We present here a unified, easily adaptable, open-source NMR exchange format (NEF) for NMR restraints and associated data.
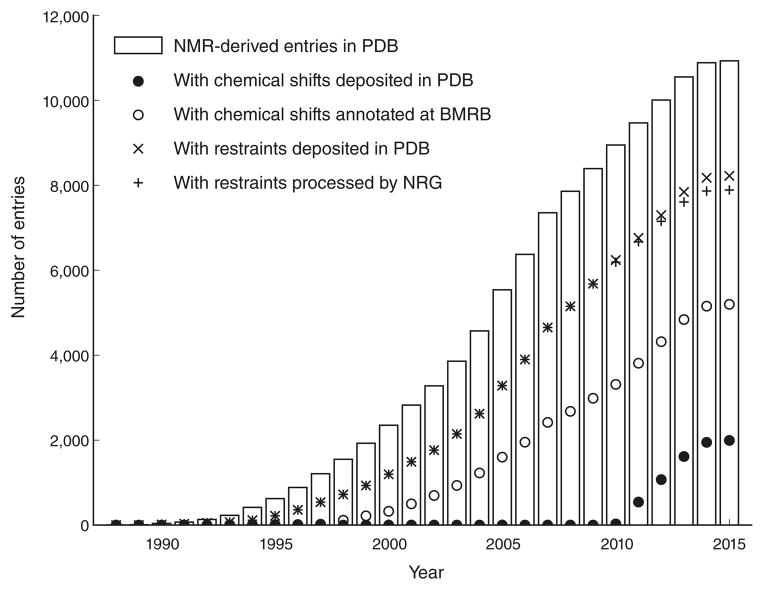
Atomic-resolution, three-dimensional structures of macromolecules have been determined by NMR spectroscopy since the late 1980s. In 2013, the number of NMR-derived structures in the Protein Data Bank (PDB)1 passed the milestone of 10,000 entries (Fig. 1), and they currently account for approximately 10% of the total number of structures in the PDB. To improve the quality and integrity of the archive, the Worldwide Protein Data Bank (wwPDB)2, the consortium that manages the PDB archive, made the deposition of the underlying experimental data mandatory and established expert validation task forces (VTFs) to provide consensus recommendations for validating the structures and accompanying experimental data for entries determined by X-ray, NMR or cryo-EM techniques. The initial recommendations of the NMR VTF3 have been implemented in a software pipeline that will be used to produce validation reports during structure deposition and annotation.
Figure 1.
Growth in the number of NMR entries in the PDB archive.
NMR data and restraints are diverse in their nature: they are typically derived from various kinds of NMR experiments, and they may be interpreted differently by different software programs, even when the same spectral data are used as input. In addition, almost all NMR programs rely on a variety of formats, thus necessitating conversions when multiple programs are used in structure determination and analysis, with a concomitant risk of information loss or misinterpretation. Two software projects, NMR-STAR4, developed at the Biological Magnetic Resonance Bank (BMRB)5 with input from the NMR community, and the Collaborative Computational Project for NMR (CCPN)6, provide systematic and comprehensive data models for storing and accessing NMR data. Unfortunately, neither of these two approaches has been widely adopted by the developers of popular software tools for NMR structure determination, refinement and validation, partly because both data models suffer from substantial and similar drawbacks: their data structures are large—more extensive and more complex than any single program would typically require—and they are not easily and independently adapted and extended for any specific program.
NMR restraint data are currently deposited in a variety of software-specific formats that have to be curated by the BMRB into a common format for deposition in the NMR Restraints Grid (NRG)7, thus enabling many useful applications. Unfortunately, efforts to develop universal restraint converters have been challenged because some restraint formats omit information required by other restraint formats8, and full parsing of each software-specific format has proven to be impossible. The current situation hampers the proper archiving and use of bio-molecular NMR data, and prevents the routine inclusion of NMR restraint validation in the wwPDB NMR validation pipeline.
For these reasons, the wwPDB partners, together with CCPN, organized a series of consultations and two workshops that included developers of key software packages used for NMR structure determination and refinement (Table 1), with the aim of attaining a unified approach to represent NMR restraints and associated data. Together, they agreed on and successfully implemented and tested an NMR data representation, denoted the NEF, and devised a governance structure for its maintenance and further development. Importantly, the different program developers committed to the ambitious goal of making their software capable of both reading and writing NEF-compliant files.
Table 1.
