]
- 42.Horswill AR, Savinov SN, Benkovic SJ. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 2004;101:15591–15596. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naumann TA, Tavassoli A, Benkovic SJ. Genetic selection of cyclic peptide Dam methyltransferase inhibitors. Chembiochem. 2008;9:194–197. doi: 10.1002/cbic.200700561. [DOI] [PubMed] [Google Scholar]
- 44.Young TS, Young DD, Ahmad I, Louis JM, Benkovic SJ, Schultz PG. Evolution of cyclic peptide protease inhibitors. Proc. Natl. Acad. Sci. USA. 2011;108:11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng L, Naumann TA, Horswill AR, Hong SJ, Venters BJ, Tomsho JW, Benkovic SJ, Keiler KC. Discovery of antibacterial cyclic peptides that inhibit the ClpXP protease. Protein Sci. 2007;16:1535–1542. doi: 10.1110/ps.072933007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tavassoli A, Benkovic SJ. Genetically selected cyclic-peptide inhibitors of AICAR transformylase homodimerization. Angew. Chem. Int. Ed. 2005;44:2760–2763. doi: 10.1002/anie.200500417. [DOI] [PubMed] [Google Scholar]
- 47.Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS Chem. Biol. 2008;3:757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 48.Kritzer JA, Hamamichi S, McCaffery JM, Santagata S, Naumann TA, Caldwell KA, Caldwell GA, Lindquist S. Rapid selection of cyclic peptides that reduce alpha-synuclein toxicity in yeast and animal models. Nat. Chem. Biol. 2009;5:655–663. doi: 10.1038/nchembio.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bionda N, Cryan AL, Fasan R. Bioinspired strategy for the ribosomal synthesis of thioether-bridged macrocyclic peptides in bacteria. ACS Chem. Biol. 2014;9:2008–2013. doi: 10.1021/cb500311k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith JM, Vitali F, Archer SA, Fasan R. Modular Assembly of Macrocyclic Organo-Peptide Hybrids Using Synthetic and Genetically Encoded Precursors. Angew. Chem. Int. Ed. 2011;50:5075–5080. doi: 10.1002/anie.201101331. [DOI] [PubMed] [Google Scholar]
- 52.Satyanarayana M, Vitali F, Frost JR, Fasan R. Diverse organo-peptide macrocycles via a fast and catalyst-free oxime/intein-mediated dual ligation. Chem. Commun. 2012;48:1461–1463. doi: 10.1039/c1cc13533c. [DOI] [PubMed] [Google Scholar]
- 53.Frost JR, Vitali F, Jacob NT, Brown MD, Fasan R. Macrocyclization of Organo-Peptide Hybrids through a Dual Bio-orthogonal Ligation: Insights from Structure-Reactivity Studies. Chembiochem. 2013;14:147–160. doi: 10.1002/cbic.201200579. [DOI] [PubMed] [Google Scholar]
- 54.Smith JM, Hill NC, Krasniak PJ, Fasan R. Synthesis of bicyclic organo-peptide hybrids via oxime/intein-mediated macrocyclization followed by disulfide bond formation. Org. Biomol. Chem. 2014;12:1135–1142. doi: 10.1039/c3ob42222d. [DOI] [PubMed] [Google Scholar]
- 55.Wang L, Schultz PG. Expanding the genetic code. Angew. Chem. Int. Ed. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 56.Stokes AL, Miyake-Stoner SJ, Peeler JC, Nguyen DP, Hammer RP, Mehl RA. Enhancing the utility of unnatural amino acid synthetases by manipulating broad substrate specificity. Mol. Biosyst. 2009;5:1032–1038. doi: 10.1039/b904032c. [DOI] [PubMed] [Google Scholar]
- 57.Young DD, Young TS, Jahnz M, Ahmad I, Spraggon G, Schultz PG. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry. 2011;50:1894–1900. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seyedsayamdost MR, Xie J, Chan CT, Schultz PG, Stubbe J. Site-specific insertion of 3-aminotyrosine into subunit alpha2 of E. coli ribonucleotide reductase: direct evidence for involvement of Y730 and Y731 in radical propagation. J. Am. Chem. Soc. 2007;129:15060–15071. doi: 10.1021/ja076043y. [DOI] [PubMed] [Google Scholar]
- 59.Santoro SW, Wang L, Herberich B, King DS, Schultz PG. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol. 2002;20:1044–1048. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- 60.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 61.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deiters A, Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg. Med. Chem. Lett. 2005;15:1521–1524. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 63.Telenti A, Southworth M, Alcaide F, Daugelat S, Jacobs WR, Jr, Perler FB. The Mycobacterium xenopi GyrA protein splicing element: characterization of a minimal intein. J. Bacteriol. 1997;179:6378–6382. doi: 10.1128/jb.179.20.6378-6382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 65.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gentle IE, De Souza DP, Baca M. Direct production of proteins with N-terminal cysteine for site-specific conjugation. Bioconjug. Chem. 2004;15:658–663. doi: 10.1021/bc049965o. [DOI] [PubMed] [Google Scholar]
- 67.Villain M, Vizzavona J, Rose K. Covalent capture: a new tool for the purification of synthetic and recombinant polypeptides. Chem. Biol. 2001;8:673–679. doi: 10.1016/s1074-5521(01)00044-8. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen DP, Elliott T, Holt M, Muir TW, Chin JW. Genetically encoded 1,2-aminothiols facilitate rapid and site-specific protein labeling via a bio-orthogonal cyanobenzothiazole condensation. J. Am. Chem. Soc. 2011;133:11418–11421. doi: 10.1021/ja203111c. [DOI] [PubMed] [Google Scholar]
- 69.Perrin DD. Dissociation constants of organic bases in aqueous solution. London: Butterworth; 1965. [Google Scholar]
- 70.Kreevoy MM, Harper ET, Duvall RE, Wilgus HS, Ditsch LT. Inductive Effects on the Acid Dissociation Constants of Mercaptans. J. Am. Chem. Soc. 1960;82:4899–4902. [Google Scholar]
- 71.Canne LE, Bark SJ, Kent SBH. Extending the applicability of native chemical ligation. J. Am. Chem. Soc. 1996;118:5891–5896. [Google Scholar]
- 72.Naumann TA, Savinov SN, Benkovic SJ. Engineering an affinity tag for genetically encoded cyclic peptides. Biotechnol. Bioeng. 2005;92:820–830. doi: 10.1002/bit.20644. [DOI] [PubMed] [Google Scholar]
- 73.Katz BA. Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence. Biochemistry. 1995;34:15421–15429. doi: 10.1021/bi00047a005. [DOI] [PubMed] [Google Scholar]
- 74.Giebel LB, Cass RT, Milligan DL, Young DC, Arze R, Johnson CR. Screening of cyclic peptide phage libraries identifies ligands that bind streptavidin with high affinities. Biochemistry. 1995;34:15430–15435. doi: 10.1021/bi00047a006. [DOI] [PubMed] [Google Scholar]
- 75.Katz BA. Binding to protein targets of peptidic leads discovered by phage display: crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence. Biochemistry. 1995;34:15421–15429. doi: 10.1021/bi00047a005. [DOI] [PubMed] [Google Scholar]
- 76.Dove SL, Hochschild A. A bacterial two-hybrid system based on transcription activation. Methods Mol. Biol. 2004;261:231–246. doi: 10.1385/1-59259-762-9:231. [DOI] [PubMed] [Google Scholar]
- 77.Lofblom J. Bacterial display in combinatorial protein engineering. Biotechnol. J. 2011;6:1115–1129. doi: 10.1002/biot.201100129. [DOI] [PubMed] [Google Scholar]
- 78.Fasan R, Dias RL, Moehle K, Zerbe O, Obrecht D, Mittl PR, Grutter MG, Robinson JA. Structure-activity studies in a family of beta-hairpin protein epitope mimetic inhibitors of the p53-HDM2 protein-protein interaction. Chembiochem. 2006;7:515–526. doi: 10.1002/cbic.200500452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.