Software packages implementing the NEF
| Software package | Category | Principal investigator or representative |
|---|---|---|
| AMBER | Molecular dynamics (with NMR restraints) | D.A. Case |
| CYANA | Automated assignment and structure determination | P. Güntert |
| UNIO | Automation from spectral acquisition to structure | T. Herrmann |
| CS-ROSETTA | Structure determination from chemical shifts | O. Lange |
| NMR-STAR converter | Format conversion | J.L. Markley, E.L. Ulrich |
| ASDP | Automated NOESY cross-peak assignment | G.T. Montelione, Y.J. Huang |
| PSVS and PDBStat | Structure validation | G.T. Montelione, R. Tejero, Y.J. Huang |
| ARIA and CNS | Structure determination and refinement | M. Nilges, B. Bardiaux |
| XPLOR-NIH | Structure determination and refinement | C.D. Schwieters |
| CCPN FormatConverter | Format conversion | W.F. Vranken |
| CCPN | Data modeling, spectral analysis, format conversion, integration of other NMR software | G.W. Vuister, R.H. Fogh |
| CING | Structure validation | G.W. Vuister |
| CS23D | Structure determination from chemical shifts | D. Wishart |
| PROSESS and RESPROX | Structure validation | D. Wishart |
The detailed specifications of the NEF (https://github.com/NMRExchangeFormat/NEF/) are based on the consensus that emerged during the consultations and workshops: the format accommodates a variety of restraint types and is extensible beyond the common agreed-upon elements, so that new science can be easily incorporated. The NEF format is self-contained, so that unambiguous interpretation of the data does not require any auxiliary software-specific files, and is readable by both machines and humans. In addition to the restraints data, NEF requires polymer sequence information and chemical-shift assignments, and allows inclusion of peak lists. A compliant NEF file contains all the data in a single, appropriately sectioned file, implemented with the STAR syntax9 and controlled by a versioned dictionary of tag names. Developers can extend the standard dictionary to accommodate their own new data or experimental practices, which need not be supported by other software packages, by simply registering an individual dictionary namespace. Thus, the NEF is inherently flexible and extensible, and it allows for unlimited program-specific additional data without the need for any adaptation of the format. Importantly, it has been anticipated that such initially nonstandard additions might evolve into the general practice and be adopted by other programs. A mechanism to incorporate such developments is part of the management of the NEF specification.
All authors of this Correspondence have been involved in the planning and development of the NEF, and they include representatives of all major packages for NMR structure determination, refinement and validation (Table 1). The program developers have agreed to release updated versions of their software capable of handling the NEF format by the end of September 2015. After a transition period, the wwPDB partners are expected to accept only NEF-formatted NMR data for deposition into the PDB.
The efforts presented here show that the biological NMR community is ready to resolve the issues of representation and exchange of experimental NMR data. We encourage developers of current and future NMR software to support the NEF, and we invite the wider community of NMR-software developers and other stakeholders to participate in its development and maintenance.
Acknowledgments
The European Bioinformatics Institute and Rutgers University workshops were made possible by the generous support of the Wellcome Trust (grant 088944 to G.J.K.), the European Molecular Biology Laboratory, the UK Biotechnology and Biological Sciences Research Council (grants BB/J007471 to G.J.K., BB/J007897/1 to G.W.V. and BB/K021249/1 to G.W.V. and G.T.M.), the UK Medical Research Council (grant MR/L000555/1 to G.W.V.), the US National Science Foundation (grant DBI-1338415 to H.M.B.), the Japan Science and Technology Agency–National Bioscience Database Center, the US National Institutes of Health (NIH; grants and P41LM05799 and R01GM109046 to J.L.M.). C.D.S. is supported by the Intramural Research Program of the NIH Center for Information Technology. P.G. is supported by the Lichtenberg program of the Volkswagen Foundation and by a Grant-in-Aid for Scientific Research by the Japan Society for the Promotion of Science. W.F.V. is supported by the Brussels Institute for Research and Innovation (Innoviris, grant BB2B 2010-1-12).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bernstein FC, et al. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 2.Berman H, et al. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montelione GT, et al. Structure. 2013;21:1563–1570. doi: 10.1016/j.str.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markley JL, et al. Methods Biochem Anal. 2003;44:89–113. [PubMed] [Google Scholar]
- 5.Ulrich EL, et al. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vranken WF, et al. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 7.Doreleijers JF, et al. J Biomol NMR. 2009;45:389–396. doi: 10.1007/s10858-009-9378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tejero R, et al. J Biomol NMR. 2013;56:337–351. doi: 10.1007/s10858-013-9753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall SRJ. Chem Inf Comput Sci. 1991;31:326–333. [Google Scholar]